Study of SH-SY5Y Cancer Cell Response to Treatment with Polyphenol Extracts Using FT-IR Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
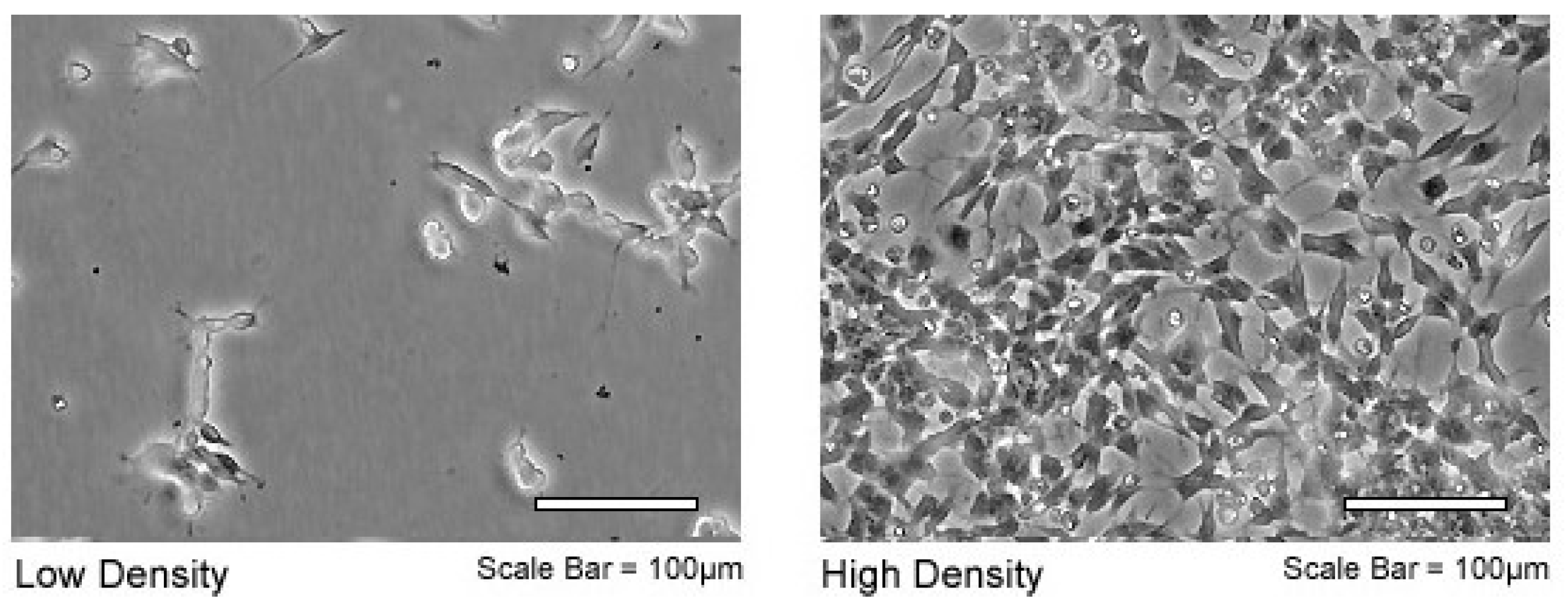
2.1. Cell Line
2.2. Preparation of Cherry Derived Polyphenol Extract
2.3. Preparation of Cherry Derived Polyphenol Extract
2.4. Cell Fixing Protocol
2.5. Spectra Acquisition
2.6. Data Analysis
3. Results and Discussion
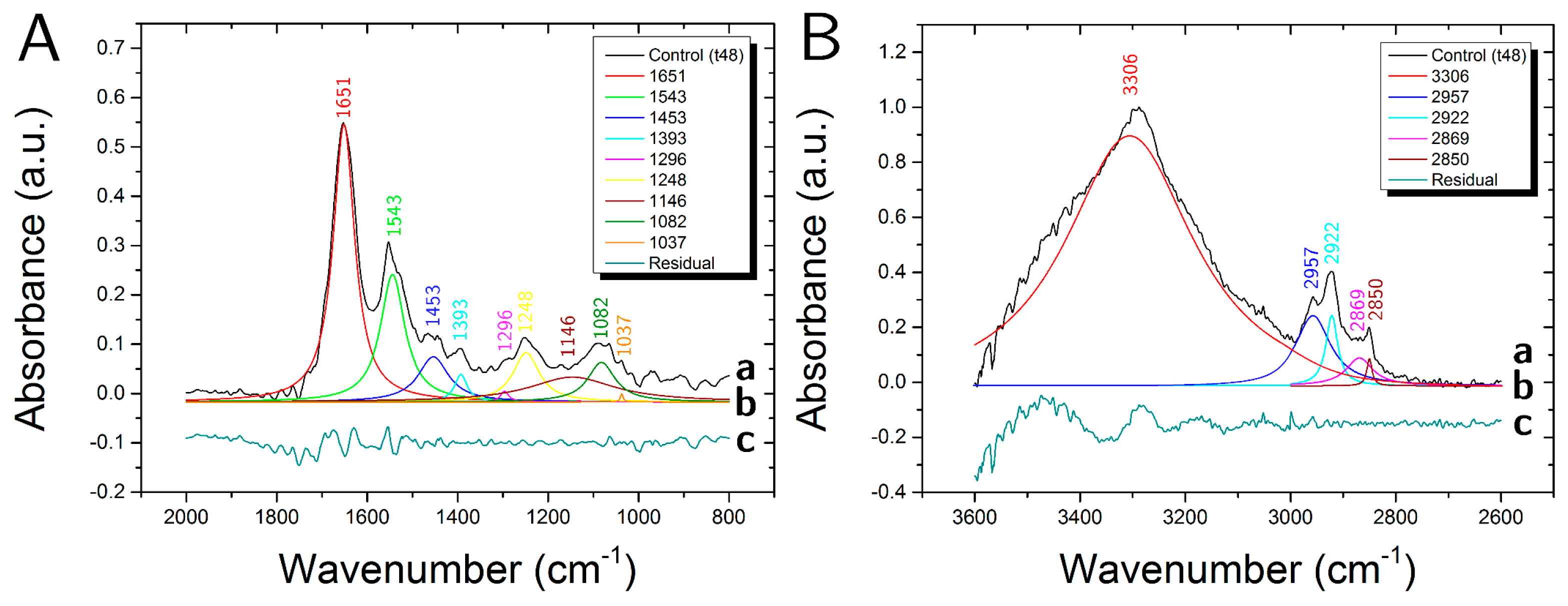
3.1. Control Samples
3.2. Cells Exposed to PaDRw-25 Dose
3.3. Cells Exposed to PaDRw-500 Dose
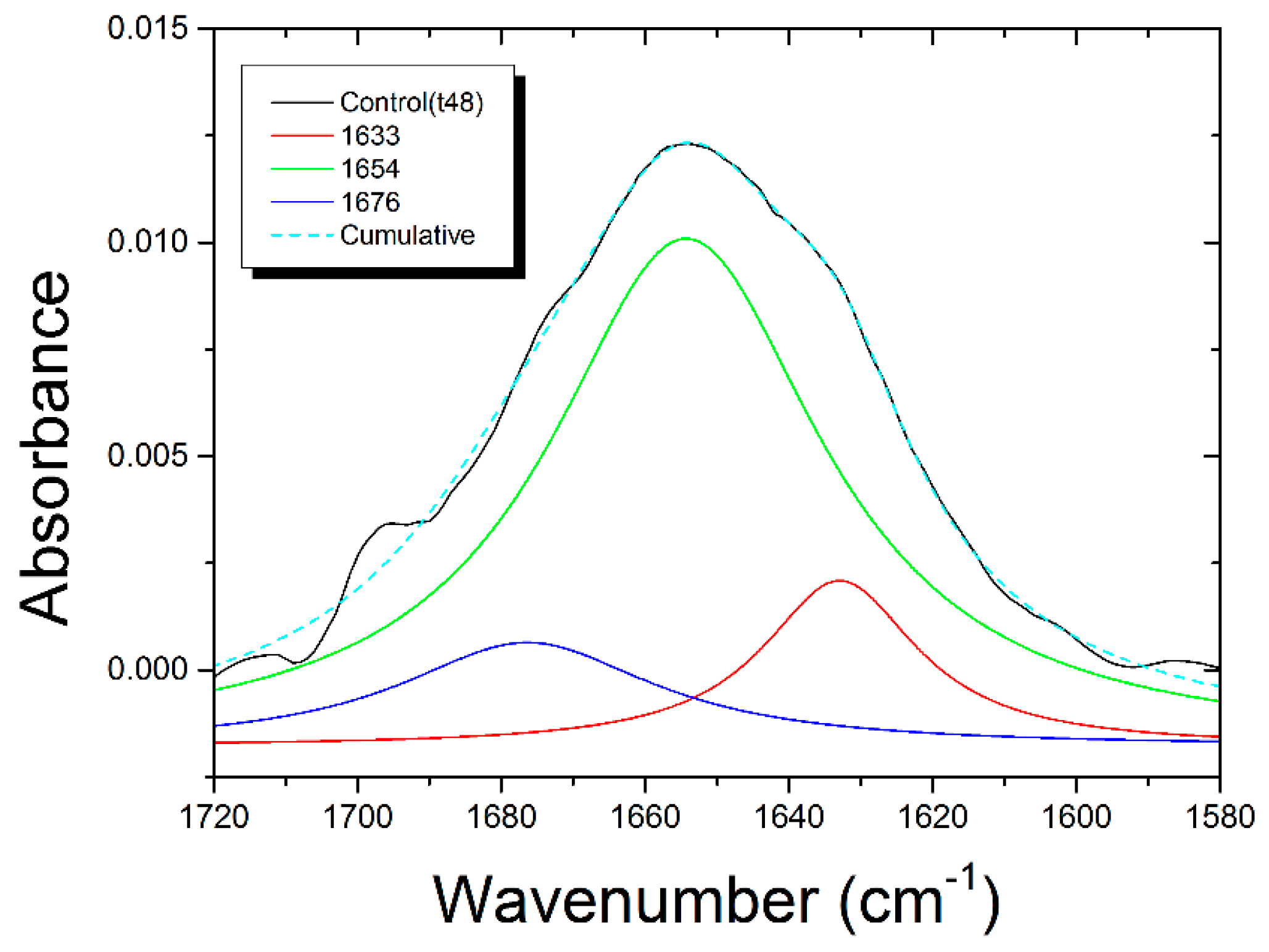
3.4. Deconvolution of Amide I Band
4. Conclusions
Supplementary Materials
Author Contributions
Conflicts of Interest
References
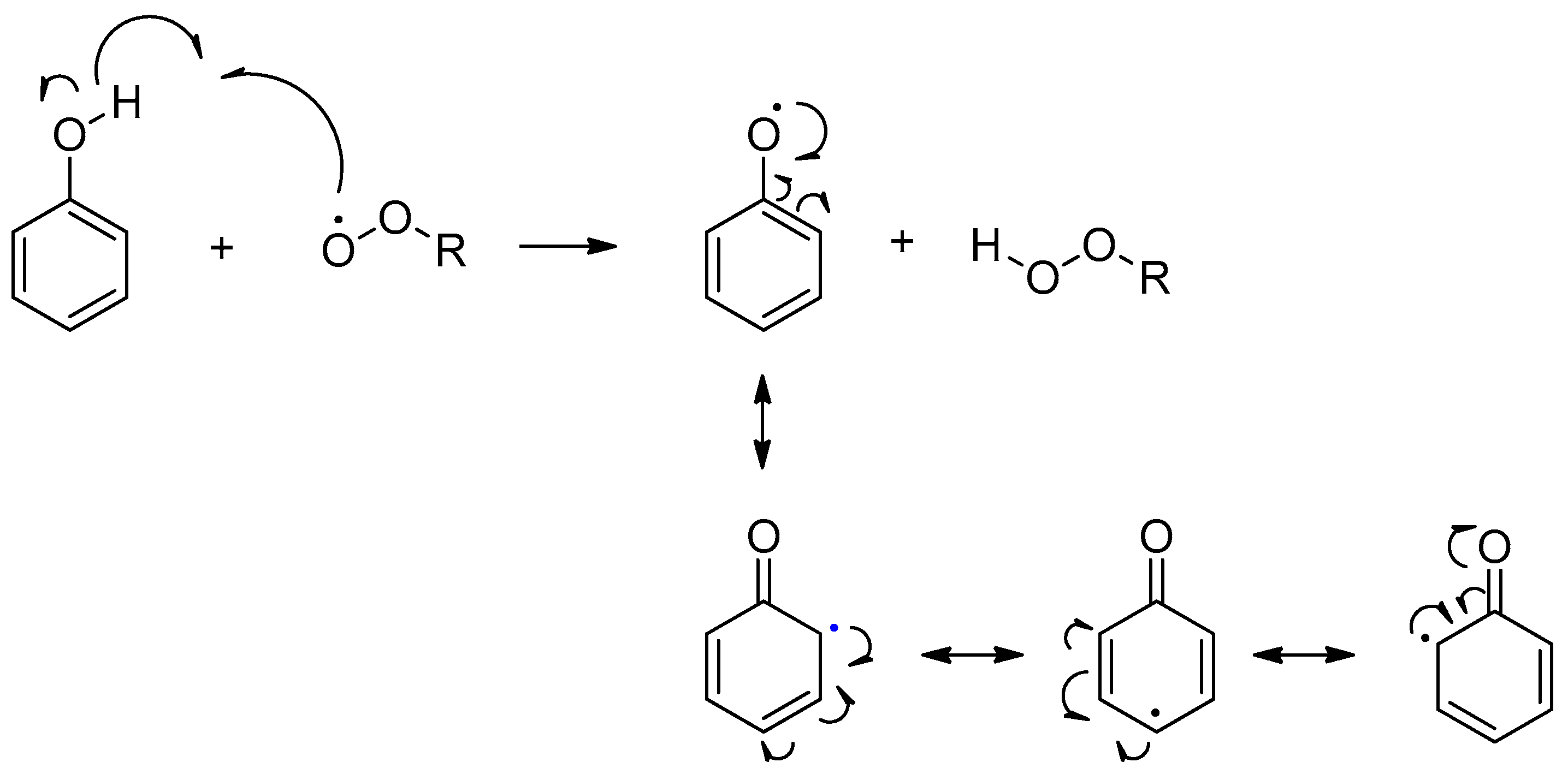
- Hadi, S.M.; Bhat, S.H.; Azmi, A.S.; Hanif, S.; Shamim, U.; Ullah, M.F. Oxidative break-age of cellular DNA by plant polyphenols: A putative mechanism for anticancerproperties. Semin. Cancer Biol. 2007, 17, 370–376. [Google Scholar] [CrossRef] [PubMed]
- Perron, N.R.; García, C.R.; Pinzón, J.R.; Chaur, M.N.; Brumaghim, J.L. Antioxidant and pro-oxidant effects of polyphenol compounds on copper-mediated DNA damage. J. Inorg. Biochem. 2011, 105, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Sebastia, N.; Montoro, A.; Hervas, D.; Pantelias, G.; Hatzi, V.; Soriano, J.; Villaescusa, J.; Terzoudi, G. Curcumin and trans-resveratrol exert cell cycle-dependent radioprotective or radiosensitizing effects as elucidated by the PCC and g2-assay. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2014, 766–767, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, S.; Piccolella, S.; Papale, F.; Nocera, P.; Lettieri, A.; Catauro, M. A polyphenol complex from thymus vulgaris l. plants cultivated in the Campania region (Italy): New perspectives against neuroblastoma. J. Funct. Foods 2016, 20, 253–266. [Google Scholar] [CrossRef]
- Pacifico, S.; Capozzi, V.; Delfino, I.; Lasalvia, M.; Lepore, M.; Perna, G.; Perozziello, F.M.; Piccolella, S.; Ricciardi, V.; Manti, L. Radiomodulating activity of cherry polyphenol-enriched extracts. In Proceedings of the 42nd Annual Meeting of the European Radiation Research Society (ERRS2016), Amsterdam, The Netherlands, 4–8 September 2016. [Google Scholar]
- Abbas, M.; Saeed, F.; Anjum, F.M.; Afzaal, M.; Tufail, T.; Bashir, M.S.; Ishtiaq, A.; Hussain, S.; Suleria, H.A.R. Natural polyphenols: An overview. Int. J. Food Prop. 2017, 20, 1689–1699. [Google Scholar] [CrossRef]
- Yordi, E.G.; Pérez, E.M.; Matos, M.J.; Villares, E.U. Antioxidant and Pro-Oxidant Effects of Polyphenolic Compounds and Structure-Activity Relationship Evidence. In Nutrition, Well-Being and Health; InTech: London, UK, 2012; pp. 23–48. [Google Scholar]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Clemens, G.; Hands, J.R.; Dorling, K.M.; Baker, M.J. Vibrational spectroscopic methods for cytology and cellular research. Analyst 2014, 139, 4411–4444. [Google Scholar] [CrossRef] [PubMed]
- Mourant, J.; Yamada, Y.; Carpenter, S.; Dominique, L.; Freyer, J. FTIR spectroscopy demonstrates biochemical differences in mammalian cell cultures at different growth stages. Biophys. J. 2003, 85, 1938–1947. [Google Scholar] [CrossRef]
- Gianoncelli, A.; Vaccari, L.; Kourousias, G.; Cassese, D.; Bedolla, D.E.; Kenig, S.; Storici, P.; Lazzarino, M.; Kiskinova, M. Soft X-ray microscopy radiation damage on fixed cells investigated with synchrotron radiation FTIR microscopy. Sci. Rep. 2015, 5, 10250. [Google Scholar] [CrossRef] [PubMed]
- Vileno, B.; Jeney, S.; Sienkiewicz, A.; Marcoux, P.R.; Miller, L.M.; Forró, L. Evidence of lipid peroxidation and protein phosphorylation in cells upon oxidative stress photo-generated by fullerols. Biophys. Chem. 2010, 152, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Olesko, A.; Olsztyńska-Janus, S.; Walski, T.; Grzeszckuz-Kuc, K.; Bujok, J.; Galecka, K.; Czerski, A.; Witkiewicz, W.; Komorowska, M. Application of FTIR-ATR Spectroscopy to determinate the extent of lipid peroxidation in plasma during haemodialysis. BioMed Res. Int. 2015, 2015, 245607. [Google Scholar]
- Cakmak, G.; Togan, I.; Uğut, C.; Severcan, F. FT-IR Spectroscopic Analysis of Rainbow Trout Liver Exposed to Nonylphenol. Appl. Spectrosc. 2003, 57, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Derenne, A.; Van Hemelryck, V.; Lamoral-Theys, D.; Kiss, R.; Goormaghtigh, E. FT-IR spectroscopy: A new valuable tool to classify the effects of polyphenolic compounds on cancer cells. Biochim. Biophys. Acta 2013, 1832, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Barraza-Garza, G.; Castillo-Michel, H.; De la Rosa, L.A.; Martinez-Martinez, A.; Pérez-León, J.A.; Cotte, M.; Alvarez-Parrilla, E. Infrared spectroscopy as a tool to study the antioxidant activity of polyphenolic compounds in isolated rat enterocytes. Oxid. Med. Cell. Longev. 2016, 2016, 9245150. [Google Scholar] [CrossRef] [PubMed]
- Ulrih, N.P. Analytical techniques for the study of polyphenol–protein interactions. Crit. Rev. Food Sci. Nutr. 2017, 57, 2144–2161. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Verma, T.; Mukherjee, R.; Ariese, F.; Somasundaram, K.; Umapathy, S. Raman and infra-red microspectroscopy: Towards quantitative evaluation for clinical research by ratiometric analysis. Chem. Soc. Rev. 2016, 45, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Nguyen, X.; Muzammil, S.; Towers, E.; Gabrielson, J.; Narhi, L. Qualification of FT-IR spectroscopic method for protein secondary structural analysis. J. Pharm. Sci. 2011, 100, 4631–4641. [Google Scholar] [CrossRef] [PubMed]
- Agholme, L.; Lindström, T.; Kågedal, K.; Marcusson, J.; Hallbeck, M. An in vitro model for neuroscience: Differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J. Alzheimers Dis. 2010, 20, 1069–1082. [Google Scholar] [CrossRef] [PubMed]
- Jämsä, A.; Hasslund, K.; Cowburn, R.F.; Bäckström, A.; Vasänge, M. The retinoic acid and brain-derived neurotrophic factor differentiated SH-SY5Y cell line as a model for Alzheimer’s disease-like tau phosphorylation. Biochem. Biophys. Res. Commun. 2004, 319, 993–1000. [Google Scholar] [CrossRef] [PubMed]
- Koriyama, Y.; Furukawa, A.; Muramatsu, M.; Takino, J.; Takeuchi, M. Glyceraldehyde caused Alzheimer’s disease-like alterations in diagnostic marker levels in SH-SY5Y human neuroblastoma cells. Sci. Rep. 2015, 5, 13313. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, S.; Di Maro, A.; Petriccione, M.; Galasso, S.; Piccolella, S.; Di Giuseppe, A.M.A.; Scortichini, M.; Monaco, P. Chemical composition, nutritional value and antioxidant properties of autochthonous Prunus avium cultivars from Campania Region. Food Res. Int. 2014, 64, 188–199. [Google Scholar] [CrossRef]
- Bouayed, J.; Bohn, T. Exogenous antioxidants-Double-edged swords in cellular redox state: Health beneficial effects at physiologic doses versus deleterious effects at high doses. Oxid. Med. Cell. Longev. 2010, 3, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Lyng, F.; Gazi, E.; Gardner, P. Preparation of tissues and cells for infrared and Raman spectroscopy and imaging. RSC Anal. Spectrosc. Monogr. 2011, 11, 145–189. [Google Scholar]
- Meade, A.D.; Lyng, F.M.; Knief, P.; Byrne, H.J. Growth substrate induced functional changes elucidated by FT-IR and Raman spectroscopy in in-vitro cultured human keratinocytes. Anal. Bioanal. Chem. 2007, 387, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Gazi, E.; Dwyer, E.; Lockyer, N.P.; Miyan, J.; Gardner, P.; Hart, C.; Brown, M.; Clarke, N.W. Fixation protocols for subcellular imaging by synchrotron-based Fourier transform infrared microspectroscopy. Biopolymers 2005, 77, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Portaccio, M.; Menale, C.; Diano, N.; Mita, D.G.; Lepore, M. FT-IR microspectroscopy for monitoring production process of cisplatin-loaded PLGA nanoparticles for drug delivery. J. Appl. Polym. Sci. 2015, 132, 41305. [Google Scholar] [CrossRef]
- Delfino, I.; Perna, G.; Lasalvia, M.; Capozzi, V.; Manti, L.; Camerlingo, C.; Lepore, M. Visible micro-Raman spectroscopy of single human mammary epithelial cells exposed to X-ray radiation. J. Biomed. Opt. 2015, 20, 035003. [Google Scholar] [CrossRef] [PubMed]
- Barth, A. Infrared spectroscopy of proteins. Biochim. Biophys. Acta 2007, 1767, 1073–1101. [Google Scholar] [CrossRef] [PubMed]
- Coe, J.V.; Nystrom, S.V.; Chen, Z.; Li, R.; Verreault, D.; Hitchcock, C.L.; Allen, H.C. Extracting Infrared Spectra of Protein Secondary Structures Using a Library of Protein Spectra and the Ramachandran Plot. J. Phys. Chem. B 2015, 119, 13079–13092. [Google Scholar] [CrossRef] [PubMed]
- Mei, Y.; Miller, L.; Gao, W.; Gross, R.A. Imaging the distribution and secondary structure of immobilized enzymes using infrared microspectroscopy. Biomacromolecules 2003, 4, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Delfino, I.; Portaccio, M.; Della Ventura, B.; Mita, D.G.; Lepore, M. Enzyme distribution and secondary structure of sol-gel immobilized glucose oxidase by microattenuated total reflection FT-IR spectroscopy. Mater. Sci. Eng. C 2013, 33, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Gault, N.; Lefaix, J.L. Infrared microspectroscopic characteristics of radiation-induced apoptosis in human lymphocytes. Radiat. Res. 2003, 160, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Meade, A.; Clarke, C.; Byrne, H.; Lyng, F. Fourier transform infrared microspectroscopy and multivariate methods for radiobiological dosimetry. Radiat. Res. 2010, 173, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Gault, N.; Rigaud, O.; Poncy, L.; Lefaix, L. Infrared microspectroscopy study of g–irradiated and H2O2 -treated human cells. Int. J. Radiat. Biol. 2005, 81, 767–779. [Google Scholar] [CrossRef] [PubMed]
- Zelig, U.; Kapelushnik, J.; Moreh, R.; Mordechai, S.; Nathan, I. Diagnosis of cell death by means of Infrared Spectroscopy. Biophys. J. 2009, 97, 2107–2114. [Google Scholar] [CrossRef] [PubMed]
- Nagata, S. Apoptotic DNA fragmentation. Exp. Cell Res. 2000, 256, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.K.; Dasgupta, A.; Adayen, T.; Shahmehdi, S.A.; Hammond, D.; Banerjee, P. Apoptosis is associated with an increase in saturated fatty acid containing phospholipids in the neuronal cell line, HN2–5. Biochim. Biophys. Acta 1996, 1304, 171–178. [Google Scholar] [CrossRef]
- Shibata, T.; Iio, K.; Kawai, Y.; Shibata, N.; Kawaguchi, M.; Toi, S.; Kobayashi, M.; Kobayashi, M.; Yamamoto, K.; Uchida, K. Identification of a Lipid peroxidation product as a potential trigger of the p53 pathway. J. Biol. Chem. 2006, 281, 1196–1204. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Yamada, M.; Ohsawa, T.; Morisawa, H.; Nishine, T.; Nishimura, O.; Toda, T. Phosphoproteomic profiling of human SH-SY5Y neuroblastoma cells during response to 6-hydroxydopamine-induced oxidative stress. Biochim. Biophys. Acta 2006, 1763, 977–989. [Google Scholar] [CrossRef] [PubMed]

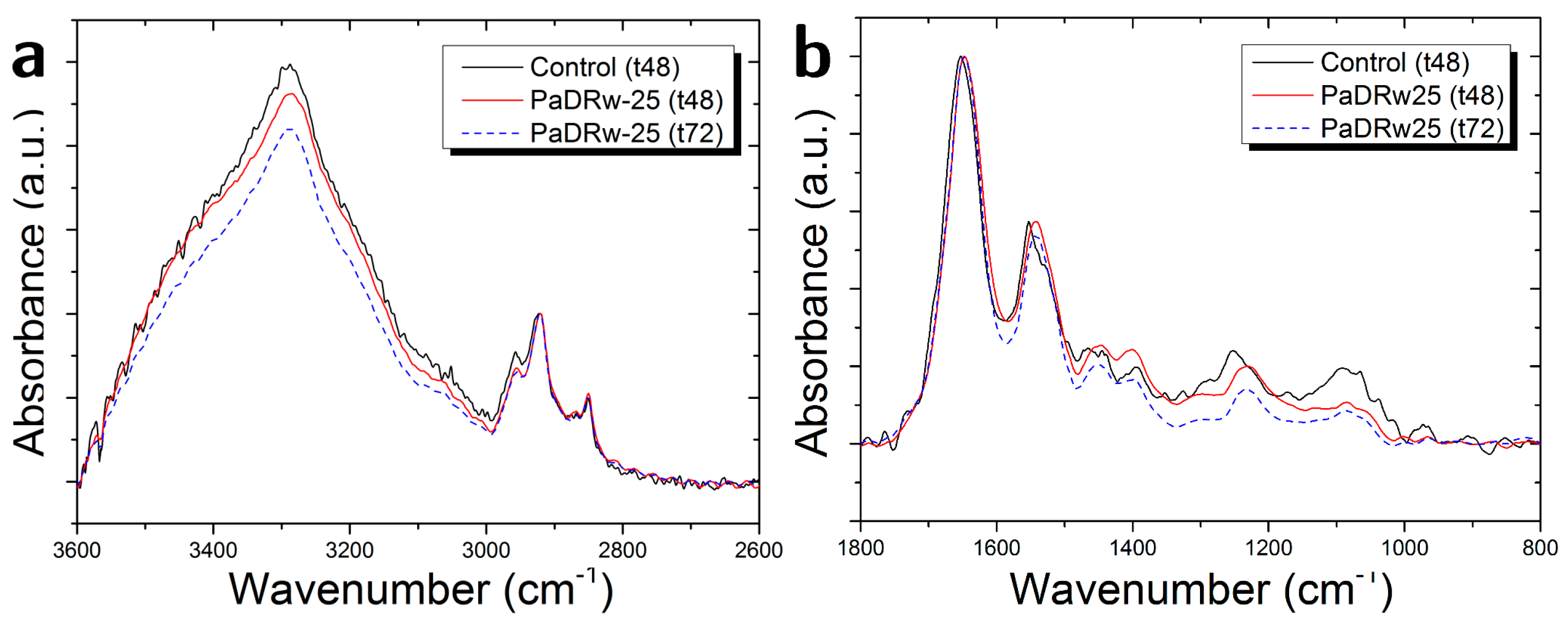
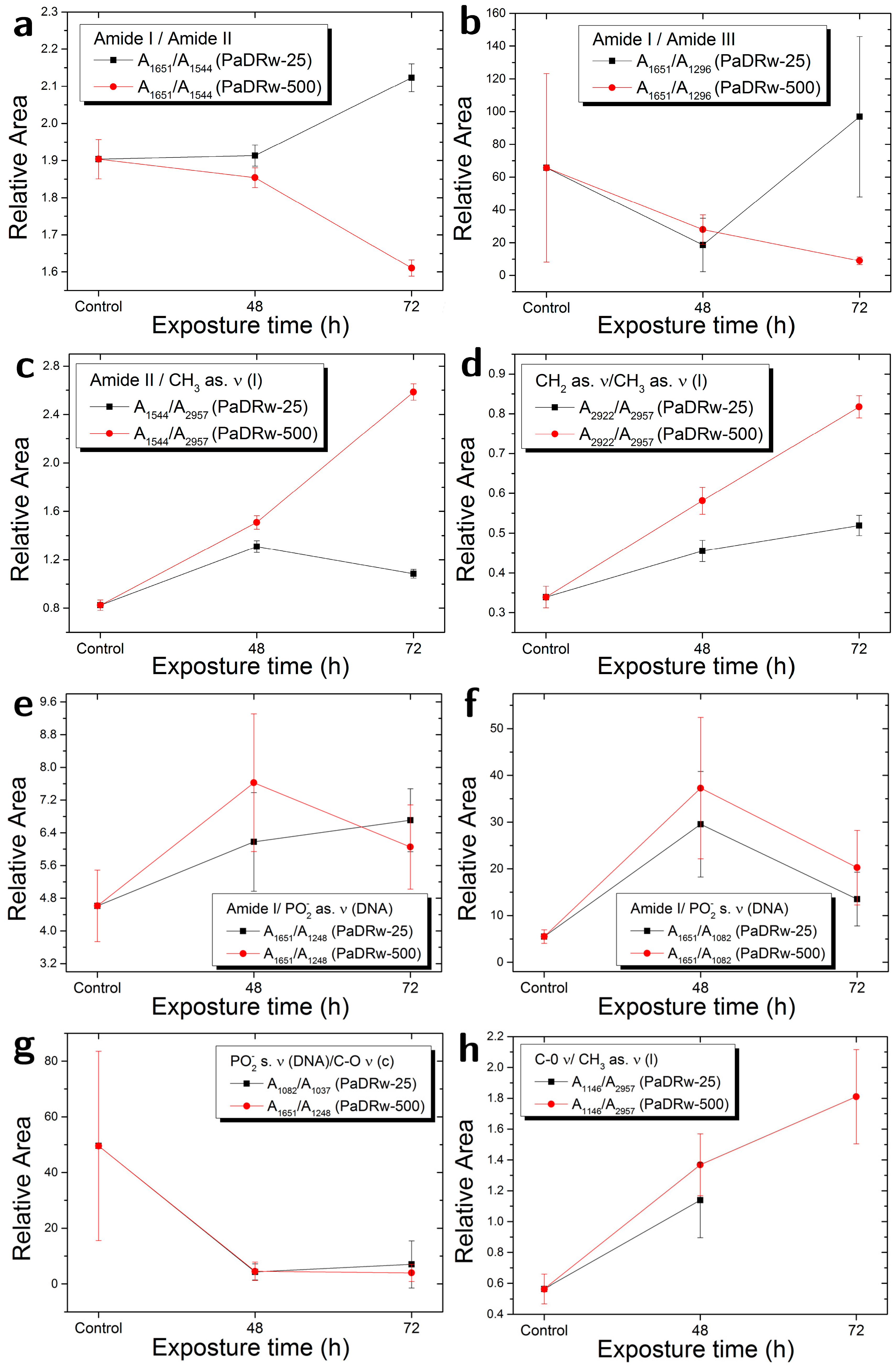
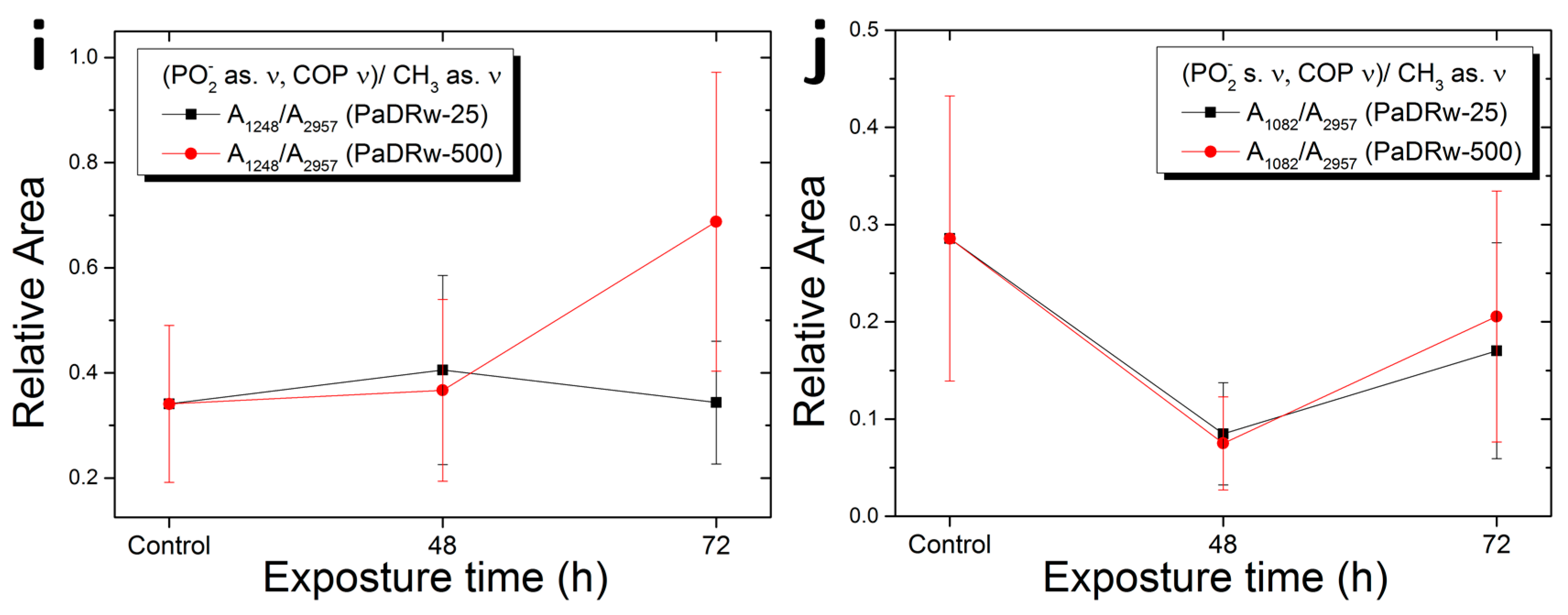
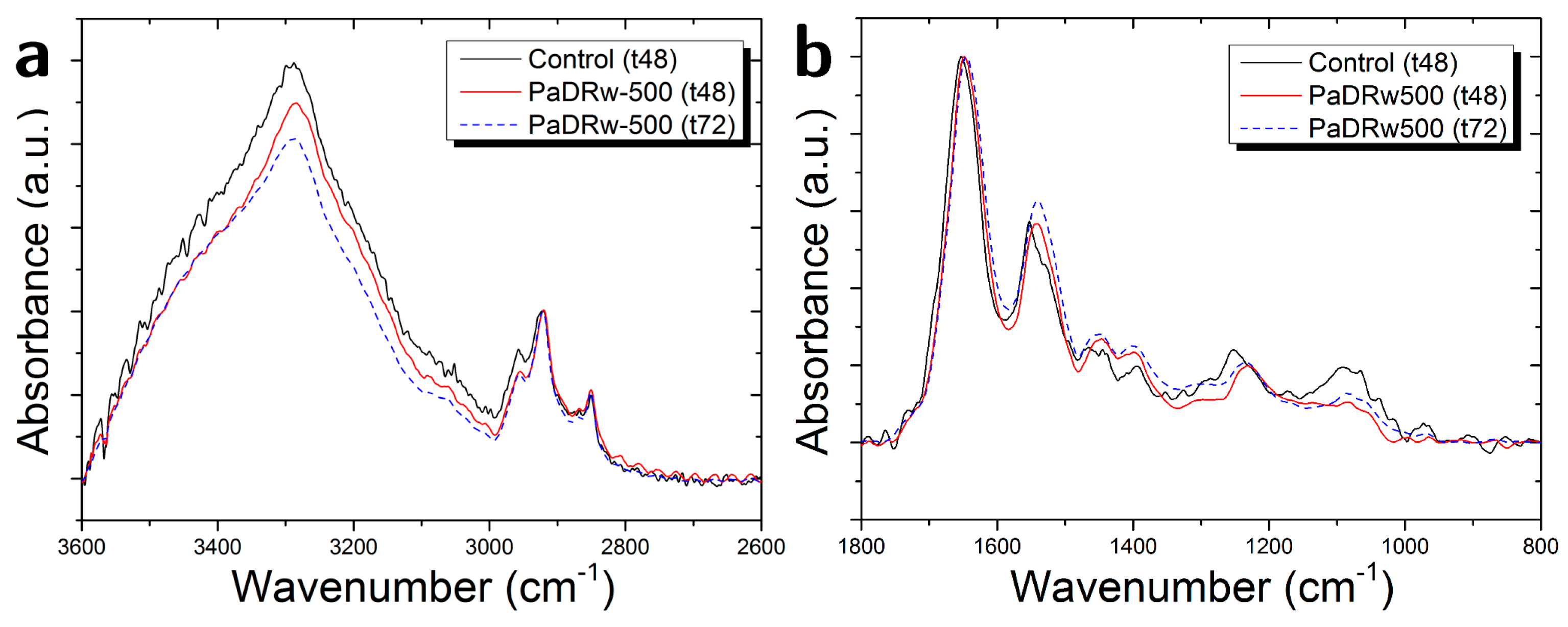
| Ratio | Biomolecular Origin | Indication |
|---|---|---|
| AX (cm−1)/AY (cm−1) | ||
| A1651/A1544 | Amide I/Amide II | Protein rearrangement,DNA/Protein content |
| A1651/A1296 | Amide I/Amide III | Protein rearrangement |
| A1544/A2957 | Amide II/CH3 as. ν | Protein/Lipid content |
| A2922/A2957 | CH2 as. ν/CH3 as. ν | Lipid saturation |
| A1651/A1248 | Amide I/PO2− as. ν | Protein/DNA content |
| A1651/A1082 | Amide I/PO2− s. ν | Protein/DNA content |
| A1082/A1037 | PO2− s. ν/C-O ν | DNA/Carbohydrate content |
| A1146/A2957 | C-O ν/CH3 as. ν | Lipid peroxidation |
| A1248/A2957 | PO2− as. Ν, C-O-P ν/CH3 as. ν | Protein phosphorylation |
| A1082/A2957 | PO2− s. ν, C-O-P ν/CH3 as. ν | Protein phosphorylation |
| Peak | Assignment | |||
|---|---|---|---|---|
| cm−1 | DNA/RNA | Protein | Lipid | Carbohydrate |
| 3306 | Amide A (N-H str.) | O-H str. | ||
| 2957 | CH3 asym. str. | CH3 asym. str. | ||
| 2922 | CH2 asym. str. | |||
| 2869 | CH3 sym. str. | CH3 sym. str. | ||
| 2850 | CH2 sym. str. | |||
| 1651 | Amide I (C=O str., C-N str.) | |||
| 1544 | Amide II (C-N str., C-NH ben.) | |||
| 1453 | CH3 asym. ben., CH2 sc. | CH3 asym. ben., CH2 sc. | ||
| 1393 | COO− sym. str. | COO− sym. str. | ||
| 1296 | Amide III | |||
| 1248 | PO2− asym. str. | C-O-P str. | C-O-P str. | |
| 1146 | CO-O-C str. | C-O str. | C-O str., CO-O-C str. | |
| 1082 | PO2− sym. str. | C-O-P str. | C-O-P str. | |
| 1037 | C-O str. | C-O str. | ||
| Control | PaDRw-25 (t48) | PaDRw-25 (t72) |
|---|---|---|
| Peak (cm−1) | Peak (cm−1) | Peak (cm−1) |
| 3306 (p, c) | 3308 (+2) | 3308 (+2) |
| 2957 (p, l) | 2958 (+1) | 2958 (+1) |
| 2922 (l) | 2921 (−1) | 2922 |
| 2869 (p, l) | 2874 (+5) | 2875 (+6) |
| 2850 (l) | 2850 | 2851 (+1) |
| 1651 (p) | 1646 (−5) | 1648 (−3) |
| 1544 (p) | 1538 (−6) | 1538 (−6) |
| 1453 (p, l) | 1450 (−3) | 1449 (−4) |
| 1393 (p, l) | 1399 (+6) | 1398 (+5) |
| 1296 (p) | 1305 (+9) | 1304 (+8) |
| 1248 (DNA) | 1231 (+17) | 1230 (+18) |
| 1146 (DNA, c) | 1150 (+4) | - |
| 1082 (DNA) | 1084 (+2) | 1096 (+14) |
| 1037 (DNA, c) | 1057 (+20) | 1067 (+30) |
| Control | PaDRw-500 (t48) | PaDRw-500 (t72) |
|---|---|---|
| Peak (cm−1) | Peak (cm−1) | Peak (cm−1) |
| 3306 (p, c) | 3301 (−5) | 3313 (+7) |
| 2957 (p, l) | 2958 (+1) | 2959 (+2) |
| 2922 (l) | 2921 (−1) | 2922 |
| 2869 (p, l) | 2878 (+9) | 2874 (+5) |
| 2850 (l) | 2851 (+1) | 2851 (+1) |
| 1651 (p) | 1646 (−5) | 1644 (−7) |
| 1544 (p) | 1537 (−7) | 1536 (−8) |
| 1453 (p, l) | 1448 (−5) | 1450 (−3) |
| 1393 (p, l) | 1397 (+4) | 1396 (+3) |
| 1296 (p) | 1303 (+7) | 1307 (+11) |
| 1248 (DNA) | 1231 (+17) | 1233 (+15) |
| 1146 (DNA, c) | 1157 (+11) | 1129 (−17) |
| 1082 (DNA) | 1084 (+2) | 1085 (+3) |
| 1037 (DNA, c) | 1056 (+19) | 1056 (+19) |
| Control | Assignments | PaDRw-25 (t48) | PaDRw-25 (t72) | PaDRw-500 (t48) | PaDRw-500 (t72) |
|---|---|---|---|---|---|
| Peak (cm−1) | Peak (cm−1) | Peak (cm−1) | Peak (cm−1) | Peak (cm−1) | |
| 1633 | Β-sheets | 1634 (+1) | 1634 (+1) | 1634 (+1) | 1630 (−3) |
| %A = 10 ± 3 | %A = 34 ± 2 | %A = 31 ± 5 | %A = 38 ± 4 | %A = 26 ± 3 | |
| 1654 | Α-helix | 1654 | 1654 | 1654 | 1652 (−2) |
| %A = 52 ± 13 | %A = 30 ± 3 | %A = 31 ± 7 | %A = 22 ± 5 | %A = 42 ± 5 | |
| 1676 | Turn | 1678 (+2) | 1675 (−1) | 1677 (+1) | 1678 (+2) |
| %A = 10 ± 4 | %A = 6.1 ± 0.9 | %A = 8 ± 2 | %A = 11 ± 2 | %A = 3.8 ± 1.0 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricciardi, V.; Portaccio, M.; Piccolella, S.; Manti, L.; Pacifico, S.; Lepore, M. Study of SH-SY5Y Cancer Cell Response to Treatment with Polyphenol Extracts Using FT-IR Spectroscopy. Biosensors 2017, 7, 57. https://doi.org/10.3390/bios7040057
Ricciardi V, Portaccio M, Piccolella S, Manti L, Pacifico S, Lepore M. Study of SH-SY5Y Cancer Cell Response to Treatment with Polyphenol Extracts Using FT-IR Spectroscopy. Biosensors. 2017; 7(4):57. https://doi.org/10.3390/bios7040057
Chicago/Turabian StyleRicciardi, Valerio, Marianna Portaccio, Simona Piccolella, Lorenzo Manti, Severina Pacifico, and Maria Lepore. 2017. "Study of SH-SY5Y Cancer Cell Response to Treatment with Polyphenol Extracts Using FT-IR Spectroscopy" Biosensors 7, no. 4: 57. https://doi.org/10.3390/bios7040057
APA StyleRicciardi, V., Portaccio, M., Piccolella, S., Manti, L., Pacifico, S., & Lepore, M. (2017). Study of SH-SY5Y Cancer Cell Response to Treatment with Polyphenol Extracts Using FT-IR Spectroscopy. Biosensors, 7(4), 57. https://doi.org/10.3390/bios7040057










