An Aptamer-Based gFET-Sensor for Specific Quantification of Gene Therapeutic Human Adenovirus Type 5
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Adenoviral Particles
2.3. Immobilization of Adenoviral Particles on NHS-Activated Magnetic Beads
2.4. SDS-PAGE and Silver Staining
2.5. In Vitro Selection of Aptamer Libraries Against HAdV-5
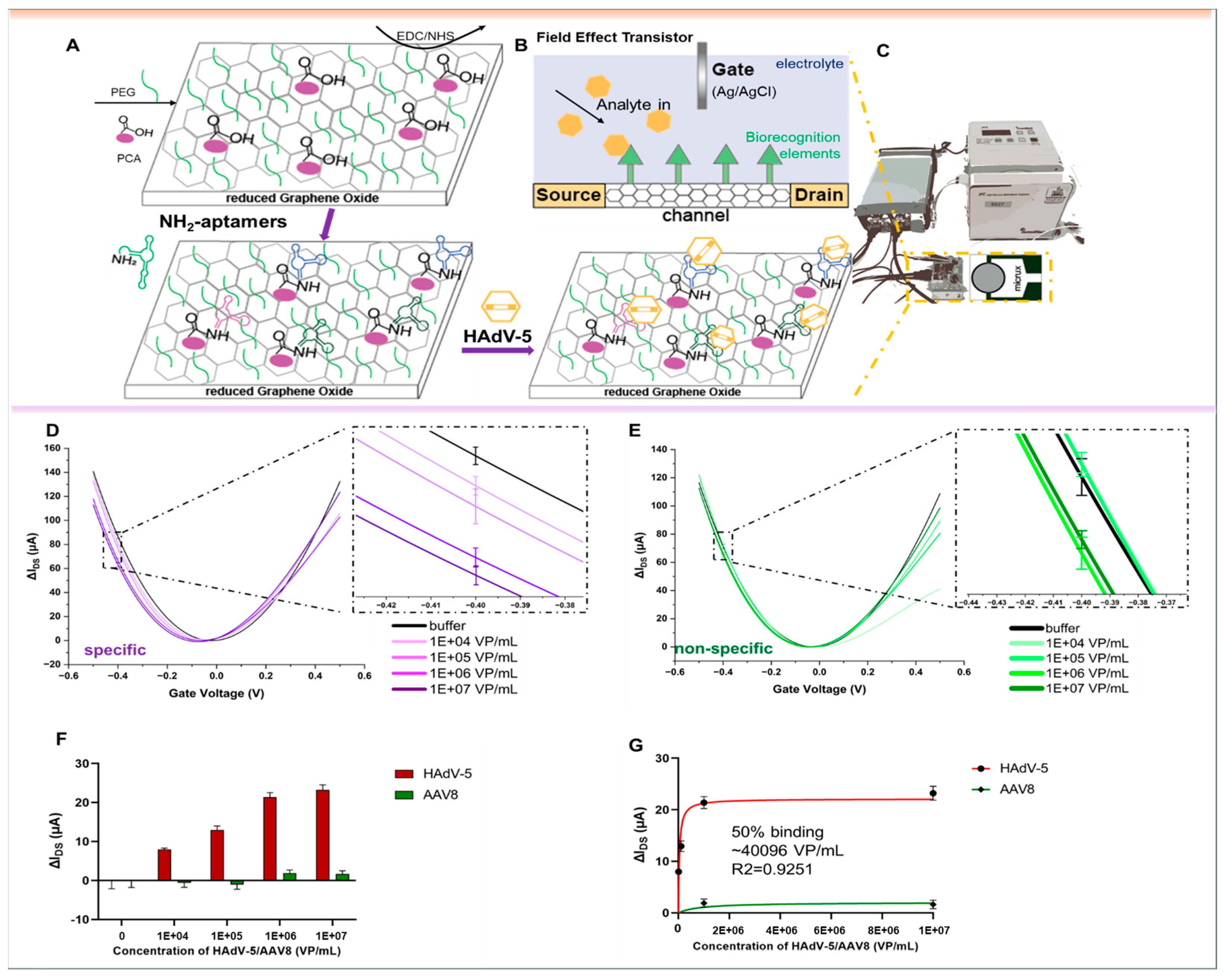
2.6. rGO-FET-Based Quantitative Measurements
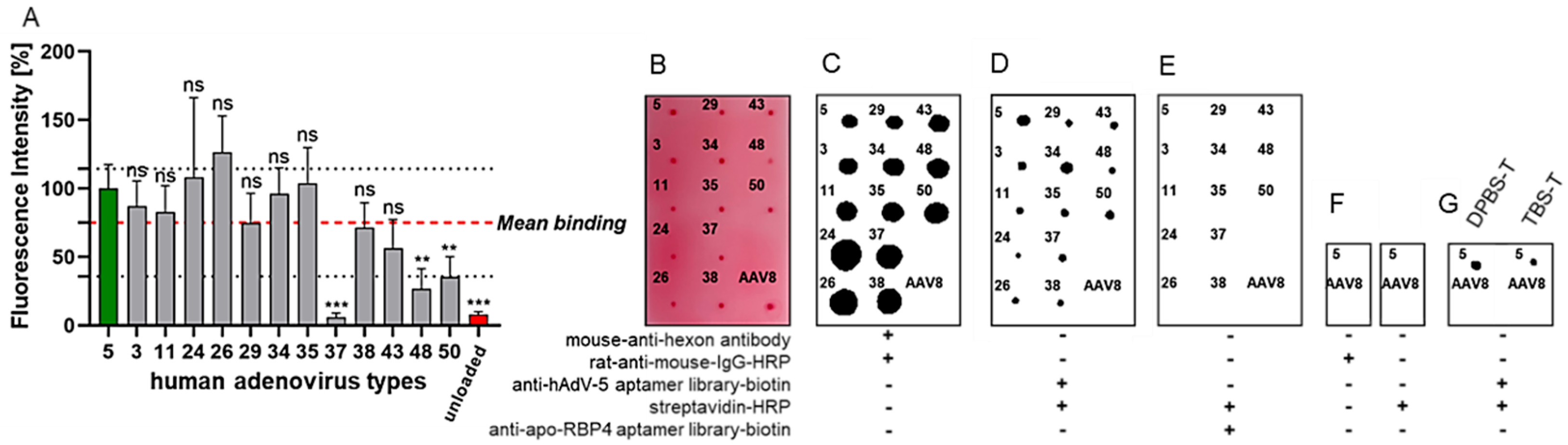
2.7. Dot-Blot Analysis of Aptamer-Binding to Adenoviral Particles
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ellington, A.D.; Szostak, J.W. In Vitro Selection of RNA Molecules That Bind Specific Ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.-H.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic Acid Aptamers: An Emerging Tool for Biotechnology and Biomedical Sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in Analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef]
- Bunka, D.H.J.; Stockley, P.G. Aptamers Come of Age—At Last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Chopra, A.; Shukla, R.; Sharma, T.K. Aptamers as an Emerging Player in Biology. Front. Genet. 2014, 5, 99. [Google Scholar] [CrossRef]
- Chen, A.; Yang, S. Replacing Antibodies with Aptamers in Lateral Flow Immunoassay. Biosens. Bioelectron. 2015, 71, 230–242. [Google Scholar] [CrossRef]
- Guo, W.; Zhang, C.; Ma, T.; Liu, X.; Chen, Z.; Li, S.; Deng, Y. Advances in Aptamer Screening and Aptasensors’ Detection of Heavy Metal Ions. J. Nanobiotechnol. 2021, 19, 166. [Google Scholar] [CrossRef]
- Kusumawati, A.; Mustopa, A.Z.; Wibawan, I.W.T.; Setiyono, A.; Sudarwanto, M.B. A Sequential Toggle Cell-SELEX DNA Aptamer for Targeting Staphylococcus Aureus, Streptococcus Agalactiae, and Escherichia Coli Bacteria. J. Genet. Eng. Biotechnol. 2022, 20, 95. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wan, J.; Wen, X.; Guo, Q.; Jiang, H.; Wang, J.; Ren, Y.; Wang, K. Identification of a New DNA Aptamer by Tissue-SELEX for Cancer Recognition and Imaging. Anal. Chem. 2021, 93, 7369–7377. [Google Scholar] [CrossRef]
- Murakami, K.; Izuo, N.; Bitan, G. Aptamers Targeting Amyloidogenic Proteins and Their Emerging Role in Neurodegenerative Diseases. J. Biol. Chem. 2022, 298, 101478. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA Aptamers Using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef]
- Yang, J.; Bowser, M.T. Capillary Electrophoresis-SELEX Selection of Catalytic DNA Aptamers for a Small-Molecule Porphyrin Target. Anal. Chem. 2013, 85, 1525–1530. [Google Scholar] [CrossRef]
- Zhong, W.; Pu, Y.; Tan, W.; Liu, J.; Liao, J.; Liu, B.; Chen, K.; Yu, B.; Hu, Y.; Deng, Y.; et al. Identification and Application of an Aptamer Targeting Papillary Thyroid Carcinoma Using Tissue-SELEX. Anal. Chem. 2019, 91, 8289–8297. [Google Scholar] [CrossRef]
- Tan, S.Y.; Acquah, C.; Sidhu, A.; Ongkudon, C.M.; Yon, L.S.; Danquah, M.K. SELEX Modifications and Bioanalytical Techniques for Aptamer–Target Binding Characterization. Crit. Rev. Anal. Chem. 2016, 46, 521–537. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. FluMag-SELEX as an Advantageous Method for DNA Aptamer Selection. Anal. Bioanal. Chem. 2005, 383, 83–91. [Google Scholar] [CrossRef]
- Kubiczek, D.; Raber, H.; Bodenberger, N.; Oswald, T.; Sahan, M.; Mayer, D.; Wiese, S.; Stenger, S.; Weil, T.; Rosenau, F. The Diversity of a Polyclonal FluCell-SELEX Library Outperforms Individual Aptamers as Emerging Diagnostic Tools for the Identification of Carbapenem Resistant Pseudomonas Aeruginosa. Chemistry 2020, 26, 14536–14545. [Google Scholar] [CrossRef]
- Raber, H.F.; Kubiczek, D.H.; Bodenberger, N.; Kissmann, A.K.; D’souza, D.; Hu, X.; Mayer, D.; Xu, P.; Knippschild, U.; Spellerberg, B.; et al. FluCell-SELEX Aptamers as Specific Binding Molecules for Diagnostics of the Health Relevant Gut Bacterium Akkermansia Muciniphila. Int. J. Mol. Sci. 2021, 22, 10425. [Google Scholar] [CrossRef]
- Xing, H.; Zhang, Y.; Krämer, M.; Kissmann, A.K.; Henkel, M.; Weil, T.; Knippschild, U.; Rosenau, F. A Polyclonal Selex Aptamer Library Directly Allows Specific Labelling of the Human Gut Bacterium Blautia Producta without Isolating Individual Aptamers. Molecules 2022, 27, 5693. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Kissmann, A.K.; Raber, H.F.; Krämer, M.; Amann, V.; Kohn, K.; Weil, T.; Rosenau, F. Polyclonal Aptamers for Specific Fluorescence Labeling and Quantification of the Health Relevant Human Gut Bacterium Parabacteroides Distasonis. Microorganisms 2021, 9, 2284. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xing, H.; Bolotnikov, G.; Krämer, M.; Gotzmann, N.; Knippschild, U.; Kissmann, A.K.; Rosenau, F. Enriched Aptamer Libraries in Fluorescence-Based Assays for Rikenella Microfusus-Specific Gut Microbiome Analyses. Microorganisms 2023, 11, 2266. [Google Scholar] [CrossRef]
- Xing, H.; Zhang, Y.; Krämer, M.; Kissmann, A.K.; Amann, V.; Raber, H.F.; Weil, T.; Stieger, K.R.; Knippschild, U.; Henkel, M.; et al. A Polyclonal Aptamer Library for the Specific Binding of the Gut Bacterium Roseburia Intestinalis in Mixtures with Other Gut Microbiome Bacteria and Human Stool Samples. Int. J. Mol. Sci. 2022, 23, 7744. [Google Scholar] [CrossRef]
- Kneißle, K.; Krämer, M.; Kissmann, A.K.; Xing, H.; Müller, F.; Amann, V.; Noschka, R.; Gottschalk, K.E.; Bozdogan, A.; Andersson, J.; et al. A Polyclonal SELEX Aptamer Library Allows Differentiation of Candida albicans, C. auris and C. parapsilosis Cells from Human Dermal Fibroblasts. J. Fungi 2022, 8, 856. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Bolotnikov, G.; Krämer, M.; Bozdogan, A.; Kissmann, A.K.; Weil, T.; Spellerberg, B.; Stenger, S.; Rosenau, F. Robust Fluorometric Aptamer Assay for Direct and Rapid Detection of Clinical Isolates of Candida Spec. Int. J. Mol. Sci. 2024, 25, 3444. [Google Scholar] [CrossRef]
- Kissmann, A.K.; Wolf, D.; Krämer, M.; Müller, F.; Amann, V.; Xing, H.; Gottschalk, K.E.; Weil, T.; Eichmann, R.; Schäfer, P.; et al. Polyclonal Aptamer Libraries from a FluRoot-SELEX for the Specific Labeling of the Apical and Elongation/Differentiation Zones of Arabidopsis Thaliana Roots. Int. J. Mol. Sci. 2022, 23, 12220. [Google Scholar] [CrossRef] [PubMed]
- Alemany, R. Oncolytic Adenoviruses in Cancer Treatment. Biomedicines 2014, 2, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Farrera-Sal, M.; Moya-Borrego, L.; Bazan-Peregrino, M.; Alemany, R. Evolving Status of Clinical Immunotherapy with Oncolytic Adenovirus. Clin. Cancer Res. 2021, 27, 2979–2988. [Google Scholar] [CrossRef]
- Suleman, S.; Schrubaji, K.; Filippou, C.; Ignatova, S.; Hewitson, P.; Huddleston, J.; Karda, R.; Waddington, S.N.; Themis, M. Rapid and Inexpensive Purification of Adenovirus Vectors Using an Optimised Aqueous Two-Phase Technology. J. Virol. Methods 2022, 299, 114305. [Google Scholar] [CrossRef]
- Dicks, M.D.J.; Spencer, A.J.; Edwards, N.J.; Wadell, G.; Bojang, K.; Gilbert, S.C.; Hill, A.V.S.; Cottingham, M.G. A Novel Chimpanzee Adenovirus Vector with Low Human Seroprevalence: Improved Systems for Vector Derivation and Comparative Immunogenicity. PLoS ONE 2012, 7, e40385. [Google Scholar] [CrossRef] [PubMed]
- Bos, R.; Rutten, L.; van der Lubbe, J.E.M.; Bakkers, M.J.G.; Hardenberg, G.; Wegmann, F.; Zuijdgeest, D.; de Wilde, A.H.; Koornneef, A.; Verwilligen, A.; et al. Ad26 Vector-Based COVID-19 Vaccine Encoding a Prefusion-Stabilized SARS-CoV-2 Spike Immunogen Induces Potent Humoral and Cellular Immune Responses. NPJ Vaccines 2020, 5, 91. [Google Scholar] [CrossRef]
- Wu, S.; Zhong, G.; Zhang, J.; Shuai, L.; Zhang, Z.; Wen, Z.; Wang, B.; Zhao, Z.; Song, X.; Chen, Y.; et al. A Single Dose of an Adenovirus-Vectored Vaccine Provides Protection against SARS-CoV-2 Challenge. Nat. Commun. 2020, 11, 4081. [Google Scholar] [CrossRef]
- Pilely, K.; Johansen, M.R.; Lund, R.R.; Kofoed, T.; Jørgensen, T.K.; Skriver, L.; Mørtz, E. Monitoring Process-Related Impurities in Biologics–Host Cell Protein Analysis. Anal. Bioanal. Chem. 2021, 414, 747. [Google Scholar] [CrossRef] [PubMed]
- Madisch, I.; Wölfel, R.; Harste, G.; Pommer, H.; Heim, A. Molecular Identification of Adenovirus Sequences: A Rapid Scheme for Early Typing of Human Adenoviruses in Diagnostic Samples of Immunocompetent and Immunodeficient Patients. J. Med. Virol. 2006, 78, 1210–1217. [Google Scholar] [CrossRef]
- Heim, A.; Ebnet, C.; Harste, G.; Pring-Åkerblom, P. Rapid and Quantitative Detection of Human Adenovirus DNA by Real-Time PCR. J. Med. Virol 2003, 70, 228–239. [Google Scholar] [CrossRef]
- Ban, D.K.; Hajian, R.; Chan, M.; Abdolrahimi, S.; Barron, F.; Datta, S.; Aran, K. Real-Time Monitoring in Biomanufacturing with Graphene Field-Effect Transistor Sensors: Detection of PH, Glucose, and Antibodies. GEN Biotechnol. 2024, 3, 341–349. [Google Scholar] [CrossRef]
- Saiki, P.Y.; Rufino, F.C.; de Almeida, C.R.; Sales, G.M.; de Oliveira, A.N.; Larrude, D.R.G.; Teixeira, R.C.; Diniz, J.A.; Catharino, R.R. Sensor Detection of Shelf Life: A Multi-Technique Food Analytical Platform for Mushroom Analysis. Food Res. Int. 2025, 217, 116801. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, D.; Meyerbroeker, N.; Purschke, W.; Sell, S.; Neumann, C.; Winter, A.; Tang, Z.; Hüger, D.; Maasch, C.; Bethge, L.; et al. Ultrasensitive Detection of Chemokines in Clinical Samples with Graphene-Based Field-Effect Transistors. Adv. Mater. 2024, 36, e2407487. [Google Scholar] [CrossRef]
- Geiwitz, M.; Page, O.R.; Marello, T.; Nichols, M.E.; Kumar, N.; Hummel, S.; Belosevich, V.; Ma, Q.; van Opijnen, T.; Batten, B.; et al. Graphene Multiplexed Sensor for Point-of-Need Viral Wastewater-Based Epidemiology. ACS Appl. Bio Mater. 2024, 7, 4622–4632. [Google Scholar] [CrossRef] [PubMed]
- Fomin, M.; Jorde, L.; Steinbach, F.; You, C.; Meyer, C. Liquid-Gated Graphene Field-Effect Transistors for Biosensing on Lipid Monolayers. Phys. Status Solidi B Basic Res. 2023, 260, 2300324. [Google Scholar] [CrossRef]
- Selvarajan, R.S.; Rahim, R.A.; Majlis, B.Y.; Gopinath, S.C.B.; Hamzah, A.A. Ultrasensitive and Highly Selective Graphene-Based Field-Effect Transistor Biosensor for Anti-Diuretic Hormone Detection. Sensors 2020, 20, 2642. [Google Scholar] [CrossRef]
- Danielson, E.; Sontakke, V.A.; Porkovich, A.J.; Wang, Z.; Kumar, P.; Ziadi, Z.; Yokobayashi, Y.; Sowwan, M. Graphene Based Field-Effect Transistor Biosensors Functionalized Using Gas-Phase Synthesized Gold Nanoparticles. Sens. Actuators B Chem. 2020, 320, 128432. [Google Scholar] [CrossRef]
- Béraud, A.; Sauvage, M.; Bazán, C.M.; Tie, M.; Bencherif, A.; Bouilly, D. Graphene Field-Effect Transistors as Bioanalytical Sensors: Design, Operation and Performance. Analyst 2021, 146, 403–428. [Google Scholar] [CrossRef] [PubMed]
- Reiner-Rozman, C.; Larisika, M.; Nowak, C.; Knoll, W. Graphene-Based Liquid-Gated Field Effect Transistor for Biosensing: Theory and Experiments. Biosens. Bioelectron. 2015, 70, 21–27. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, M.; Zhu, M.; Zhang, T.; Qian, X.; Liu, Y.; Ma, X.; Dai, C.; Wei, D.; Zhu, Z.; et al. Ultraprecise Detection of Influenza Virus by Antibody-Modified Graphene Transistors. Sensors 2025, 25, 959. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhu, B.; Zhu, C.; Lai, P.; Taylor, J.; Honney, C.; Nutsford, A.; Ma, C.; Chen, H.; Aw, K.C.; et al. Ultrasensitive, Real-Time Detection of Viral Antigens and RNA Enabled by Scalable Graphene-Based FET Sensors for Pathogen Detection: A Case Study on COVID-19. ACS Sens. 2025, 10, 1909–1921. [Google Scholar] [CrossRef] [PubMed]
- Kissmann, A.K.; Andersson, J.; Bozdogan, A.; Amann, V.; Krämer, M.; Xing, H.; Raber, H.F.; Kubiczek, D.H.; Aspermair, P.; Knoll, W.; et al. Polyclonal Aptamer Libraries as Binding Entities on a Graphene FET Based Biosensor for the Discrimination of Apo- and Holo-Retinol Binding Protein 4. Nanoscale Horiz. 2022, 7, 770–778. [Google Scholar] [CrossRef]
- Ono, T.; Kanai, Y.; Inoue, K.; Watanabe, Y.; Nakakita, S.I.; Kawahara, T.; Suzuki, Y.; Matsumoto, K. Electrical Biosensing at Physiological Ionic Strength Using Graphene Field-Effect Transistor in Femtoliter Microdroplet. Nano Lett. 2019, 19, 4004–4009. [Google Scholar] [CrossRef]
- Wang, S.; Hossain, M.Z.; Shinozuka, K.; Shimizu, N.; Kitada, S.; Suzuki, T.; Ichige, R.; Kuwana, A.; Kobayashi, H. Graphene Field-Effect Transistor Biosensor for Detection of Biotin with Ultrahigh Sensitivity and Specificity. Biosens. Bioelectron. 2020, 165, 112363. [Google Scholar] [CrossRef]
- Vu, C.-A.; Chen, W.-Y. Predicting Future Prospects of Aptamers in Field-Effect Transistor Biosensors. Molecules 2020, 25, 680. [Google Scholar] [CrossRef]
- Shaban, S.M.; Kim, D.-H. Recent Advances in Aptamer Sensors. Sensors 2021, 21, 979. [Google Scholar] [CrossRef]
- Sánchez-Tirado, E.; Agüí, L.; González-Cortés, A.; Campuzano, S.; Yáñez-Sedeño, P.; Pingarrón, J.M. Electrochemical (Bio)Sensing Devices for Human-Microbiome-Related Biomarkers. Sensors 2023, 23, 837. [Google Scholar] [CrossRef]
- Kissmann, A.K.; Bolotnikov, G.; Li, R.; Müller, F.; Xing, H.; Krämer, M.; Gottschalk, K.E.; Andersson, J.; Weil, T.; Rosenau, F. IMPATIENT-QPCR: Monitoring SELEX Success during in Vitro Aptamer Evolution. Appl. Microbiol. Biotechnol. 2024, 108, 284. [Google Scholar] [CrossRef] [PubMed]
- Herdina, A.N.; Bozdogan, A.; Aspermair, P.; Dostalek, J.; Klausberger, M.; Lingg, N.; Cserjan-Puschmann, M.; Aguilar, P.P.; Auer, S.; Demirtas, H.; et al. Bridging Basic Science and Applied Diagnostics: Comprehensive Viral Diagnostics Enabled by Graphene-Based Electronic Biosensor Technology Advancements. Biosens. Bioelectron. 2025, 267, 116807. [Google Scholar] [CrossRef] [PubMed]
- Reiner-Rozman, C.; Hasler, R.; Andersson, J.; Rodrigues, T.; Bozdogan, A.; Bintinger, J.; Aspermair, P. The Top Performer: Towards Optimized Parameters for Reduced Graphene Oxide Uniformity by Spin Coating. Micro Nano Lett. 2021, 16, 436–442. [Google Scholar] [CrossRef]
- Leikas, A.J.; Ylä-Herttuala, S.; Hartikainen, J.E.K. Adenoviral Gene Therapy Vectors in Clinical Use—Basic Aspects with a Special Reference to Replication-Competent Adenovirus Formation and Its Impact on Clinical Safety. Int. J. Mol. Sci. 2023, 24, 16519. [Google Scholar] [CrossRef]
- Michalik, S.; Siegerist, F.; Palankar, R.; Franzke, K.; Schindler, M.; Reder, A.; Seifert, U.; Cammann, C.; Wesche, J.; Steil, L.; et al. Comparative Analysis of ChAdOx1 NCoV-19 and Ad26.COV2.S SARS-CoV-2 Vector Vaccines. Haematologica 2022, 107, 947–957. [Google Scholar] [CrossRef] [PubMed]
- Xing, H.; Zhang, Y.; Li, R.; Ruzicka, H.M.; Hain, C.; Andersson, J.; Bozdogan, A.; Henkel, M.; Knippschild, U.; Hasler, R.; et al. A Blautia Producta Specific GFET-Based Aptasensor for Quantitative Monitoring of Microbiome Quality. Nanoscale Horiz. 2024, 10, 124–134. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Li, R.; Andersson, J.; Bozdogan, A.; Strassl, R.; Draphoen, B.; Lindén, M.; Henkel, M.; Knippschild, U.; et al. Specific GFET-Based Aptasensors for Monitoring of Microbiome Quality: Quantification of the Enteric Health-Relevant Bacterium Roseburia Intestinalis. Adv. Heal. Mater. 2025, 14, e2403827. [Google Scholar] [CrossRef]
- Li, K.; Tu, J.; Zhang, Y.; Jin, D.; Li, T.; Li, J.; Ni, W.; Xiao, M.M.; Zhang, Z.Y.; Zhang, G.J. Ultrasensitive Detection of Exosomal MiRNA with PMO-Graphene Quantum Dots-Functionalized Field-Effect Transistor Biosensor. iScience 2022, 25, 104522. [Google Scholar] [CrossRef]
- Thakur, B.; Zhou, G.; Chang, J.; Pu, H.; Jin, B.; Sui, X.; Yuan, X.; Yang, C.H.; Magruder, M.; Chen, J. Rapid Detection of Single E. Coli Bacteria Using a Graphene-Based Field-Effect Transistor Device. Biosens. Bioelectron. 2018, 110, 16–22. [Google Scholar] [CrossRef]
- Zhou, L.; Mao, H.; Wu, C.; Tang, L.; Wu, Z.; Sun, H.; Zhang, H.; Zhou, H.; Jia, C.; Jin, Q.; et al. Label-Free Graphene Biosensor Targeting Cancer Molecules Based on Non-Covalent Modification. Biosens. Bioelectron. 2017, 87, 701–707. [Google Scholar] [CrossRef]
- Thriveni, G.; Ghosh, K. Advancement and Challenges of Biosensing Using Field Effect Transistors. Biosensors 2022, 12, 647. [Google Scholar] [CrossRef]
- Forsyth, R.; Devadoss, A.; Guy, O. Graphene Field Effect Transistors for Biomedical Applications: Current Status and Future Prospects. Diagnostics 2017, 7, 45. [Google Scholar] [CrossRef] [PubMed]
- Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; San Martín, C. Adenovirus Structure: What Is New? Int. J. Mol. Sci. 2021, 22, 5240. [Google Scholar] [CrossRef]
- Chen, H.; Zheng, X.; Di, B.Y.; Wang, D.; Zhang, Y.; Xia, H.; Mao, Q. Aptamer Modification Improves the Adenoviral Transduction of Malignant Glioma Cells. J. Biotechnol. 2013, 168, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, X.; Xiao, S.; Lin, Y.; Li, C.; Hao, N.; Zhou, M.; Deng, R.; Ke, S.; Zhong, Z.; et al. Characterization of Aptamer-Mediated Gene Delivery System for Liver Cancer Therapy. Oncotarget 2017, 9, 6830–6840. [Google Scholar] [CrossRef] [PubMed]
- Kukushkin, V.; Ambartsumyan, O.; Subekin, A.; Astrakhantseva, A.; Gushchin, V.; Nikonova, A.; Dorofeeva, A.; Zverev, V.; Keshek, A.; Meshcheryakova, N.; et al. Multiplex Lithographic SERS Aptasensor for Detection of Several Respiratory Viruses in One Pot. Int. J. Mol. Sci. 2023, 24, 8081. [Google Scholar] [CrossRef]
- Peinetti, A.S.; Lake, R.J.; Cong, W.; Cooper, L.; Wu, Y.; Ma, Y.; Pawel, G.T.; Toimil-Molares, M.E.; Trautmann, C.; Rong, L.; et al. Direct Detection of Human Adenovirus or SARS-CoV-2 with Ability to Inform Infectivity Using DNA Aptamer-Nanopore Sensors. Sci. Adv. 2021, 7, eabh2848. [Google Scholar] [CrossRef]
- Szunerits, S.; Rodrigues, T.; Bagale, R.; Happy, H.; Boukherroub, R.; Knoll, W. Graphene-Based Field-Effect Transistors for Biosensing: Where Is the Field Heading To? Anal. Bioanal. Chem. 2024, 416, 2137–2150. [Google Scholar] [CrossRef]
- Nakatsuka, N.; Yang, K.-A.; Abendroth, J.M.; Cheung, K.M.; Xu, X.; Yang, H.; Zhao, C.; Zhu, B.; Rim, Y.S.; Yang, Y.; et al. Aptamer–Field-Effect Transistors Overcome Debye Length Limitations for Small-Molecule Sensing. Science 2018, 362, 319–324. [Google Scholar] [CrossRef]
- Sekhon, S.; Bayford, R.; Demosthenous, A. Capacitive Sensors for Label-Free Detection in High-Ionic-Strength Bodily Fluids: A Review. Biosensors 2025, 15, 491. [Google Scholar] [CrossRef]
- Hu, W.-P.; Wang, J.-S.; Chiu, Y.-P.; Huang, T.-C.; Yadlapalli, B.K.; Chen, W.-Y. The Optimal Ionic Concentration of Sensing Buffer in the Detection of RNA by Using the DNA Probe with the Silicon Nanowire Field-Effect Transistor (SiNW-FET). Talanta 2025, 295, 128375. [Google Scholar] [CrossRef]
- Wang, L.; Bao, L.; Qiao, L.; Wang, J.; Wang, Y.; Fu, W.; Zhang, X. Epitope-Imprinted Field-Effect Transistors Overcome Debye Length Limitations for Label-Free Protein Detection. Nano Lett. 2025, 25, 6184–6191. [Google Scholar] [CrossRef]
- Rodrigues, T.; Mishyn, V.; Leroux, Y.R.; Butruille, L.; Woitrain, E.; Barras, A.; Aspermair, P.; Happy, H.; Kleber, C.; Boukherroub, R.; et al. Highly Performing Graphene-Based Field Effect Transistor for the Differentiation between Mild-Moderate-Severe Myocardial Injury. Nano Today 2022, 43, 101391. [Google Scholar] [CrossRef]
- Xing, H.; Li, R.; Zhang, Y.; Rajpal, S.; Bolotnikov, G.; Gruber, D.; Ruzicka, H.-M.; Andersson, J.; Bozdogan, A.; Herdina, A.N.; et al. Single-Sequence Based GFET-Aptasensors for the Discrimination of Apo–and Holo-RBP4 in Human Serum. 2025. Available online: https://ssrn.com/abstract=5345344 (accessed on 28 August 2025). [CrossRef]
- Schilham, M.W.; Claas, E.C.; Van Zaane, W.; Heemskerk, B.; Vossen, J.M.; Lankester, A.C.; Toes, R.E.; Echavarria, M.; Kroes, A.C.; Van Tol, M.J. High Levels of Adenovirus DNA in Serum Correlate with Fatal Outcome of Adenovirus Infection in Children after Allogeneic Stem-Cell Transplantation. Clin. Infect. Dis. 2002, 35, 526–532. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, T.; Vijayalingam, S.; Chinnadurai, G. Genetic Identification of Adenovirus Type 5 Genes That Influence Viral Spread. J. Virol. 2006, 80, 2000–2012. [Google Scholar] [CrossRef]
- Cashdollar, J.L.; Huff, E.; Ryu, H.; Grimm, A.C. The Influence of Incubation Time on Adenovirus Quantitation in A549 Cells by Most Probable Number. J. Virol. Methods 2016, 237, 200–203. [Google Scholar] [CrossRef]
- Li, X.; Zhao, C.; Hou, G.; Sun, Z.; Liu, X.; Ding, Y.; Fang, Y.; Liu, Q. Simultaneously Ultrasensitive and Differential Detection of SARS-CoV-2, Adenovirus and Influenza a Virus Using Multiplex Fluorescence Lateral Flow Immunoassay. Front. Immunol. 2025, 16, 1540676. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, Y.; Lyu, J.; Ren, X.; Cheng, W. Development and Application of a Method to Detect 27 Respiratory Pathogens Using Multiplex RT-PCR Combined with MassARRAY Technology. BMC Infect. Dis. 2021, 21, 870. [Google Scholar] [CrossRef] [PubMed]
- EZERTM ADV Antigen Rapid Test. Available online: https://genesis-ivd.com/index/respiratory-pathogen-rapid-test/13.html (accessed on 28 August 2025).
- Roggendorf, M.; Wigand, R.; Deinhardt, F.; Frösner, G.G. Enzyme-Linked Immunosorbent Assay for Acute Adenovirus Infection. J. Virol. Methods 1982, 4, 27–35. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, L.; Chen, K.; Gu, S.; Wang, J.; Meng, Z.; Li, Y.; Wang, P. Intraperitoneal Oncolytic Virotherapy for Patients with Malignant Ascites: Characterization of Clinical Efficacy and Antitumor Immune Response. Mol. Ther. Oncolytics 2022, 25, 31–42. [Google Scholar] [CrossRef] [PubMed]
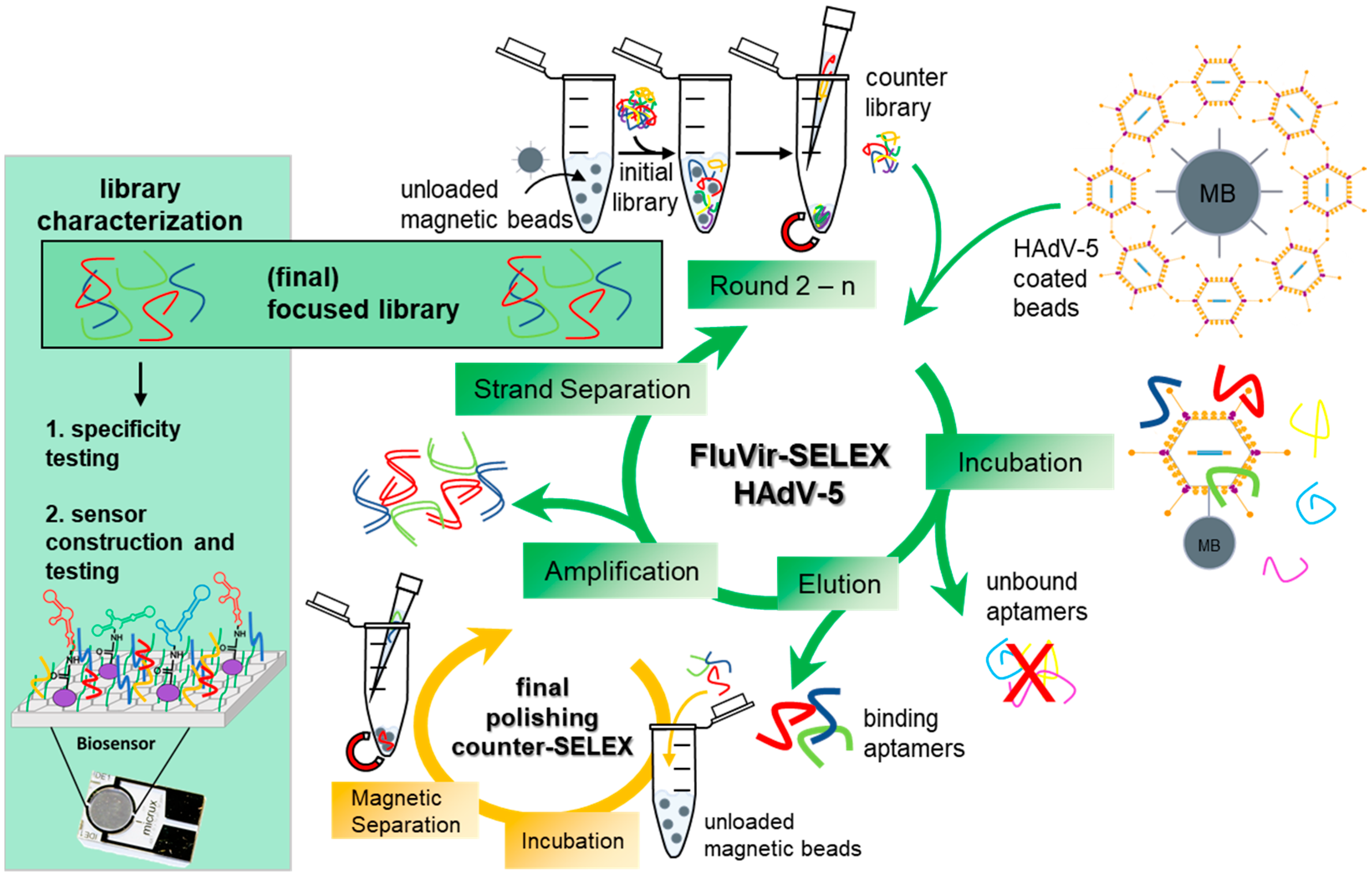
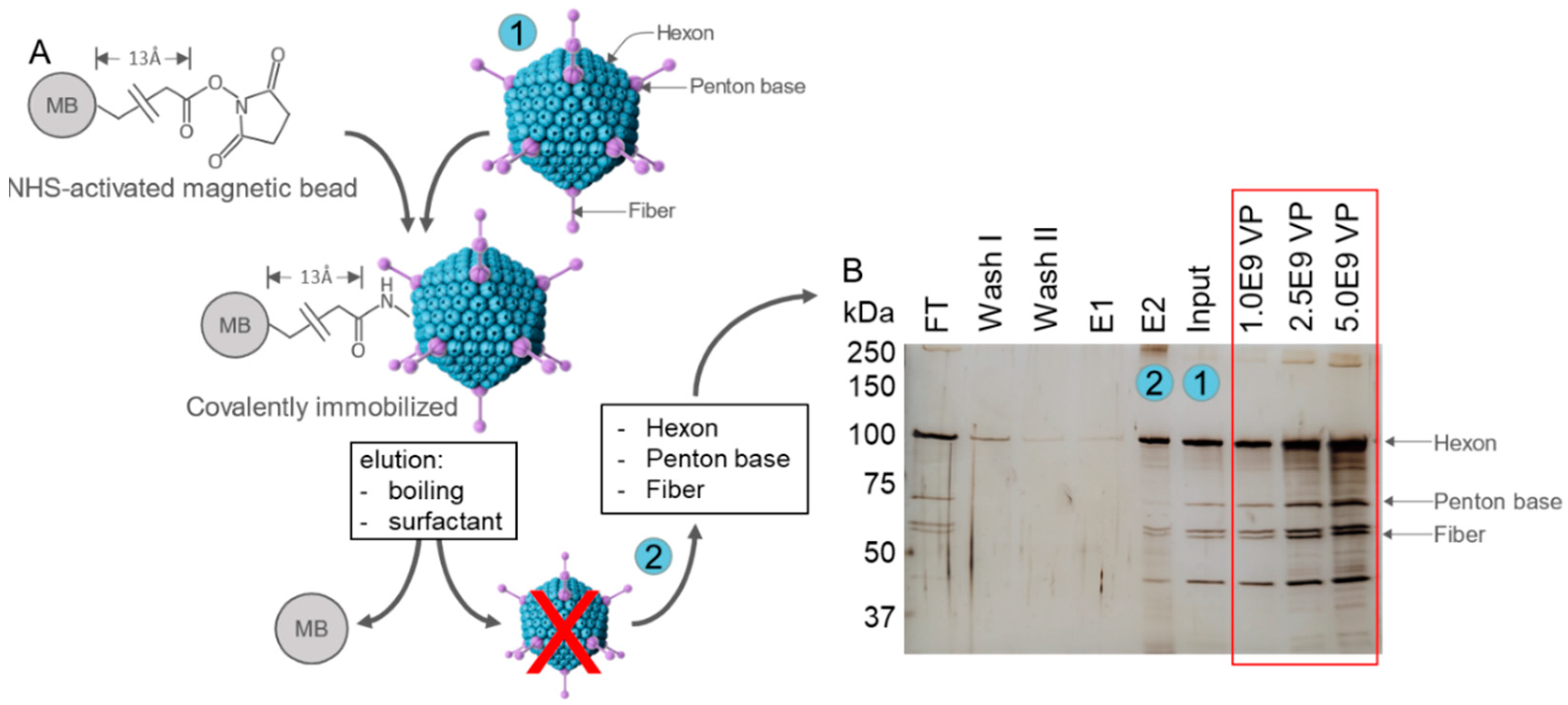
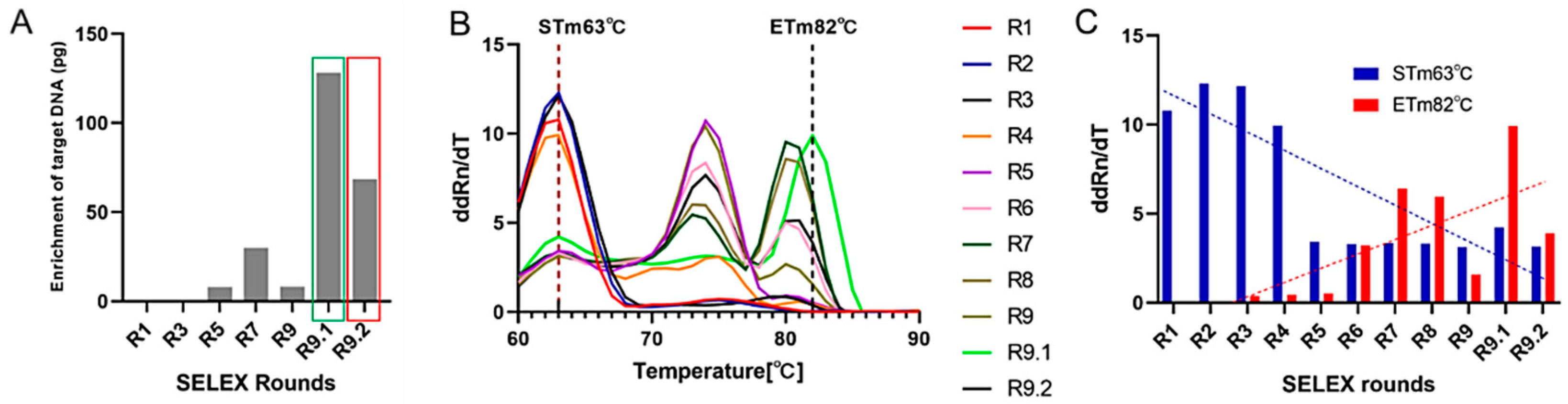
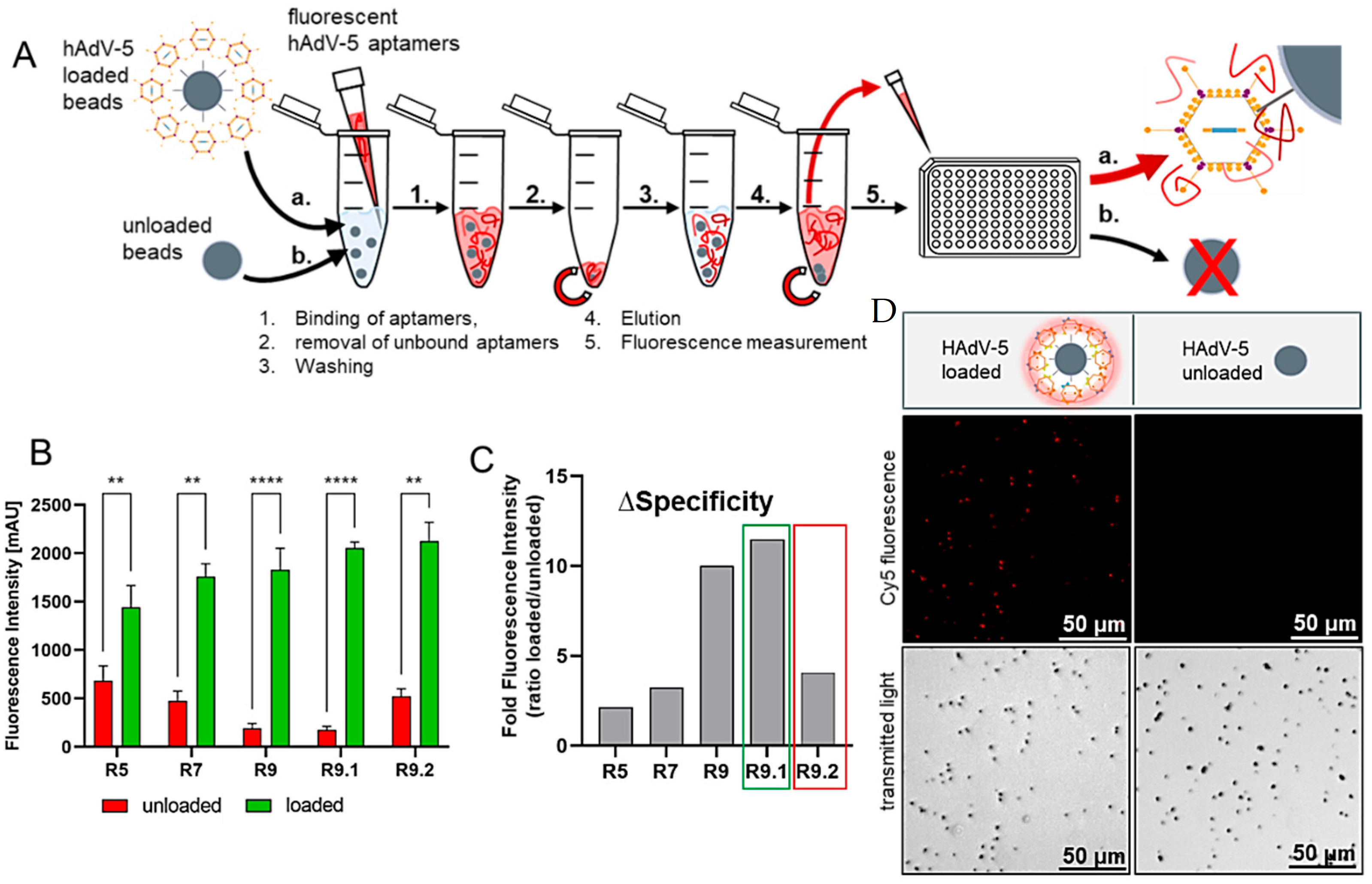
| SELEX Round | Aptamer [pmol] | Counter SELEX | Target SELEX | BSA/tRNA [pmol] | Washing Steps |
|---|---|---|---|---|---|
| 1 | 100 | 1 | 1 | 600 | 3 × HEPES |
| 2 | 10 | 1 | 1 | 900 | 3 × HEPES |
| 3 | 10 | 1 | 1 | 1200 | 3 × HEPES |
| 4 | 10 | 1 | 1 | 1500 | 3 × HEPES |
| 5 | 1 | 1 | 1 | 1800 | 3 × HEPES + 0.1% BSA |
| 6 | 1 | 1 | 1 | 2100 | 4 × HEPES + 0.1% BSA |
| 7 | 1 | 1 | 1 | 2400 | 5 × HEPES + 0.1% BSA |
| 8 | 1 | 1 | 1 | 2700 | 6 × HEPES + 0.1% BSA |
| 9 | 0.1 | 1 | 1 | 3000 | 7 × HEPES + 0.1% BSA |
| 9.1 | 0.1 | 2 | / | / | / |
| 9.2 | 0.01 | 2 | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Kissmann, A.-K.; Xing, H.; Hasler, R.; Kleber, C.; Knoll, W.; Schmietendorf, H.; Engler, T.; Krutzke, L.; Kochanek, S.; et al. An Aptamer-Based gFET-Sensor for Specific Quantification of Gene Therapeutic Human Adenovirus Type 5. Biosensors 2025, 15, 605. https://doi.org/10.3390/bios15090605
Li R, Kissmann A-K, Xing H, Hasler R, Kleber C, Knoll W, Schmietendorf H, Engler T, Krutzke L, Kochanek S, et al. An Aptamer-Based gFET-Sensor for Specific Quantification of Gene Therapeutic Human Adenovirus Type 5. Biosensors. 2025; 15(9):605. https://doi.org/10.3390/bios15090605
Chicago/Turabian StyleLi, Runliu, Ann-Kathrin Kissmann, Hu Xing, Roger Hasler, Christoph Kleber, Wolfgang Knoll, Hannes Schmietendorf, Tatjana Engler, Lea Krutzke, Stefan Kochanek, and et al. 2025. "An Aptamer-Based gFET-Sensor for Specific Quantification of Gene Therapeutic Human Adenovirus Type 5" Biosensors 15, no. 9: 605. https://doi.org/10.3390/bios15090605
APA StyleLi, R., Kissmann, A.-K., Xing, H., Hasler, R., Kleber, C., Knoll, W., Schmietendorf, H., Engler, T., Krutzke, L., Kochanek, S., & Rosenau, F. (2025). An Aptamer-Based gFET-Sensor for Specific Quantification of Gene Therapeutic Human Adenovirus Type 5. Biosensors, 15(9), 605. https://doi.org/10.3390/bios15090605





