Protein-Flavonoid Interaction Studies by a Taylor Dispersion Surface Plasmon Resonance (SPR) Technique: A Novel Method to Assess Biomolecular Interactions
Abstract
:1. Introduction
2. Experimental Section
2.1. Materials
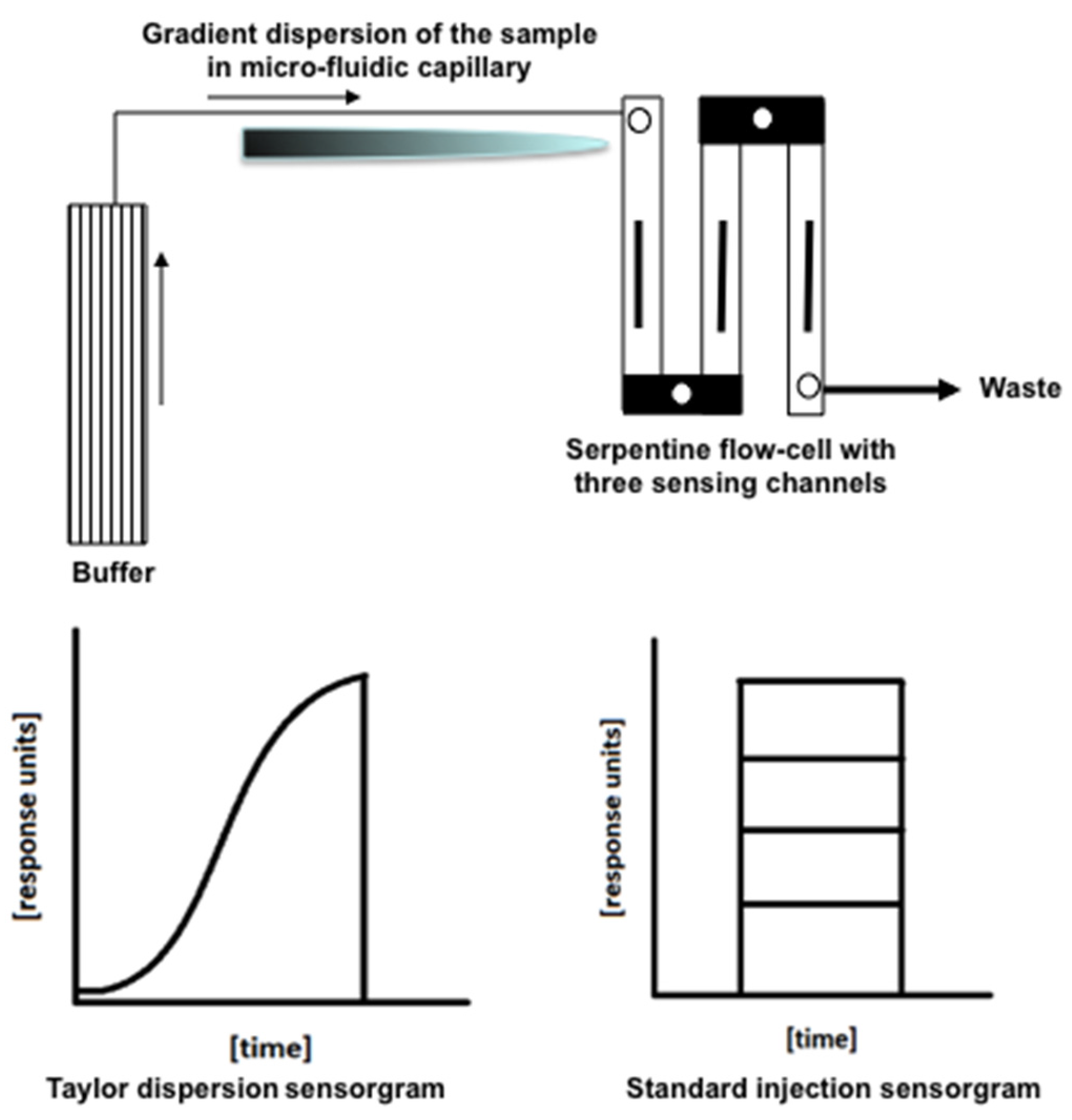
2.2. SPR Measurements
2.3. Data Processing
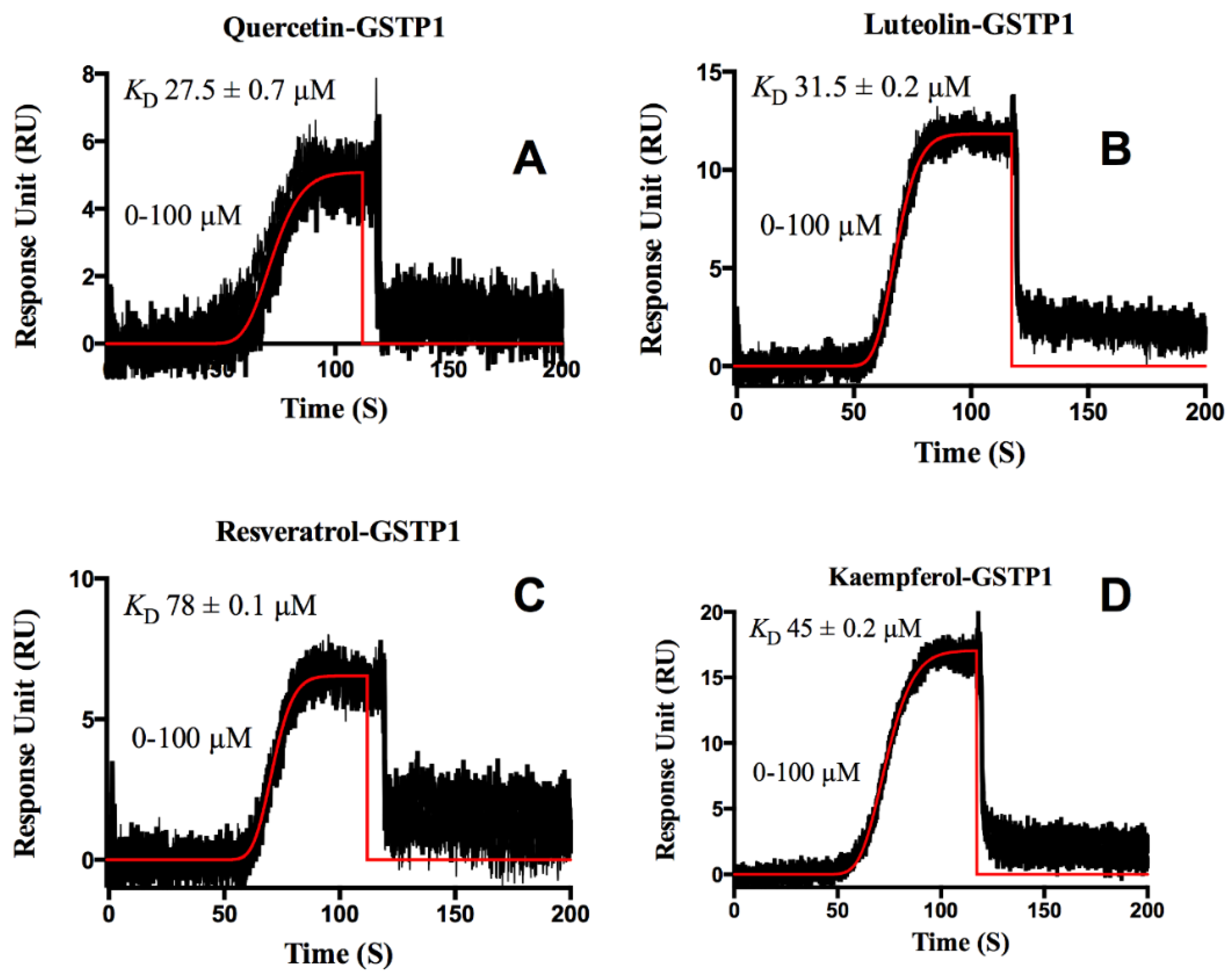
3. Results and Discussion
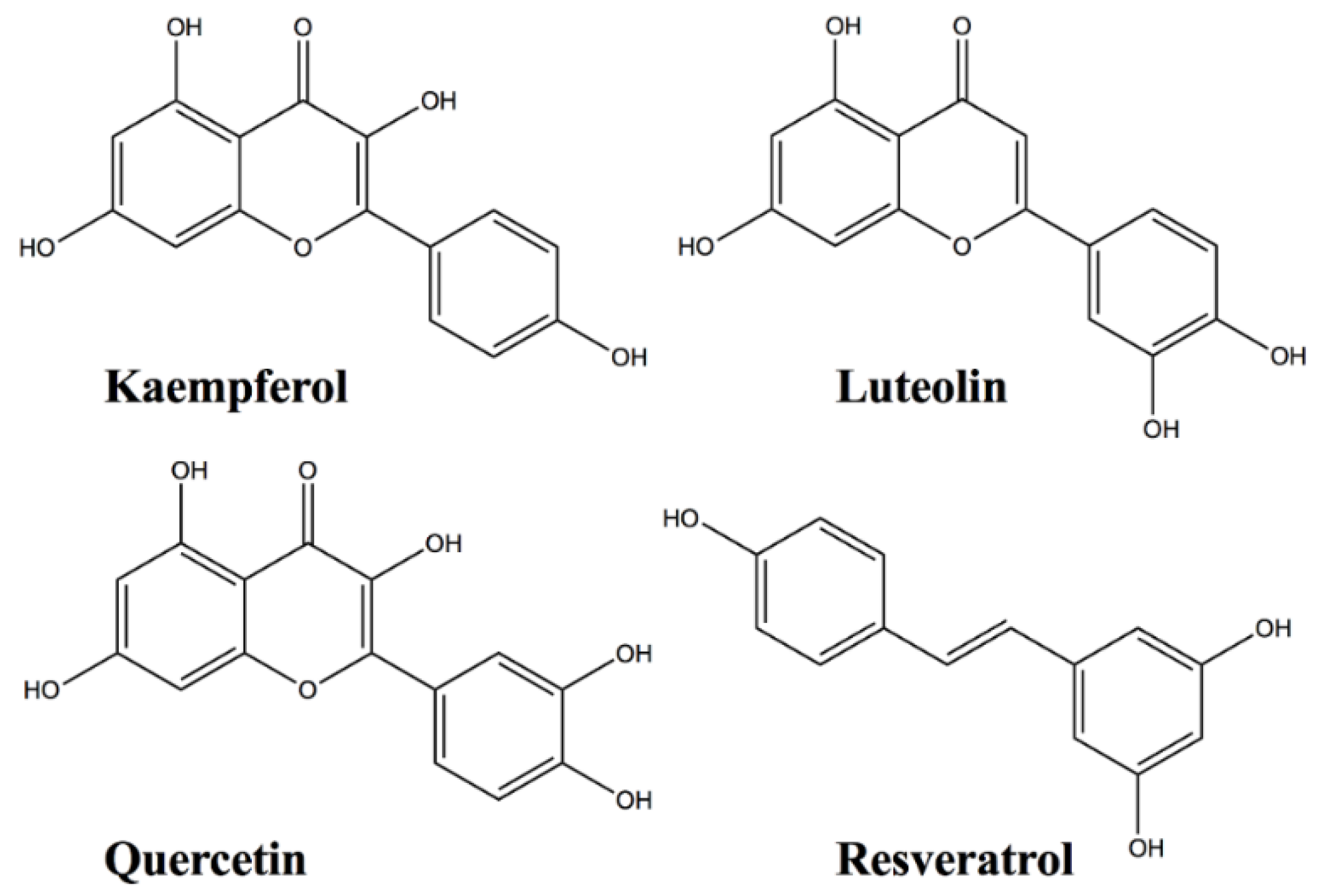
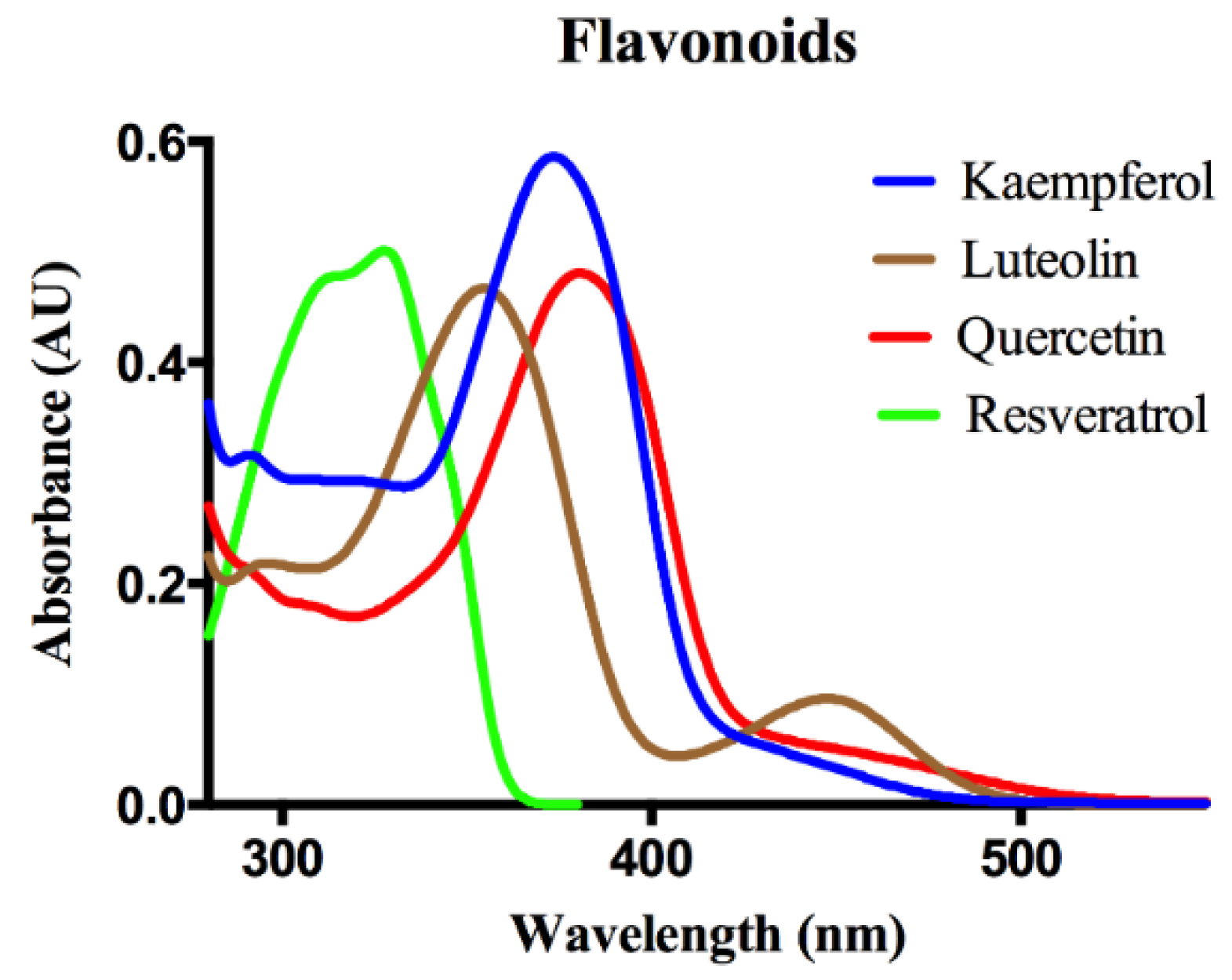
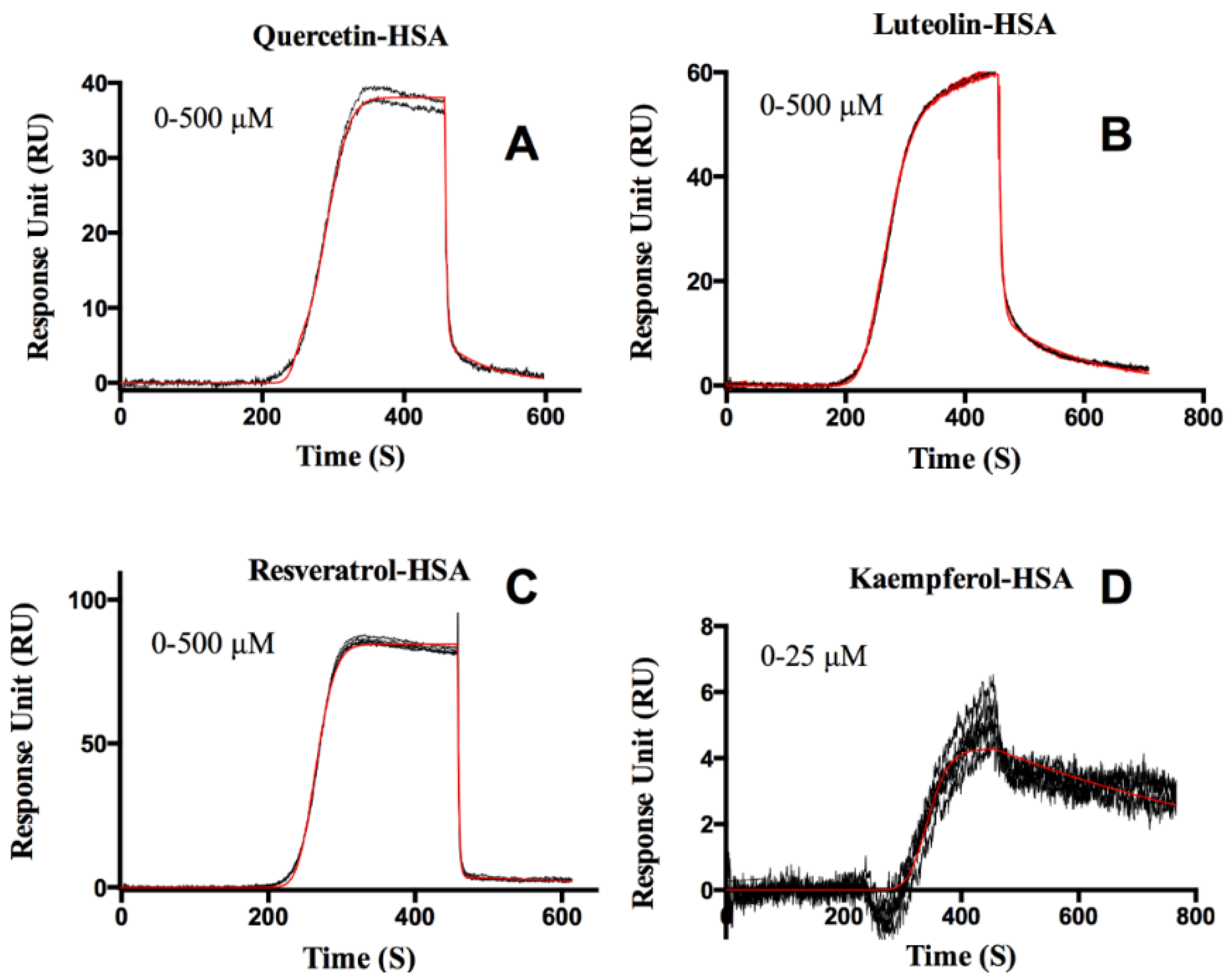
Protein-Flavonoid Interactions
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Michale, G.; Hertog, L. Epidemiological evidence on potential health properties of flavonoids. Proc. Nutr. Soc. 1996, 55, 385–397. [Google Scholar]
- Cook, N.C.; Samman, S. Flavonoids—Chemistry, metabolism, cardioprotective effects, and dietary sources. J. Nutr. Biochem. 1996, 7, 66–76. [Google Scholar] [CrossRef]
- Yang, C.S.; Landau, J.M.; Huang, M.-T.; Newmark, H.L. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu. Rev. Nutr. 2001, 21, 381–406. [Google Scholar] [CrossRef] [PubMed]
- Middleton, E., Jr.; Kandaswami, C.; Theoharides, T.C. The effects of plant flavonoids on mammalian cells: Implications for Inflammation, Heart Disease, and Cancer. Pharmacol. Rev. 2000, 52, 673–751. [Google Scholar] [PubMed]
- Dugas, B.; Charbonnier, S.; Baarine, M.; Ragot, K.; Delmas, D.; Ménétrier, F.; Lherminier, J.; Malvitte, L.; Khalfaoui, T.; Bron, A.; et al. Effects of oxysterols on cell viability, inflammatory cytokines, VEGF, and reactive oxygen species production on human retinal cells: Cytoprotective Effects and Prevention of VEGF Secretion by Resveratrol. Eur. J. Nutr. 2010, 49, 435–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Head, K.A. Natural therapies for ocular disorders, part two: Cataracts and Glaucoma. Altern. Med. Rev. 2001, 6, 141–166. [Google Scholar] [PubMed]
- Hanneken, A.; Lin, F.F.; Johnson, J.; Maher, P. Flavonoids protect human retinal pigment epithelial cells from oxidative-stress-induced death. Invest. Ophthalmol. Vis. Sci. 2006, 47, 3164–3177. [Google Scholar] [CrossRef] [PubMed]
- Maher, P.; Hanneken, A. Flavonoids protect retinal ganglion cells from oxidative stress-induced death. Invest. Ophthalmol. Vis. Sci. 2005, 46, 4796–4803. [Google Scholar] [CrossRef] [PubMed]
- Beatty, S.; Koh, H.; Phil, M.; Henson, D.; Boulton, M. The role of oxidative stress in the pathogenesis of age-related macular degeneration. Surv. Ophthalmol. 2000, 45, 115–134. [Google Scholar] [CrossRef]
- Kampkotter, A.; Gombitang Nkwonkam, C.; Zurawski, R.F.; Timpel, C.; Chovolou, Y.; Watjen, W.; Kahl, R. Effects of the flavonoids kaempferol and fisetin on thermotolerance, oxidative stress and FoxO transcription factor DAF-16 in the model organism Caenorhabditis elegans. Arch. Toxicol. 2007, 81, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Laabich, A.; Manmoto, C.C.; Kuksa, V.; Leung, D.W.; Vissvesvaran, G.P.; Karliga, I.; Kamat, M.; Scott, I.L.; Fawzi, A.; Kubota, R. Protective effects of myricetin and related flavonols against A2E and light mediated-cell death in bovine retinal primary cell culture. Exp. Eye Res. 2007, 85, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Theodore, P., Jr. All About Albumin Biochemistry, Genetics, and Medical Applications; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- van Zanden, J.J.; Ben Hamman, O.; van Iersel, M.L.; Boeren, S.; Cnubben, N.H.; Lo Bello, M.; Vervoort, J.; van Bladeren, P.J.; Rietjens, I.M. Inhibition of human glutathione S-transferase P1-1 by the flavonoid quercetin. Chem. Biol. Interact. 2003, 145, 139–148. [Google Scholar] [CrossRef]
- Bhosale, P.; Larson, A.J.; Frederick, J.M.; Southwick, K.; Thulin, C.D.; Bernstein, P.S. Identification and characterization of a Pi isoform of glutathione S-transferase (GSTP1) as a zeaxanthin-binding protein in the macula of the human eye. J. Biol. Chem. 2004, 279, 49447–49454. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.G. Modeling Taylor dispersion injections: Determination of Kinetic/Affinity Interaction Constants and Diffusion Coefficients in Label-Free Biosensing. Anal. Biochem. 2012, 421, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Quinn, J.G.; Morton, T.; Stepp, J.D.; Myszka, D.G. Biosensor-based fragment screening using FastStep injections. Anal. Biochem. 2010, 407, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Vachali, P.; Li, B.; Nelson, K.; Bernstein, P.S. Surface plasmon resonance (SPR) studies on the interactions of carotenoids and their binding proteins. Arch. Biochem. Biophys. 2012, 519, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, B.; Löfås, S.; Lindquist, G. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal. Biochem. 1991, 198, 268–277. [Google Scholar] [CrossRef]
- Myszka, D.G. Improving biosensor analysis. J. Mol. Recognit. 1999, 12, 279–284. [Google Scholar] [CrossRef]
- Myszka, D.G. Analysis of small-molecule interactions using Biacore S51 technology. Anal. Biochem. 2004, 329, 316–323. [Google Scholar] [CrossRef] [PubMed]
- Day, Y.S.N.; Myszka, D.G. Characterizing a drug's primary binding site on albumin. J. Pharm. Sci. 2003, 92, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Rich, R.L.; Day, Y.S.; Morton, T.A.; Myszka, D.G. High-resolution and high-throughput protocols for measuring drug/human serum albumin interactions using BIACORE. Anal. Biochem. 2001, 296, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.G. Evaluation of Taylor dispersion injections: Determining Kinetic/Affinity Interaction Constants and Diffusion Coefficients in Label-Free Biosensing. Anal. Biochem. 2012, 421, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, L.; Pan, J. Probing the binding of the flavonoid diosmetin to human serum albumin by multispectroscopic techniques. J. Agric. Food Chem. 2012, 60, 2721–2729. [Google Scholar] [CrossRef] [PubMed]
- Bolli, A.; Marino, M.; Rimbach, G.; Fanali, G.; Fasano, M.; Ascenzi, P. Flavonoid binding to human serum albumin. Biochem. Biophys. Res. Commun. 2010, 398, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Jurasekova, Z.; Marconi, G.; Sanchez-Cortes, S.; Torreggiani, A. Spectroscopic and molecular modeling studies on the binding of the flavonoid luteolin and human serum albumin. Biopolymers 2009, 91, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Kaldas, M.I.; Walle, U.K.; van der Woude, H.; McMillan, J.M.; Walle, T. Covalent binding of the flavonoid quercetin to human serum albumin. J. Agric. Food Chem. 2005, 53, 4194–4197. [Google Scholar] [CrossRef] [PubMed]
- Zsila, F.; Bikádi, Z.; Simonyi, M. Probing the binding of the flavonoid, quercetin to human serum albumin by circular dichroism, electronic absorption spectroscopy and molecular modelling methods. Biochem. Pharmacol. 2003, 65, 447–456. [Google Scholar] [CrossRef]
- Di Bari, L.; Ripoli, S.; Pradhan, S.; Salvadori, P. Interactions between quercetin and Warfarin for albumin binding: A New Eye on Food/Drug Interference. Chirality 2010, 22, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Kanakis, C.D.; Tarantilis, P.A.; Polissiou, M.G.; Diamantoglou, S.; Tajmir-Riahi, H.A. Antioxidant flavonoids bind human serum albumin. J. Mol. Struct. 2006, 798, 69–74. [Google Scholar] [CrossRef]
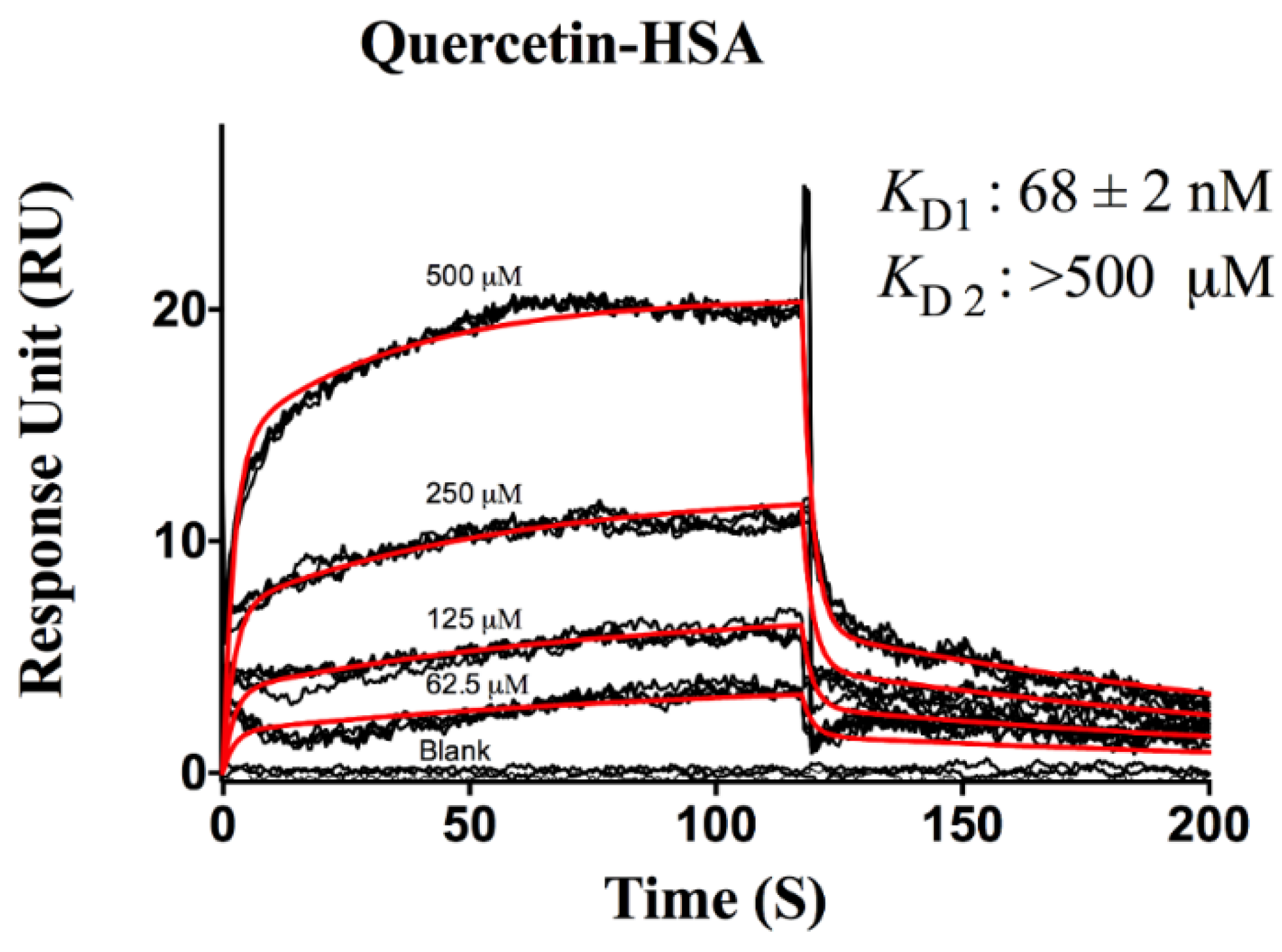
| Flavonoids | ka (M−1s−1) | kd (s−1) | KD (M) | |
|---|---|---|---|---|
| Quercetin | Site 1 | 2.40 ± 0.01 × 105 | 0.016 ± 0.003 | 6.30 ± 0.03 × 10−8 |
| Site 2 | 1.00 ± 0.10 × 102 | 0.405 ± 0.004 | >5.00 × 10−4 | |
| Luteolin | Site 1 | 3.20 ± 0.30 × 103 | 0.205 ± 0.005 | 6.34 ± 0.01 × 10−5 |
| Site 2 | 1.71 ± 0.01 × 101 | 0.007 ± 0.001 | 4.06 ± 0.02 × 10−4 | |
| Resveratrol | Site 1 | 7.00 ± 0.20 × 103 | 0.003± 0.002 | 4.00 ± 0.10 × 10−7 |
| Site 2 | 2.90 ± 0.60 × 103 | 0.520 ± 0.003 | 1.80 ± 0.40 × 10−4 | |
| Kaempferol | Site 1 | 4.84 ± 0.06 × 104 | 0.002 ± 0.002 | 3.70 ± 0.07 × 10−8 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vachali, P.P.; Li, B.; Besch, B.M.; Bernstein, P.S. Protein-Flavonoid Interaction Studies by a Taylor Dispersion Surface Plasmon Resonance (SPR) Technique: A Novel Method to Assess Biomolecular Interactions. Biosensors 2016, 6, 6. https://doi.org/10.3390/bios6010006
Vachali PP, Li B, Besch BM, Bernstein PS. Protein-Flavonoid Interaction Studies by a Taylor Dispersion Surface Plasmon Resonance (SPR) Technique: A Novel Method to Assess Biomolecular Interactions. Biosensors. 2016; 6(1):6. https://doi.org/10.3390/bios6010006
Chicago/Turabian StyleVachali, Preejith P., Binxing Li, Brian M. Besch, and Paul S. Bernstein. 2016. "Protein-Flavonoid Interaction Studies by a Taylor Dispersion Surface Plasmon Resonance (SPR) Technique: A Novel Method to Assess Biomolecular Interactions" Biosensors 6, no. 1: 6. https://doi.org/10.3390/bios6010006
APA StyleVachali, P. P., Li, B., Besch, B. M., & Bernstein, P. S. (2016). Protein-Flavonoid Interaction Studies by a Taylor Dispersion Surface Plasmon Resonance (SPR) Technique: A Novel Method to Assess Biomolecular Interactions. Biosensors, 6(1), 6. https://doi.org/10.3390/bios6010006





