Accuracy Enhancement in Refractive Index Sensing via Full-Spectrum Machine Learning Modeling
Abstract
1. Introduction
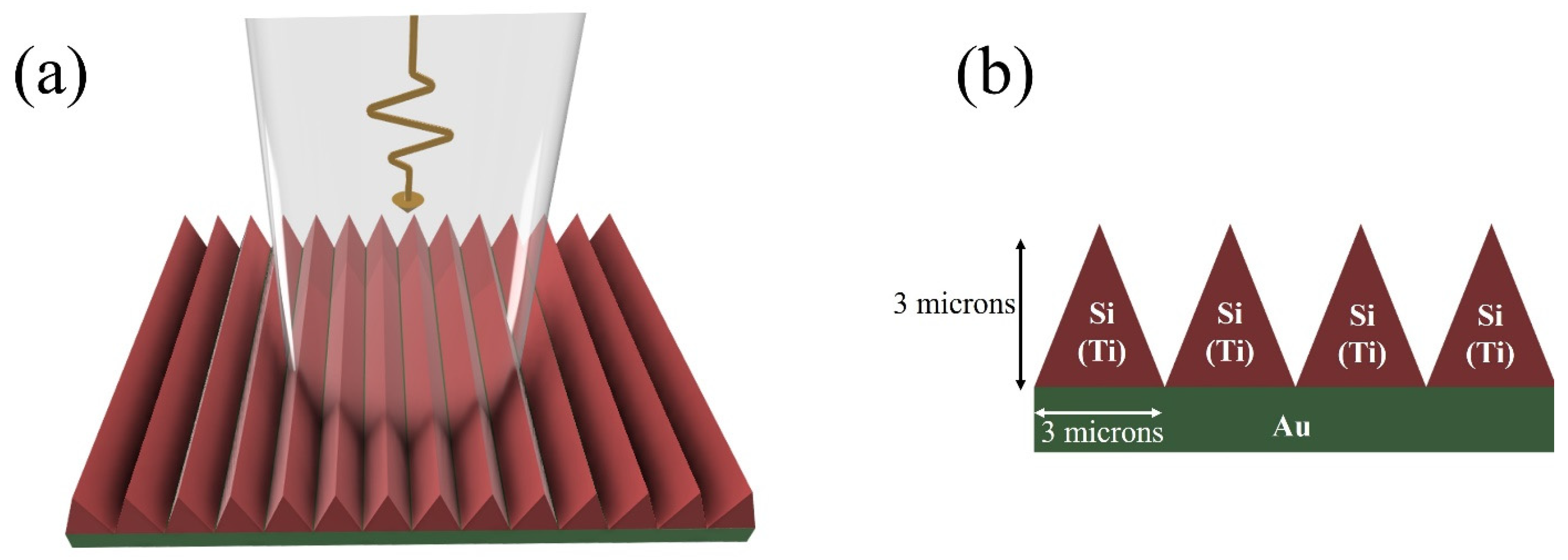
2. Proposed Structure
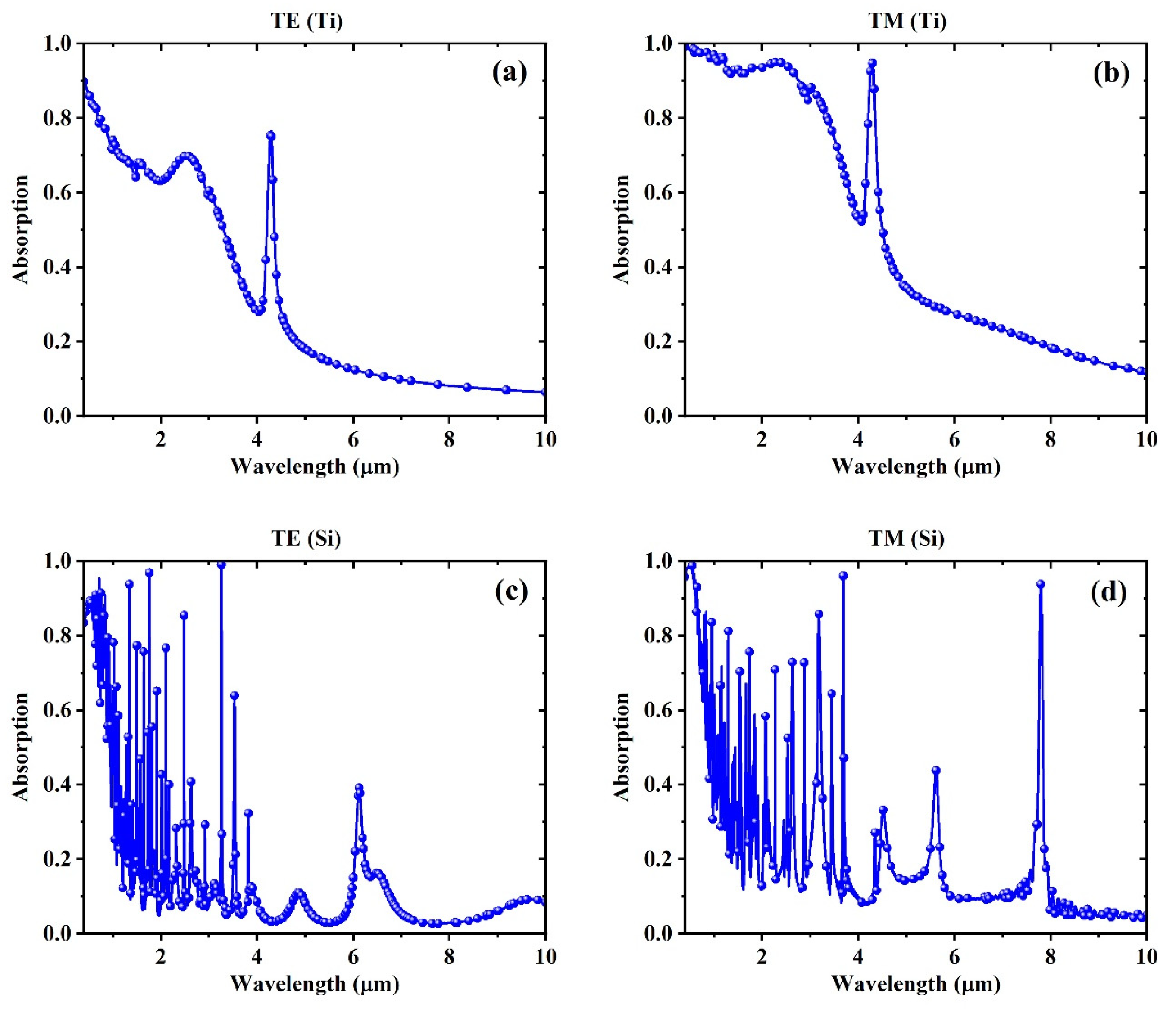
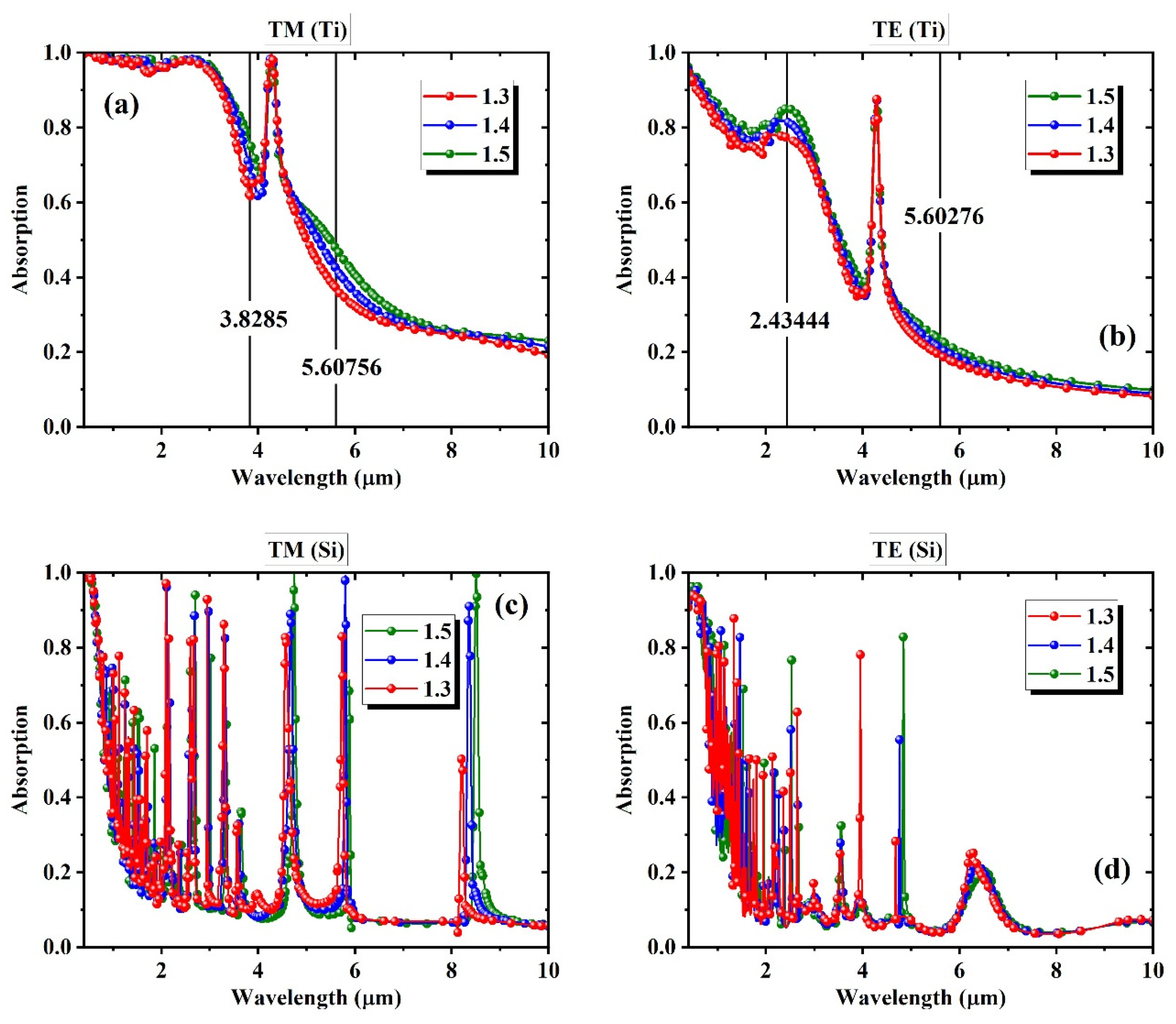
3. Simulation Results
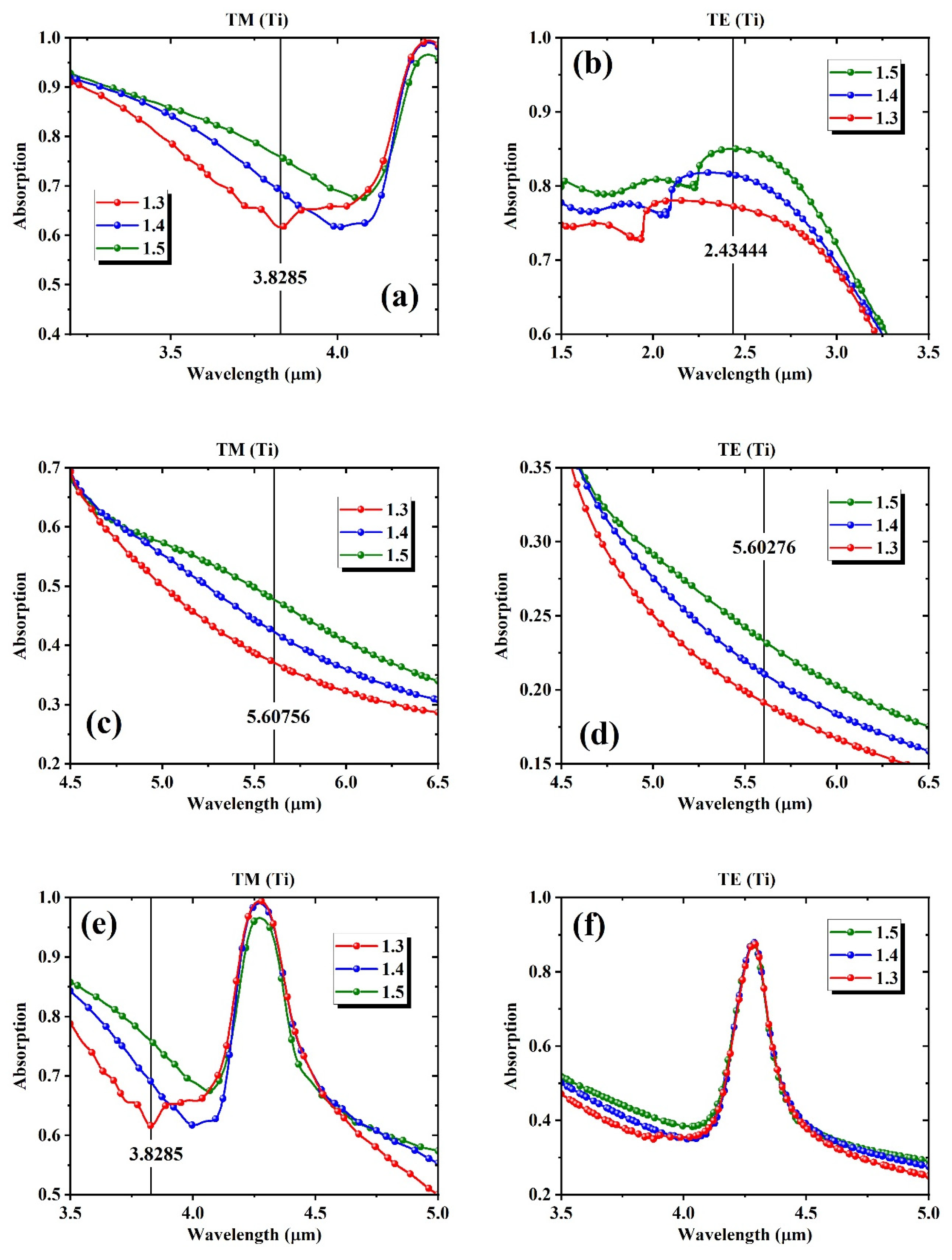
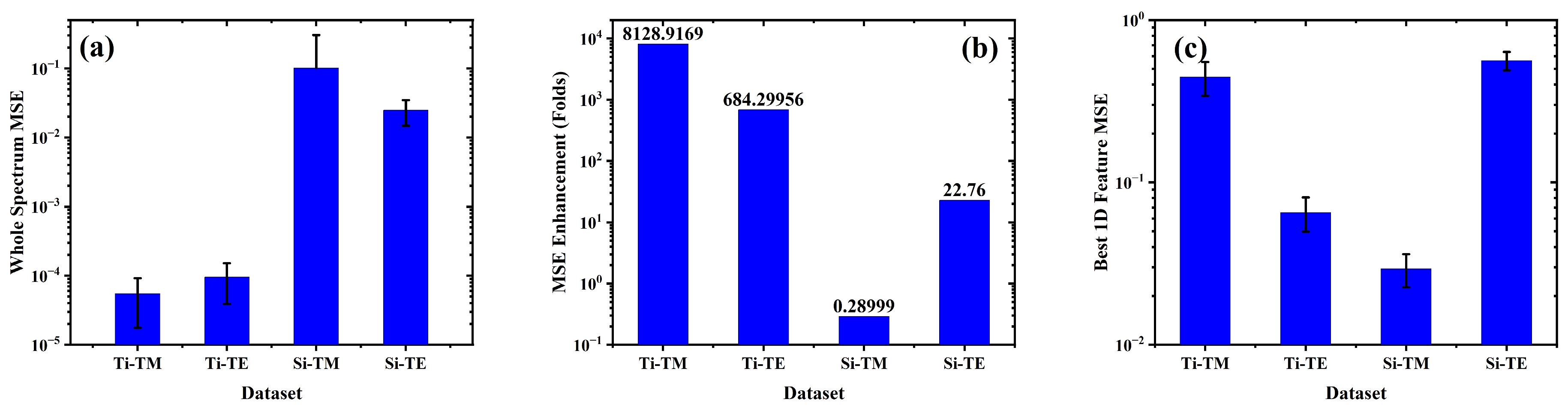
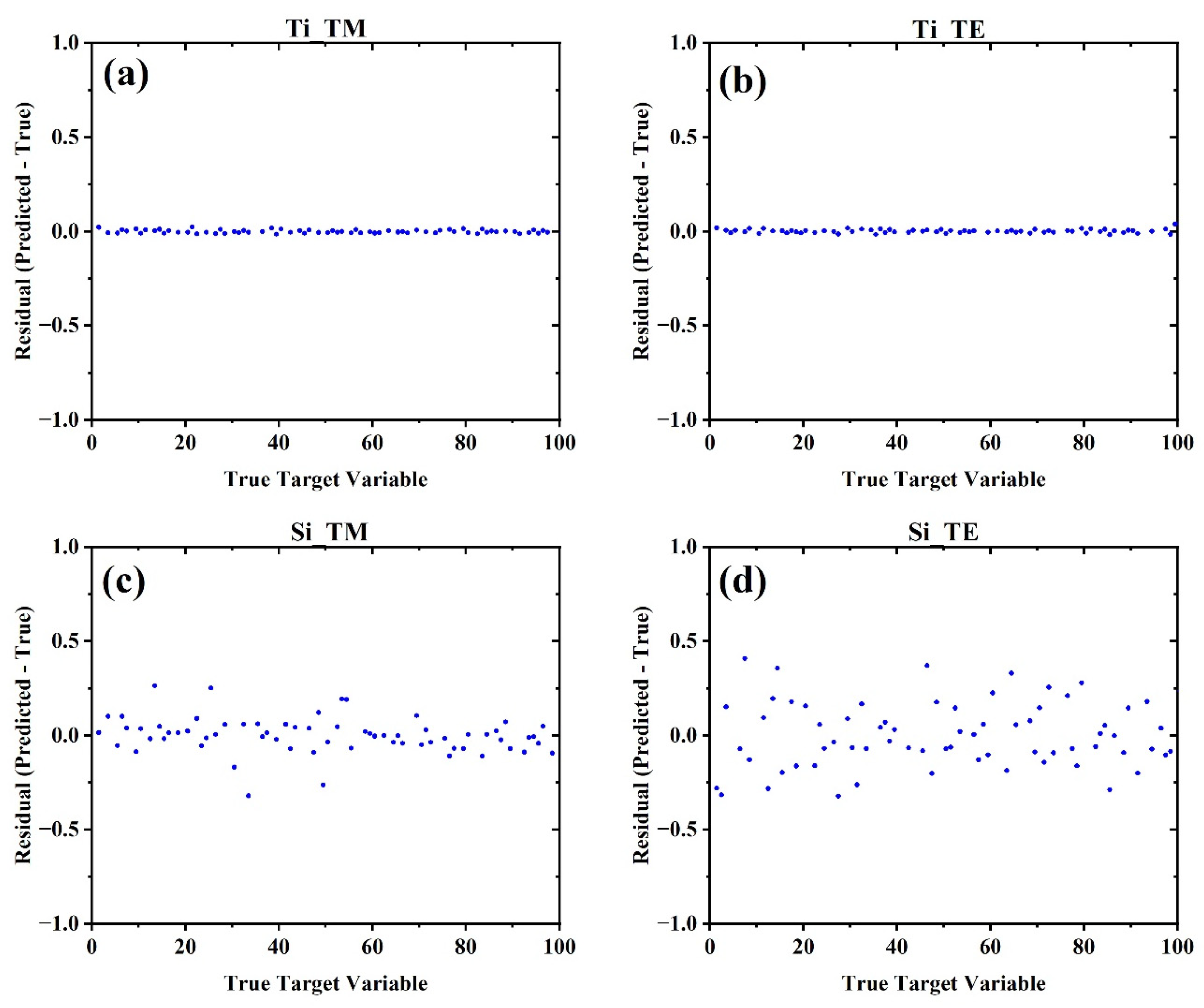
4. Machine Learning Based Full Spectrum Modeling for Precision Enhancement
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, X.; White, I.M.; Shopova, S.I.; Zhu, H.; Suter, J.D.; Sun, Y. Sensitive optical biosensors for unlabeled targets: A review. Anal. Chim. Acta 2008, 620, 8–26. [Google Scholar] [CrossRef]
- Akkilic, N.; Geschwindner, S.; Höök, F. Single-molecule biosensors: Recent advances and applications. Biosens. Bioelectron. 2020, 151, 111944. [Google Scholar] [CrossRef]
- Chen, C.; Wang, J. Optical biosensors: An exhaustive and comprehensive review. Analyst 2020, 145, 1605–1628. [Google Scholar] [CrossRef] [PubMed]
- Haque, E.; Hossain, A.; Namihira, Y.; Ahmed, F. Microchannel-based plasmonic refractive index sensor for low refractive index detection. Appl. Opt. 2019, 58, 1547–1554. [Google Scholar] [CrossRef] [PubMed]
- Homola, J. Present and future of surface plasmon resonance biosensors. Anal. Bioanal. Chem. 2003, 377, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Jágerská, J.; Zhang, H.; Diao, Z.; Le Thomas, N.; Houdré, R. Refractive index sensing with an air-slot photonic crystal nanocavity. Opt. Lett. 2010, 35, 2523–2525. [Google Scholar] [CrossRef]
- Jakšić, Z.; Vuković, S.; Matovic, J.; Tanasković, D. Negative Refractive Index Metasurfaces for Enhanced Biosensing. Materials 2010, 4, 1–36. [Google Scholar] [CrossRef]
- Kaur, B.; Kumar, S.; Kaushik, B.K. Recent advancements in optical biosensors for cancer detection. Biosens. Bioelectron. 2022, 197, 113805. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. A biosensor based on photonic crystal surface waves with an independent registration of the liquid refractive index. Biosens. Bioelectron. 2010, 25, 1212–1216. [Google Scholar] [CrossRef]
- Lavín, Á.; Vicente, J.; Holgado, M.; Laguna, M.; Casquel, R.; Santamaría, B.; Maigler, M.; Hernández, A.; Ramírez, Y.; Lavín, Á.; et al. On the Determination of Uncertainty and Limit of Detection in Label-Free Biosensors. Sensors 2018, 18, 2038. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, W.; Sun, Y. Optical Refractive Index Sensing Based on High-Q Bound States in the Continuum in Free-Space Coupled Photonic Crystal Slabs. Sensors 2017, 17, 1861. [Google Scholar] [CrossRef] [PubMed]
- Lodewijks, K.; Van Roy, W.; Borghs, G.; Lagae, L.; Van Dorpe, P. Boosting the Figure-Of-Merit of LSPR-Based Refractive Index Sensing by Phase-Sensitive Measurements. Nano Lett. 2012, 12, 1655–1659. [Google Scholar] [CrossRef] [PubMed]
- Maksimov, D.N.; Gerasimov, V.S.; Romano, S.; Polyutov, S.P. Refractive index sensing with optical bound states in the continuum. Opt. Express 2020, 28, 38907–38916. [Google Scholar] [CrossRef] [PubMed]
- Perera, C.; Vernon, K.; Cheng, E.; Sathian, J.; Jaatinen, E.; Davis, T. Highly compact refractive index sensor based on stripe waveguides for lab-on-a-chip sensing applications. Beilstein J. Nanotechnol. 2016, 7, 751–757. [Google Scholar] [CrossRef]
- Shangguan, Q.; Chen, Z.; Yang, H.; Cheng, S.; Yang, W.; Yi, Z.; Wu, X.; Wang, S.; Yi, Y.; Wu, P. Design of Ultra-Narrow Band Graphene Refractive Index Sensor. Sensors 2022, 22, 6483. [Google Scholar] [CrossRef]
- Wang, Q.; Ren, Z.-H.; Zhao, W.-M.; Wang, L.; Yan, X.; Zhu, A.-S.; Qiu, F.-M.; Zhang, K.-K. Research advances on surface plasmon resonance biosensors. Nanoscale 2021, 14, 564–591. [Google Scholar] [CrossRef]
- White, I.M.; Fan, X. On the performance quantification of resonant refractive index sensors. Opt. Express 2008, 16, 1020–1028. [Google Scholar] [CrossRef]
- Yan, Z.; Yan, S.; Xu, Z.; Chen, C.; Cao, Y.; Yan, X.; Wang, C.; Wu, T. Multi-Structure-Based Refractive Index Sensor and Its Application in Temperature Sensing. Sensors 2025, 25, 412. [Google Scholar] [CrossRef]
- Ahn, J.-Y.; Lee, K.-A.; Lee, M.-J.; Sekhon, S.S.; Rhee, S.-K.; Cho, S.-J.; Ko, J.H.; Lee, L.; Han, J.; Kim, S.Y.; et al. Surface Plasmon Resonance Aptamer Biosensor for Discriminating Pathogenic Bacteria Vibrio parahaemolyticus. J. Nanosci. Nanotechnol. 2018, 18, 1599–1605. [Google Scholar] [CrossRef]
- Cao, S.; Shao, Y.; Wang, Y.; Wu, T.; Zhang, L.; Huang, Y.; Zhang, F.; Liao, C.; He, J.; Wang, Y. Highly sensitive surface plasmon resonance biosensor based on a low-index polymer optical fiber. Opt. Express 2018, 26, 3988–3994. [Google Scholar] [CrossRef]
- Castiello, F.R.; Tabrizian, M. Multiplex Surface Plasmon Resonance Imaging-Based Biosensor for Human Pancreatic Islets Hormones Quantification. Anal. Chem. 2018, 90, 3132–3139. [Google Scholar] [CrossRef]
- Culver, H.R.; Wechsler, M.E.; Peppas, N.A. Label-Free Detection of Tear Biomarkers Using Hydrogel-Coated Gold Nanoshells in a Localized Surface Plasmon Resonance-Based Biosensor. ACS Nano 2018, 12, 9342–9354. [Google Scholar] [CrossRef]
- Zhu, J.; Gan, S.; Ruan, B.; Wu, L.; Cai, H.; Dai, X.; Xiang, Y. Fano Resonance in Waveguide Coupled Surface Exciton Polaritons: Theory and Application in Biosensor. Sensors 2018, 18, 4437. [Google Scholar] [CrossRef] [PubMed]
- Cennamo, N.; Bencivenga, D.; Annunziata, M.; Arcadio, F.; Stampone, E.; Piccirillo, A.; Della Ragione, F.; Zeni, L.; Guida, L.; Borriello, A. Plasmon resonance biosensor for interleukin-1β point-of-care determination: A tool for early periodontitis diagnosis. iScience 2023, 27, 108741. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.C.; Tseng, W.-C.; Tsai, W.-T.; Huang, C.-S. Handheld Biosensor System Based on a Gradient Grating Period Guided-Mode Resonance Device. Biosensors 2023, 14, 21. [Google Scholar] [CrossRef]
- Matveeva, E.G.; Stoddard, A.K.; Zeng, H.-H.; Franke, G.; Bourne, L.; Fierke, C.A.; Thompson, R.B. Ratiometric Zinc Biosensor Based on Bioluminescence Resonance Energy Transfer: Trace Metal Ion Determination with Tunable Response. Int. J. Mol. Sci. 2022, 23, 14936. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Rahmanian, V.; Gholami, A.; Chiang, W.-H.; Lai, C.W. Biomedical Applications of an Ultra-Sensitive Surface Plasmon Resonance Biosensor Based on Smart MXene Quantum Dots (SMQDs). Biosensors 2022, 12, 743. [Google Scholar] [CrossRef]
- Qi, M.; Lv, D.; Zhang, Y.; Wang, D.; Chen, X.; Zhu, Z.; Hong, Z.; Chai, Y.; Zhang, H.; Cao, Y. Development of a surface plasmon resonance biosensor for accurate and sensitive quantitation of small molecules in blood samples. J. Pharm. Anal. 2022, 12, 929–936. [Google Scholar] [CrossRef]
- Wu, T.-H.; Chang, C.-C.; Vaillant, J.; Bruyant, A.; Lin, C.-W. DNA biosensor combining single-wavelength colorimetry and a digital lock-in amplifier within a smartphone. Lab Chip 2016, 16, 4527–4533. [Google Scholar] [CrossRef]
- Toma, M.; Namihara, S.; Kajikawa, K. Direct detection of neuron-specific enolase using a spectrometer-free colorimetric plasmonic biosensor. Anal. Methods 2023, 15, 2755–2760. [Google Scholar] [CrossRef]
- Fan, Y.; Cui, M.; Liu, Y.; Jin, M.; Zhao, H. Selection and characterization of DNA aptamers for constructing colorimetric biosensor for detection of PBP2a. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 228, 117735. [Google Scholar] [CrossRef] [PubMed]
- Kishnani, V.; Kumari, S.; Gupta, A. A Chemometric-Assisted Colorimetric-Based Inexpensive Paper Biosensor for Glucose Detection. Biosensors 2022, 12, 1008. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Jiao, L.; Cao, F.; Liu, X.; Zhou, Y.; Yang, C.; Gao, Z.; Zhang, M.; Lin, P.; Han, Y.; et al. A Real-Time Detection Method of Hg2+ in Drinking Water via Portable Biosensor: Using a Smartphone as a Low-Cost Micro-Spectrometer to Read the Colorimetric Signals. Biosensors 2022, 12, 1017. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Maity, S.; Mastrangelo, C.H. Nanostructures for Biosensing, with a Brief Overview on Cancer Detection, IoT, and the Role of Machine Learning in Smart Biosensors. Sensors 2021, 21, 1253. [Google Scholar] [CrossRef]
- Hassan, M.; Xu, Y.; Sayada, J.; Zareef, M.; Shoaib, M.; Chen, X.; Li, H.; Chen, Q. Progress of machine learning-based biosensors for the monitoring of food safety: A review. Biosens. Bioelectron. 2024, 267, 116782. [Google Scholar] [CrossRef]
- Khanal, B.; Pokhrel, P.; Khanal, B.; Giri, B. Machine-Learning-Assisted Analysis of Colorimetric Assays on Paper Analytical Devices. ACS Omega 2021, 6, 33837–33845. [Google Scholar] [CrossRef]
- Kokabi, M.; Tahir, M.N.; Singh, D.; Javanmard, M. Advancing Healthcare: Synergizing Biosensors and Machine Learning for Early Cancer Diagnosis. Biosensors 2023, 13, 884. [Google Scholar] [CrossRef]
- Pan, X.; Zhang, Z.; Yun, Y.; Zhang, X.; Sun, Y.; Zhang, Z.; Wang, H.; Yang, X.; Tan, Z.; Yang, Y.; et al. Machine Learning-Assisted High-Throughput Identification and Quantification of Protein Biomarkers with Printed Heterochains. J. Am. Chem. Soc. 2024, 146, 19239–19248. [Google Scholar] [CrossRef]
- Schackart, K.E.; Yoon, J.-Y. Machine Learning Enhances the Performance of Bioreceptor-Free Biosensors. Sensors 2021, 21, 5519. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, J.; Liu, T.; Luo, Y.; Loh, X.J.; Chen, X. Machine Learning-Reinforced Noninvasive Biosensors for Healthcare. Adv. Healthc. Mater. 2021, 10, 2100734. [Google Scholar] [CrossRef]
- Ajala, S.; Jalajamony, H.M.; Nair, M.; Marimuthu, P.; Fernandez, R.E. Comparing machine learning and deep learning regression frameworks for accurate prediction of dielectrophoretic force. Sci. Rep. 2022, 12, 11971. [Google Scholar] [CrossRef]
- Li, W.; Long, Y.; Yan, Y.; Xiao, K.; Wang, Z.; Zheng, D.; Leal-Junior, A.; Kumar, S.; Ortega, B.; Marques, C.; et al. Wearable photonic smart wristband for cardiorespiratory function assessment and biometric identification. Opto-Electron. Adv. 2025, 8, 240254. [Google Scholar] [CrossRef]
- Aalizadeh, M.; Afshar, M.A.; Fan, X. Machine Learning Enabled Multidimensional Data Utilization Through Multi-Resonance Architecture: A Pathway to Enhanced Accuracy in Biosensing. ACS Omega 2025, 10, 20713–20722. [Google Scholar] [CrossRef] [PubMed]
- Richter, F.U.; Sinev, I.; Zhou, S.; Leitis, A.; Oh, S.; Tseng, M.L.; Kivshar, Y.; Altug, H. Gradient High-Q Dielectric Metasurfaces for Broadband Sensing and Control of Vibrational Light-Matter Coupling. Adv. Mater. 2024, 36, 2314279. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, D.; Tittl, A.; Ait-Bouziad, N.; John-Herpin, A.; Limaj, O.; Kelly, C.; Yoo, D.; Wittenberg, N.J.; Oh, S.-H.; Lashuel, H.A.; et al. Resolving molecule-specific information in dynamic lipid membrane processes with multi-resonant infrared metasurfaces. Nat. Commun. 2018, 9, 2160. [Google Scholar] [CrossRef]
- Shen, Z.; Du, M. High-performance refractive index sensing system based on multiple Fano resonances in polarization-insensitive metasurface with nanorings. Opt. Express 2021, 29, 28287–28296. [Google Scholar] [CrossRef]
- Tittl, A.; John-Herpin, A.; Leitis, A.; Arvelo, E.R.; Altug, H. Metasurface-Based Molecular Biosensing Aided by Artificial Intelligence. Angew. Chem. Int. Ed. Engl. 2019, 58, 14810–14822. [Google Scholar] [CrossRef]
- Zhang, C.; Xue, T.; Zhang, J.; Liu, L.; Xie, J.; Wang, G.; Yao, J.; Zhu, W.; Ye, X. Terahertz toroidal metasurface biosensor for sensitive distinction of lung cancer cells. Nanophotonics 2021, 11, 101–109. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, M.; Yan, X.; Guo, X.; Li, J.; Yang, Y.; Wei, D.; Liu, L.; Xie, J.; Liu, Y.; et al. The Antibody-Free Recognition of Cancer Cells Using Plasmonic Biosensor Platforms with the Anisotropic Resonant Metasurfaces. ACS Appl. Mater. Interfaces 2020, 12, 11388–11396. [Google Scholar] [CrossRef]
- Aalizadeh, M.; Serebryannikov, A.E.; Ozbay, E.; Vandenbosch, G.A.E. A simple Mie-resonator based meta-array with diverse deflection scenarios enabling multifunctional operation at near-infrared. Nanophotonics 2020, 9, 4589–4600. [Google Scholar] [CrossRef]
- Wang, Z.; Dai, C.; Li, Z.; Li, Z. Free-Space Optical Merging via Meta-Grating Inverse-Design. Nano Lett. 2022, 22, 2059–2064. [Google Scholar] [CrossRef]
- Wang, X.; Tian, Z.; Pei, S.; Xu, C.; Sun, Q.; Zhang, J.; Wei, J.; Li, F.; Yun, F. Highly Efficient and Linearly Polarized Light Emission of Micro-LED Integrated with Double-Functional Meta-Grating. Nano Lett. 2024, 25, 236–243. [Google Scholar] [CrossRef]

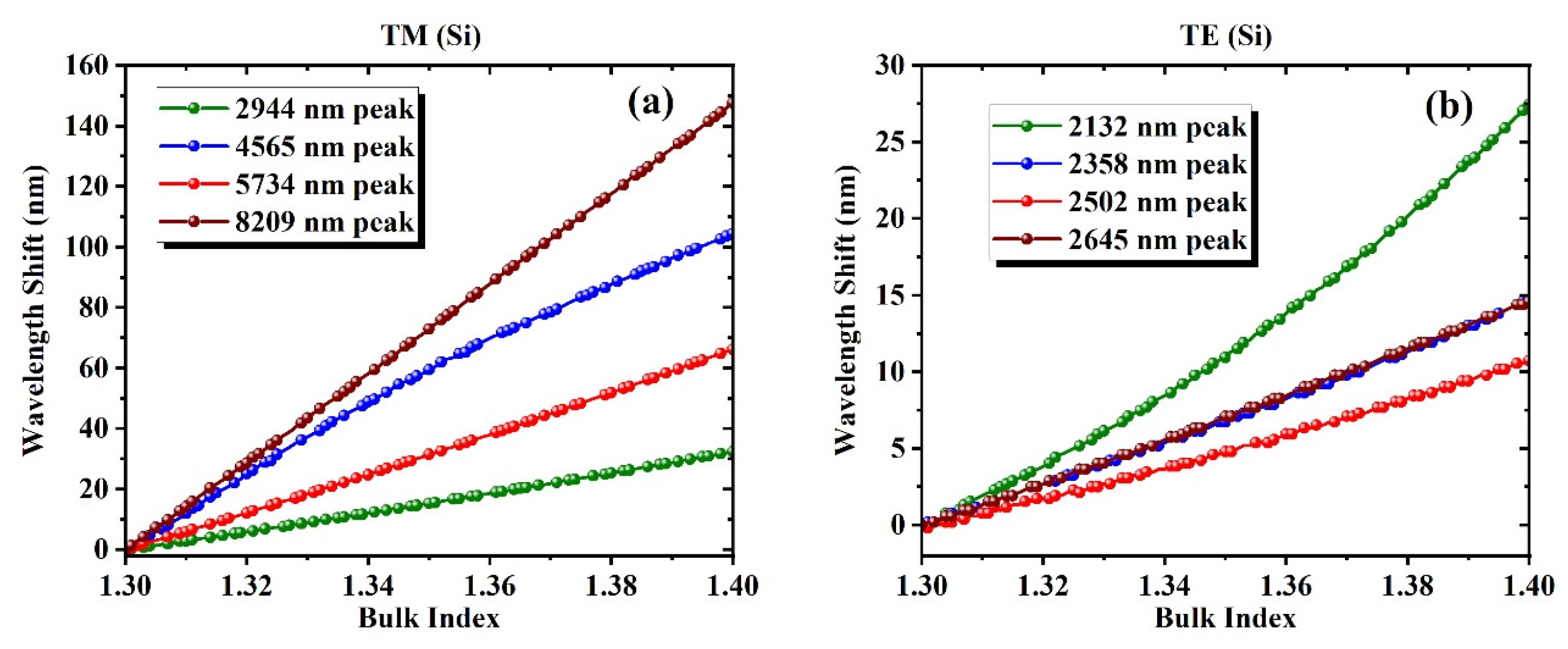
| TM_Si | TE_Si | ||||
|---|---|---|---|---|---|
| Peak Location (nm) 1 | MSE | Peak Location (nm) | MSE | ||
| Mean | std | Mean | std | ||
| 2944 | 0.79198 | 0.16526 | 2132 | 10.035007 | 1.84389 |
| 4565 | 5.89753 | 1.07706 | 2358 | 1.822854 | 0.59077 |
| 5734 | 0.75589 | 0.10943 | 2502 | 2.907157 | 0.63826 |
| 8209 | 0.02944 | 0.00683 | 2645 | 0.562897 | 0.07325 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aalizadeh, M.; Raut, C.; Azmoudeh Afshar, M.; Tabartehfarahani, A.; Fan, X. Accuracy Enhancement in Refractive Index Sensing via Full-Spectrum Machine Learning Modeling. Biosensors 2025, 15, 582. https://doi.org/10.3390/bios15090582
Aalizadeh M, Raut C, Azmoudeh Afshar M, Tabartehfarahani A, Fan X. Accuracy Enhancement in Refractive Index Sensing via Full-Spectrum Machine Learning Modeling. Biosensors. 2025; 15(9):582. https://doi.org/10.3390/bios15090582
Chicago/Turabian StyleAalizadeh, Majid, Chinmay Raut, Morteza Azmoudeh Afshar, Ali Tabartehfarahani, and Xudong Fan. 2025. "Accuracy Enhancement in Refractive Index Sensing via Full-Spectrum Machine Learning Modeling" Biosensors 15, no. 9: 582. https://doi.org/10.3390/bios15090582
APA StyleAalizadeh, M., Raut, C., Azmoudeh Afshar, M., Tabartehfarahani, A., & Fan, X. (2025). Accuracy Enhancement in Refractive Index Sensing via Full-Spectrum Machine Learning Modeling. Biosensors, 15(9), 582. https://doi.org/10.3390/bios15090582





