Synthetic Gene Circuits Enable Sensing in Engineered Living Materials
Abstract
1. Introduction
2. Synthetic Inducer–Rsponsive ELMs
3. Chemical-Responsive ELMs
4. Light-Responsive ELMs
5. Other Stimuli–Responsive ELMs
5.1. Temperature-Responsive ELMs
5.2. Mechanical Signal-Responsive ELMs
5.3. Electrical Signal-Responsive ELMs
6. Challenges and Potential Solutions
6.1. Functional Stability
6.2. Biosafety Enhancement
6.3. Control of Expression Accuracy
7. Future Perspectives
7.1. Programmable Anti-Counterfeiting Designs
7.2. Genetic Encoding and Information Storage
8. Discussion
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| aTc | anhydrotetracycline; |
| Ara | arabinose; |
| AHL | acyl-homoserine lactone; |
| AuNRs | gold nanorods; |
| BBC | biofilm@biochar; |
| BC | bacterial cellulose; |
| CAP | capsular polysaccharide; |
| CsgA-Mfp3s | curli fiber fusion protein of CsgA and mussel foot protein 3s; |
| DOMINO | DNA-based ordered memory and iteration network operator; |
| dVio | deoxyviolacein; |
| ELMs | engineered living materials; |
| EPS | extracellular polymeric substances; |
| ES | electrical stimulation; |
| GFP | green fluorescent protein; |
| hBMP-4 | human bone morphogenetic protein 4; |
| HGT | horizontal gene transfer; |
| IAct | isoamyl acetate; |
| IL-1Ra | interleukin-1 receptor antagonist; |
| IPTG | isopropyl β-D-1-thiogalactopyranoside; |
| NIR | near-infrared; |
| PAI | p-coumaric acid inducer; |
| PGA | poly-γ-glutamic acid; |
| PLA-AP | poly(l-lactic acid)-block-aniline pentamer-block-poly(l-lactic acid); |
| PLGA/HA | poly(lactic-co-glycolic acid)/hydroxyapatite; |
| Plu/PluDA | Pluronic/Pluronic diacrylate |
| PVD | peripheral vascular disease; |
| QK | VEGF-mimetic peptide QK; |
| RFP | red fluorescent protein; |
| RNAi | RNA interference; |
| SCRIBE | synthetic cellular recorders integrating biological events; |
| Tc | tetracycline; |
| VAI | vanillic acid inducer; |
| VEGF | vascular endothelial growth factor; |
| YCQ | YebF-collagen-binding domain-QK fusion protein; |
| YFP | yellow fluorescent protein. |
References
- Feng, Y.; Su, C.; Mao, G.; Sun, B.; Cai, Y.; Dai, J.; Ma, Y. When Synthetic Biology Meets Medicine. Life Med. 2024, 3, lnae010. [Google Scholar] [CrossRef]
- Lu, Y.; Wu, N.; Bai, R.; Du, W.; Zhang, J.; Min, J. Design, Optimization and Application of Whole-Cell Microbial Biosensors with Engineered Genetic Circuits. Synth. Biol. J. 2022, 3, 1061–1084. [Google Scholar] [CrossRef]
- Liu, X.; Germaine, K.J.; Ryan, D.; Dowling, D.N. Whole-Cell Fluorescent Biosensors for Bioavailability and Biodegradation of Polychlorinated Biphenyls. Sensors 2010, 10, 1377–1398. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, X.; Su, H.; Guo, M.; Liu, H. Advances in Synthetic-Biology-Based Whole-Cell Biosensors: Principles, Genetic Modules, and Applications in Food Safety. Int. J. Mol. Sci. 2023, 24, 7989. [Google Scholar] [CrossRef]
- Bereza-Malcolm, L.T.; Mann, G.; Franks, A.E. Environmental Sensing of Heavy Metals through Whole Cell Microbial Biosensors: A Synthetic Biology Approach. ACS Synth. Biol. 2015, 4, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Akboğa, D.; Saltepe, B.; Bozkurt, E.U.; Şeker, U.Ö.Ş. A Recombinase-Based Genetic Circuit for Heavy Metal Monitoring. Biosensors 2022, 12, 122. [Google Scholar] [CrossRef]
- Alhadrami, H.A. Biosensors: Classifications, Medical Applications, and Future Prospects. Biotechnol. Appl. Biochem. 2018, 65, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Mittal, D.; Kaur, G. In-Situ Monitoring of Xenobiotics Using Genetically Engineered Whole-Cell-Based Microbial Biosensors: Recent Advances and Outlook. World J. Microbiol. Biotechnol. 2021, 37, 81. [Google Scholar] [CrossRef]
- Moraskie, M.; Roshid, H.; O’Connor, G.; Dikici, E.; Zingg, J.-M.; Deo, S.; Daunert, S. Microbial Whole-Cell Biosensors: Current Applications, Challenges, and Future Perspectives. Biosens. Bioelectron. 2021, 191, 113359. [Google Scholar] [CrossRef]
- Kannappan, S.; Ramisetty, B.C.M. Engineered Whole-Cell-Based Biosensors: Sensing Environmental Heavy Metal Pollutants in Water—A Review. Appl. Biochem. Biotechnol. 2022, 194, 1814–1840. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Q.; Ge, C.; An, B.; Zhong, C. Programmable Bacterial Biofilms as Engineered Living Materials. Acc. Mater. Res. 2024, 5, 797–808. [Google Scholar] [CrossRef]
- Altin-Yavuzarslan, G.; Brooks, S.M.; Yuan, S.-F.; Park, J.O.; Alper, H.S.; Nelson, A. Additive Manufacturing of Engineered Living Materials with Bio-Augmented Mechanical Properties and Resistance to Degradation. Adv. Funct. Mater. 2023, 33, 2300332. [Google Scholar] [CrossRef]
- Liu, X.; Tang, T.C.; Tham, E.; Yuk, H.; Lin, S.; Lu, T.K.; Zhao, X. Stretchable Living Materials and Devices with Hydrogel-Elastomer Hybrids Hosting Programmed Cells. Proc. Natl. Acad. Sci. USA 2017, 114, 2200–2205. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Courchesne, N.M.D.; Duraj-Thatte, A.; Praveschotinunt, P.; Joshi, N.S. Engineered Living Materials: Prospects and Challenges for Using Biological Systems to Direct the Assembly of Smart Materials. Adv. Mater. 2018, 30, e1704847. [Google Scholar] [CrossRef]
- Luo, J.; Chen, J.; Huang, Y.; You, L.; Dai, Z. Engineering Living Materials by Synthetic Biology. Biophys. Rev. 2023, 4, 011305. [Google Scholar] [CrossRef]
- Liu, S.; Xu, W. Engineered Living Materials-Based Sensing and Actuation. Front. Sens. 2020, 1, 586300. [Google Scholar] [CrossRef]
- Sugianto, W.; Altin-Yavuzarslan, G.; Tickman, B.I.; Kiattisewee, C.; Yuan, S.-F.; Brooks, S.M.; Wong, J.; Alper, H.S.; Nelson, A.; Carothers, J.M. Gene Expression Dynamics in Input-Responsive Engineered Living Materials Programmed for Bioproduction. Mater. Today Bio 2023, 20, 100677. [Google Scholar] [CrossRef]
- Liu, X.; Inda, M.E.; Lai, Y.; Lu, T.K.; Zhao, X. Engineered Living Hydrogels. Adv. Mater. 2022, 34, 2201326. [Google Scholar] [CrossRef] [PubMed]
- Burgos-Morales, O.; Gueye, M.; Lacombe, L.; Nowak, C.; Schmachtenberg, R.; Hörner, M.; Jerez-Longres, C.; Mohsenin, H.; Wagner, H.J.; Weber, W. Synthetic Biology as Driver for the Biologization of Materials Sciences. Mater. Today Bio 2021, 11, 100115. [Google Scholar] [CrossRef]
- Srubar, W.V. Engineered Living Materials: Taxonomies and Emerging Trends. Trends Biotechnol. 2021, 39, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Huang, Y.; Cui, J.; Wu, J.; Jiang, C.; Gu, X.; Cao, Y.; Yin, S. Toward Practical Applications of Engineered Living Materials with Advanced Fabrication Techniques. ACS Synth. Biol. 2024, 13, 2295–2312. [Google Scholar] [CrossRef]
- Rivera-Tarazona, L.K.; Campbell, Z.T.; Ware, T.H. Stimuli-Responsive Engineered Living Materials. Soft Matter 2021, 17, 785–809. [Google Scholar] [CrossRef] [PubMed]
- Omer, R.; Mohsin, M.Z.; Mohsin, A.; Mushtaq, B.S.; Huang, X.; Guo, M.; Zhuang, Y.; Huang, J. Engineered Bacteria-Based Living Materials for Biotherapeutic Applications. Front. Bioeng. Biotechnol. 2022, 10, 870675. [Google Scholar] [CrossRef]
- Dong, X.; Wu, W.; Pan, P.; Zhang, X.-Z. Engineered Living Materials for Advanced Diseases Therapy. Adv. Mater. 2023, 37, e2304963. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Wang, Y.; Huang, Y.; Wang, X.; Liu, Y.; Xun, D.; Church, G.M.; Dai, Z.; Yi, X.; Tang, T.-C.; et al. Engineered Living Materials For Sustainability. Chem. Rev. 2023, 123, 2349–2419. [Google Scholar] [CrossRef]
- Jiang, S.; Li, H.; Zhang, L. Generic Diagramming Platform (GDP): A Comprehensive Database of High-Quality Biomedical Graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef] [PubMed]
- Usai, F.; Loi, G.; Scocozza, F.; Bellato, M.; Castagliuolo, I.; Conti, M.; Pasotti, L. Design and Biofabrication of Bacterial Living Materials with Robust and Multiplexed Biosensing Capabilities. Mater. Today Bio 2023, 18, 100526. [Google Scholar] [CrossRef]
- Duraj-Thatte, A.M.; Manjula-Basavanna, A.; Rutledge, J.; Xia, J.; Hassan, S.; Sourlis, A.; Rubio, A.G.; Lesha, A.; Zenkl, M.; Kan, A.; et al. Programmable Microbial Ink for 3D Printing of Living Materials Produced from Genetically Engineered Protein Nanofibers. Nat. Commun. 2021, 12, 6600. [Google Scholar] [CrossRef]
- Birnbaum, D.P.; Manjula-Basavanna, A.; Kan, A.; Tardy, B.L.; Joshi, N.S. Hybrid Living Capsules Autonomously Produced by Engineered Bacteria. Adv. Sci. 2021, 8, 2004699. [Google Scholar] [CrossRef]
- González, L.M.; Mukhitov, N.; Voigt, C.A. Resilient Living Materials Built by Printing Bacterial Spores. Nat. Chem. Biol. 2020, 16, 126–133. [Google Scholar] [CrossRef]
- Datta, D.; Weiss, E.L.; Wangpraseurt, D.; Hild, E.; Chen, S.; Golden, J.W.; Golden, S.S.; Pokorski, J.K. Phenotypically Complex Living Materials Containing Engineered Cyanobacteria. Nat. Commun. 2023, 14, 4742. [Google Scholar] [CrossRef]
- Zhu, X.; Xiang, Q.; Chen, L.; Chen, J.; Wang, L.; Jiang, N.; Hao, X.; Zhang, H.; Wang, X.; Li, Y.; et al. Engineered Bacillus Subtilis Biofilm@Biochar Living Materials for in-Situ Sensing and Bioremediation of Heavy Metal Ions Pollution. J. Hazard. Mater. 2024, 465, 133119. [Google Scholar] [CrossRef]
- Park, H.; Schwartzman, A.F.; Tang, T.-C.; Wang, L.; Lu, T.K. Ultra-Lightweight Living Structural Material for Enhanced Stiffness and Environmental Sensing. Mater. Today Bio 2022, 18, 100504. [Google Scholar] [CrossRef]
- Tang, T.-C.; Tham, E.; Liu, X.; Yehl, K.; Rovner, A.J.; Yuk, H.; De La Fuente-Nunez, C.; Isaacs, F.J.; Zhao, X.; Lu, T.K. Hydrogel-Based Biocontainment of Bacteria for Continuous Sensing and Computation. Nat. Chem. Biol. 2021, 17, 724–731. [Google Scholar] [CrossRef]
- Bhusari, S.; Kim, J.; Polizzi, K.; Sankaran, S.; Del Campo, A. Encapsulation of Bacteria in Bilayer Pluronic Thin Film Hydrogels: A Safe Format for Engineered Living Materials. Biomater. Adv. 2023, 145, 213240. [Google Scholar] [CrossRef]
- Liu, X.; Yang, Y.; Inda, M.E.; Lin, S.; Wu, J.; Kim, Y.; Chen, X.; Ma, D.; Lu, T.K.; Zhao, X. Magnetic Living Hydrogels for Intestinal Localization, Retention, and Diagnosis. Adv. Funct. Mater. 2021, 31, 2010918. [Google Scholar] [CrossRef]
- Dutto, A.; Kan, A.; Saraw, Z.; Maillard, A.; Zindel, D.; Studart, A.R. Living Porous Ceramics for Bacteria-Regulated Gas Sensing and Carbon Capture. Adv. Mater. 2025, 37, 2412555. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Tang, T.-C.; Ott, W.; Dorr, B.A.; Shaw, W.M.; Sun, G.L.; Lu, T.K.; Ellis, T. Living Materials with Programmable Functionalities Grown from Engineered Microbial Co-Cultures. Nat. Mater. 2021, 20, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Dhakane, P.; Tadimarri, V.S.; Sankaran, S. Light-Regulated Pro-Angiogenic Engineered Living Materials. Adv. Funct. Mater. 2023, 33, 2212695. [Google Scholar] [CrossRef]
- Sankaran, S.; Del Campo, A. Optoregulated Protein Release from an Engineered Living Material. Adv. Biosyst. 2019, 3, e1800312. [Google Scholar] [CrossRef] [PubMed]
- An, B.; Wang, Y.; Jiang, X.; Ma, C.; Mimee, M.; Moser, F.; Li, K.; Wang, X.; Tang, T.C.; Huang, Y.; et al. Programming Living Glue Systems to Perform Autonomous Mechanical Repairs. Matter 2020, 3, 2080–2092. [Google Scholar] [CrossRef]
- Sankaran, S.; Becker, J.; Wittmann, C.; del Campo, A. Optoregulated Drug Release from an Engineered Living Material: Self-Replenishing Drug Depots for Long-Term, Light-Regulated Delivery. Small 2019, 15, 1804717. [Google Scholar] [CrossRef] [PubMed]
- Basaran, S.; Dey, S.; Bhusari, S.; Sankaran, S.; Kraus, T. Plasmonic Stimulation of Gold Nanorods for the Photothermal Control of Engineered Living Materials. Biomater. Adv. 2023, 147, 213332. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.L.; Garrett, M.A.; Kornfield, J.A.; Shapiro, M.G. Living Material with Temperature-Dependent Light Absorption. Adv. Sci. 2023, 10, e2301730. [Google Scholar] [CrossRef]
- Nims, R.J.; Pferdehirt, L.; Ho, N.B.; Savadipour, A.; Lorentz, J.; Sohi, S.; Kassab, J.; Ross, A.K.; O’Conor, C.J.; Liedtke, W.B.; et al. A Synthetic Mechanogenetic Gene Circuit for Autonomous Drug Delivery in Engineered Tissues. Sci. Adv. 2021, 7, eabd9858. [Google Scholar] [CrossRef]
- Cui, L.; Zhang, J.; Zou, J.; Yang, X.; Guo, H.; Tian, H.; Zhang, P.; Wang, Y.; Zhang, N.; Zhuang, X.; et al. Electroactive Composite Scaffold with Locally Expressed Osteoinductive Factor for Synergistic Bone Repair upon Electrical Stimulation. Biomaterials 2020, 230, 119617. [Google Scholar] [CrossRef]
- Engstrom, M.D.; Pfleger, B.F. Transcription Control Engineering and Applications in Synthetic Biology. Synth. Syst. Biotechnol. 2017, 2, 176–191. [Google Scholar] [CrossRef]
- Vogel, A.M.; Persson, K.M.; Seamons, T.R.; Deans, T.L. Synthetic Biology for Improving Cell Fate Decisions and Tissue Engineering Outcomes. Emerg. Top. Life Sci. 2019, 3, 631–643. [Google Scholar] [CrossRef]
- Marbach, A.; Bettenbrock, K. Lac Operon Induction in Escherichia Coli: Systematic Comparison of IPTG and TMG Induction and Influence of the Transacetylase LacA. J. Biotechnol. 2012, 157, 82–88. [Google Scholar] [CrossRef]
- Oss Boll, H.; de Castro Leitão, M.; Garay, A.V.; Batista, A.C.C.; de Resende, S.G.; da Silva, L.F.; Reis, V.C.B.; Vieira, E.M.; Coelho, C.M. SynBio in 3D: The First Synthetic Genetic Circuit as a 3D Printed STEM Educational Resource. Front. Educ. 2023, 8, 1110464. [Google Scholar] [CrossRef]
- Tilley, L.; Papadopoulos, S.C.; Pende, M.; Fei, J.F.; Murawala, P. The Use of Transgenics in the Laboratory Axolotl. Dev. Dyn. 2022, 251, 942–956. [Google Scholar] [CrossRef]
- Wang, X.; Liang, Q.; Luo, Y.; Ye, J.; Yu, Y.; Chen, F. Engineering the next Generation of Theranostic Biomaterials with Synthetic Biology. Bioact. Mater. 2024, 32, 514–529. [Google Scholar] [CrossRef]
- Gossen, M.; Bujard, H. Anhydrotetracycline, a Novel Effector for Tetracycline Controlled Gene Expression Systems in Eukaryotic Cells. Nucleic Acids Res. 1993, 21, 4411–4412. [Google Scholar] [CrossRef]
- Kiattisewee, C.; Dong, C.; Fontana, J.; Sugianto, W.; Peralta-Yahya, P.; Carothers, J.M.; Zalatan, J.G. Portable Bacterial CRISPR Transcriptional Activation Enables Metabolic Engineering in Pseudomonas Putida. Metab. Eng. 2021, 66, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Fontana, J.; Dong, C.; Kiattisewee, C.; Chavali, V.P.; Tickman, B.I.; Carothers, J.M.; Zalatan, J.G. Effective CRISPRa-Mediated Control of Gene Expression in Bacteria Must Overcome Strict Target Site Requirements. Nat. Commun. 2020, 11, 1618. [Google Scholar] [CrossRef]
- Guo, S.; Dubuc, E.; Rave, Y.; Verhagen, M.; Twisk, S.A.E.; van der Hek, T.; Oerlemans, G.J.M.; van den Oetelaar, M.C.M.; van Hazendonk, L.S.; Brüls, M.; et al. Engineered Living Materials Based on Adhesin-Mediated Trapping of Programmable Cells. ACS Synth. Biol. 2020, 9, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Wu, M.; Cao, L.; Liu, B. Organic Functional Substance Engineered Living Materials for Biomedical Applications. Biomaterials 2023, 301, 122248. [Google Scholar] [CrossRef]
- Nevot, G.; Pol Cros, M.; Toloza, L.; Campamà-Sanz, N.; Artigues-Lleixà, M.; Aguilera, L.; Güell, M. Engineered Marine Biofilms for Ocean Environment Monitoring. ACS Synth. Biol. 2025, 14, 2797–2809. [Google Scholar] [CrossRef]
- Genchi, G.; Sinicropi, M.S.; Lauria, G.; Carocci, A.; Catalano, A. The Effects of Cadmium Toxicity. Int. J. Environ. Res. Public Health 2020, 17, 3782. [Google Scholar] [CrossRef] [PubMed]
- Mimee, M.; Nadeau, P.; Hayward, A.; Carim, S.; Flanagan, S.; Jerger, L.; Collins, J.; McDonnell, S.; Swartwout, R.; Citorik, R.J.; et al. An Ingestible Bacterial-Electronic System to Monitor Gastrointestinal Health. Science 2018, 360, 915–918. [Google Scholar] [CrossRef]
- Levskaya, A.; Weiner, O.D.; Lim, W.A.; Voigt, C.A. Spatiotemporal Control of Cell Signalling Using a Light-Switchable Protein Interaction. Nature 2009, 461, 997–1001. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.I.; Frey, D.; Lungu, O.I.; Jaehrig, A.; Schlichting, I.; Kuhlman, B.; Hahn, K.M. A Genetically Encoded Photoactivatable Rac Controls the Motility of Living Cells. Nature 2009, 461, 104–108. [Google Scholar] [CrossRef]
- Tan, P.; He, L.; Huang, Y.; Zhou, Y. Optophysiology: Illuminating Cell Physiology with Optogenetics. Physiol. Rev. 2022, 102, 1263–1325. [Google Scholar] [CrossRef]
- Ren, H.; Cheng, Y.; Wen, G.; Wang, J.; Zhou, M. Emerging Optogenetics Technologies in Biomedical Applications. Smart Med. 2023, 2, e20230026. [Google Scholar] [CrossRef]
- Guan, N.; Gao, X.; Ye, H. Engineering of Optogenetic Devices for Biomedical Applications in Mammalian Synthetic Biology. Eng. Biol. 2022, 6, 35–49. [Google Scholar] [CrossRef]
- Gheorghiu, M.; Polonschii, C.; Popescu, O.; Gheorghiu, E. Advanced Optogenetic-Based Biosensing and Related Biomaterials. Materials 2021, 14, 4151. [Google Scholar] [CrossRef]
- Santulli, G.; Ciccarelli, M.; Palumbo, G.; Campanile, A.; Galasso, G.; Ziaco, B.; Altobelli, G.G.; Cimini, V.; Piscione, F.; D’Andrea, L.D.; et al. In Vivo Properties of the Proangiogenic Peptide QK. J. Transl. Med. 2009, 7, 41. [Google Scholar] [CrossRef]
- Spiteri, C.; Caprettini, V.; Chiappini, C. Biomaterials-Based Approaches to Model Embryogenesis. Biomater. Sci. 2020, 8, 6992–7013. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Liao, J.; He, Y.; Zhang, T.; Xiao, Y.; Wang, H.; Shen, M.; Yu, T.; Huang, W. Recent Development of Photochromic Polymer Systems: Mechanism, Materials, and Applications. Research 2024, 7, 0392. [Google Scholar] [CrossRef]
- Shadish, J.A.; Benuska, G.M.; DeForest, C.A. Bioactive Site-Specifically Modified Proteins for 4D Patterning of Gel Biomaterials. Nat. Mater. 2019, 18, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Riedel-Kruse, I.H. Biofilm Lithography Enables High-Resolution Cell Patterning via Optogenetic Adhesin Expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- Kan, A.; Gelfat, I.; Emani, S.; Praveschotinunt, P.; Joshi, N.S. Plasmid Vectors for in Vivo Selection-Free Use with the Probiotic, E. coli Nissle 1917. ACS Synth. Biol. 2021, 10, 94–106. [Google Scholar] [CrossRef]
- Arrigoni, C.; Minor, D.L. Global versus Local Mechanisms of Temperature Sensing in Ion Channels. Pflug. Arch. 2018, 470, 733–744. [Google Scholar] [CrossRef]
- Kanchanawong, P.; Calderwood, D.A. Organization, Dynamics and Mechanoregulation of Integrin-Mediated Cell-ECM Adhesions. Nat. Rev. Mol. Cell Biol. 2023, 24, 142–161. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Schramma, N.; Wang, Z.; Qari, N.F.; Jalaal, M.; Latz, M.I.; Cai, S. Ultrasensitive and Robust Mechanoluminescent Living Composites. Sci. Adv. 2023, 9, eadi8643. [Google Scholar] [CrossRef]
- Li, C.; He, Q.; Wang, Y.; Wang, Z.; Wang, Z.; Annapooranan, R.; Latz, M.I.; Cai, S. Highly Robust and Soft Biohybrid Mechanoluminescence for Optical Signaling and Illumination. Nat. Commun. 2022, 13, 3914. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Nishio, K.; Konno, T.; Ishihara, K. The Effect of the Encapsulation of Bacteria in Redox Phospholipid Polymer Hydrogels on Electron Transfer Efficiency in Living Cell-Based Devices. Biomaterials 2012, 33, 8221–8227. [Google Scholar] [CrossRef]
- Yu, W.; Zeng, Y.; Wang, Z.; Xia, S.; Yang, Z.; Chen, W.; Huang, Y.; Lv, F.; Bai, H.; Wang, S. Solar-Powered Multi-Organism Symbiont Mimic System for beyond Natural Synthesis of Polypeptides from CO2 and N2. Sci. Adv. 2023, 9, eadf6772. [Google Scholar] [CrossRef]
- Wang, Z.; Bai, H.; Yu, W.; Gao, Z.; Chen, W.; Yang, Z.; Zhu, C.; Huang, Y.; Lv, F.; Wang, S. Flexible Bioelectronic Device Fabricated by Conductive Polymer–Based Living Material. Sci. Adv. 2022, 8, eabo1458. [Google Scholar] [CrossRef]
- Terrell, J.L.; Tschirhart, T.; Jahnke, J.P.; Stephens, K.; Liu, Y.; Dong, H.; Hurley, M.M.; Pozo, M.; McKay, R.; Tsao, C.Y.; et al. Bioelectronic Control of a Microbial Community Using Surface-Assembled Electrogenetic Cells to Route Signals. Nat. Nanotechnol. 2021, 16, 688–697. [Google Scholar] [CrossRef]
- Harimoto, T.; Hahn, J.; Chen, Y.-Y.; Im, J.; Zhang, J.; Hou, N.; Li, F.; Coker, C.; Gray, K.; Harr, N.; et al. A Programmable Encapsulation System Improves Delivery of Therapeutic Bacteria in Mice. Nat. Biotechnol. 2022, 40, 1259–1269. [Google Scholar] [CrossRef]
- Tallawi, M.; Opitz, M.; Lieleg, O. Modulation of the Mechanical Properties of Bacterial Biofilms in Response to Environmental Challenges. Biomater. Sci. 2017, 5, 887–900. [Google Scholar] [CrossRef]
- Nguyen, H.; Ybarra, A.; Başağaoğlu, H.; Shindell, O. Biofilm Viscoelasticity and Nutrient Source Location Control Biofilm Growth Rate, Migration Rate, and Morphology in Shear Flow. Sci. Rep. 2021, 11, 16118. [Google Scholar] [CrossRef]
- Lee, H.; Kim, D.-U.; Son, J.; Kim, S.-B.; Ka, J.-O. Environmental Risk Assessment of Living Modified Microorganisms (LMM) on the Indigenous Microbial Community. Sustainability 2020, 12, 5566. [Google Scholar] [CrossRef]
- Mandell, D.J.; Lajoie, M.J.; Mee, M.T.; Takeuchi, R.; Kuznetsov, G.; Norville, J.E.; Gregg, C.J.; Stoddard, B.L.; Church, G.M. Biocontainment of Genetically Modified Organisms by Synthetic Protein Design. Nature 2015, 518, 55–60. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Yan, K.; Huang, Q.; Islam, M.M.; Li, Q.; Wang, Y.; Khan, M.S.; Zhao, X.; Mir, R.R.; Li, J.; et al. Comprehensive Mechanism of Gene Silencing and Its Role in Plant Growth and Development. Front. Plant Sci. 2021, 12, 705249. [Google Scholar] [CrossRef]
- Ashfaq, M.A.; Dinesh Kumar, V.; Soma Sekhar Reddy, P.; Anil Kumar, C.; Sai Kumar, K.; Narasimha Rao, N.; Tarakeswari, M.; Sujatha, M. Post-Transcriptional Gene Silencing: Basic Concepts and Applications. J. Biosci. 2020, 45, 128. [Google Scholar] [CrossRef]
- Kunjapur, A.M.; Napolitano, M.G.; Hysolli, E.; Noguera, K.; Appleton, E.M.; Schubert, M.G.; Jones, M.A.; Iyer, S.; Mandell, D.J.; Church, G.M. Synthetic Auxotrophy Remains Stable after Continuous Evolution and in Coculture with Mammalian Cells. Sci. Adv. 2021, 7, eabf5851. [Google Scholar] [CrossRef]
- Whitford, C.M.; Dymek, S.; Kerkhoff, D.; März, C.; Schmidt, O.; Edich, M.; Droste, J.; Pucker, B.; Rückert, C.; Kalinowski, J. Auxotrophy to Xeno-DNA: An Exploration of Combinatorial Mechanisms for a High-Fidelity Biosafety System for Synthetic Biology Applications. J. Biol. Eng. 2018, 12, 13. [Google Scholar] [CrossRef]
- Rovner, A.J.; Haimovich, A.D.; Katz, S.R.; Li, Z.; Grome, M.W.; Gassaway, B.M.; Amiram, M.; Patel, J.R.; Gallagher, R.R.; Rinehart, J.; et al. Recoded Organisms Engineered to Depend on Synthetic Amino Acids. Nature 2015, 518, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Piatek, A.; Ali, Z.; Baazim, H.; Li, L.; Abulfaraj, A.; Al-Shareef, S.; Aouida, M.; Mahfouz, M.M. RNA-Guided Transcriptional Regulation in Planta via Synthetic dCas9-Based Transcription Factors. Plant Biotechnol. J. 2015, 13, 578–589. [Google Scholar] [CrossRef]
- Gentzel, I.N.; Park, C.H.; Bellizzi, M.; Xiao, G.; Gadhave, K.R.; Murphree, C.; Yang, Q.; LaMantia, J.; Redinbaugh, M.G.; Balint-Kurti, P.; et al. A CRISPR/dCas9 Toolkit for Functional Analysis of Maize Genes. Plant Methods 2020, 16, s13007–s13020. [Google Scholar] [CrossRef] [PubMed]
- Motorina, D.M.; Galimova, Y.A.; Battulina, N.V.; Omelina, E.S. Systems for Targeted Silencing of Gene Expression and Their Application in Plants and Animals. Int. J. Mol. Sci. 2024, 25, 5231. [Google Scholar] [CrossRef] [PubMed]
- Grishok, A. RNAi Mechanisms in Caenorhabditis Elegans. FEBS Lett. 2005, 579, 5932–5939. [Google Scholar] [CrossRef]
- Hernández-Soto, A.; Chacón-Cerdas, R. RNAi Crop Protection Advances. Int. J. Mol. Sci. 2021, 22, 12148. [Google Scholar] [CrossRef] [PubMed]
- Wright, O.; Delmans, M.; Stan, G.-B.; Ellis, T. GeneGuard: A Modular Plasmid System Designed for Biosafety. ACS Synth. Biol. 2015, 4, 307–316. [Google Scholar] [CrossRef]
- Bhusari, S.; Sankaran, S.; del Campo, A. Regulating Bacterial Behavior within Hydrogels of Tunable Viscoelasticity. Adv. Sci. 2022, 9, 2106026. [Google Scholar] [CrossRef]
- Kato, Y. Extremely Low Leakage Expression Systems Using Dual Transcriptional-Translational Control for Toxic Protein Production. Int. J. Mol. Sci. 2020, 21, 705. [Google Scholar] [CrossRef]
- Ho, J.M.L.; Miller, C.A.; Parks, S.E.; Mattia, J.R.; Bennett, M.R. A Suppressor tRNA-Mediated Feedforward Loop Eliminates Leaky Gene Expression in Bacteria. Nucleic Acids Res. 2020, 49, e25. [Google Scholar] [CrossRef]
- Liu, C.; Steppert, A.K.; Liu, Y.; Weis, P.; Hu, J.; Nie, C.; Xu, W.C.; Kuehne, A.J.C.; Wu, S. A Photopatternable Conjugated Polymer with Thermal-Annealing-Promoted Interchain Stacking for Highly Stable Anti-Counterfeiting Materials. Adv. Mater. 2023, 35, 2303120. [Google Scholar] [CrossRef]
- Yuan, J.; Zhou, Y.; Chen, G.; Xiao, K.; Lu, J. Materials vs Digits: A Review of Embedded Anti-Counterfeiting Fingerprints in Three-Dimensional Printing. Mater. Sci. Eng. R. Rep. 2024, 160. [Google Scholar] [CrossRef]
- Xu, C.; Shi, X.; Guo, Y.; Yu, K.; Chen, K. Environmentally Responsive Dual-Compartment Microcapsules with Full Spectrum Color-Changing Performance for Anti-Counterfeiting Applications. Matter 2025, 8, 101925. [Google Scholar] [CrossRef]
- Schwarz, N.; Enke, M.; Gruschwitz, F.V.; Winkler, D.; Franzmann, S.; Jescheck, L.; Hanf, F.; Schneeberger, A. 3D Screen Printing Offers Unprecedented Anticounterfeiting Strategies for Oral Solid Dosage Forms Feasible for Large Scale Production. Pharmaceutics 2024, 16, 368. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Huan, X.; Liu, Y.; Lee, H.; Chen, M.; Hu, S.; Cao, S.; Kim, J.T. Three-Dimensional Printing of Dipeptides with Spatioselective Programming of Crystallinity for Multilevel Anticounterfeiting. Nano. Lett. 2022, 22, 7776–7783. [Google Scholar] [CrossRef] [PubMed]
- Sheth, R.U.; Wang, H.H. DNA-Based Memory Devices for Recording Cellular Events. Nat. Rev. Genet. 2018, 19, 718–732. [Google Scholar] [CrossRef]
- Chen, W.; Choi, J. Molecular Circuits for Genomic Recording of Cellular Events. Trends Genet. 2025, 41, 647–659. [Google Scholar] [CrossRef]
- Tang, W.; Liu, D.R. Rewritable Multi-Event Analog Recording in Bacterial and Mammalian Cells. Science 2018, 360, eaap8992. [Google Scholar] [CrossRef]
- Farzadfard, F.; Lu, T.K. Genomically Encoded Analog Memory with Precise In Vivo DNA Writing in Living Cell Populations. Science 2014, 346, 1256272. [Google Scholar] [CrossRef]
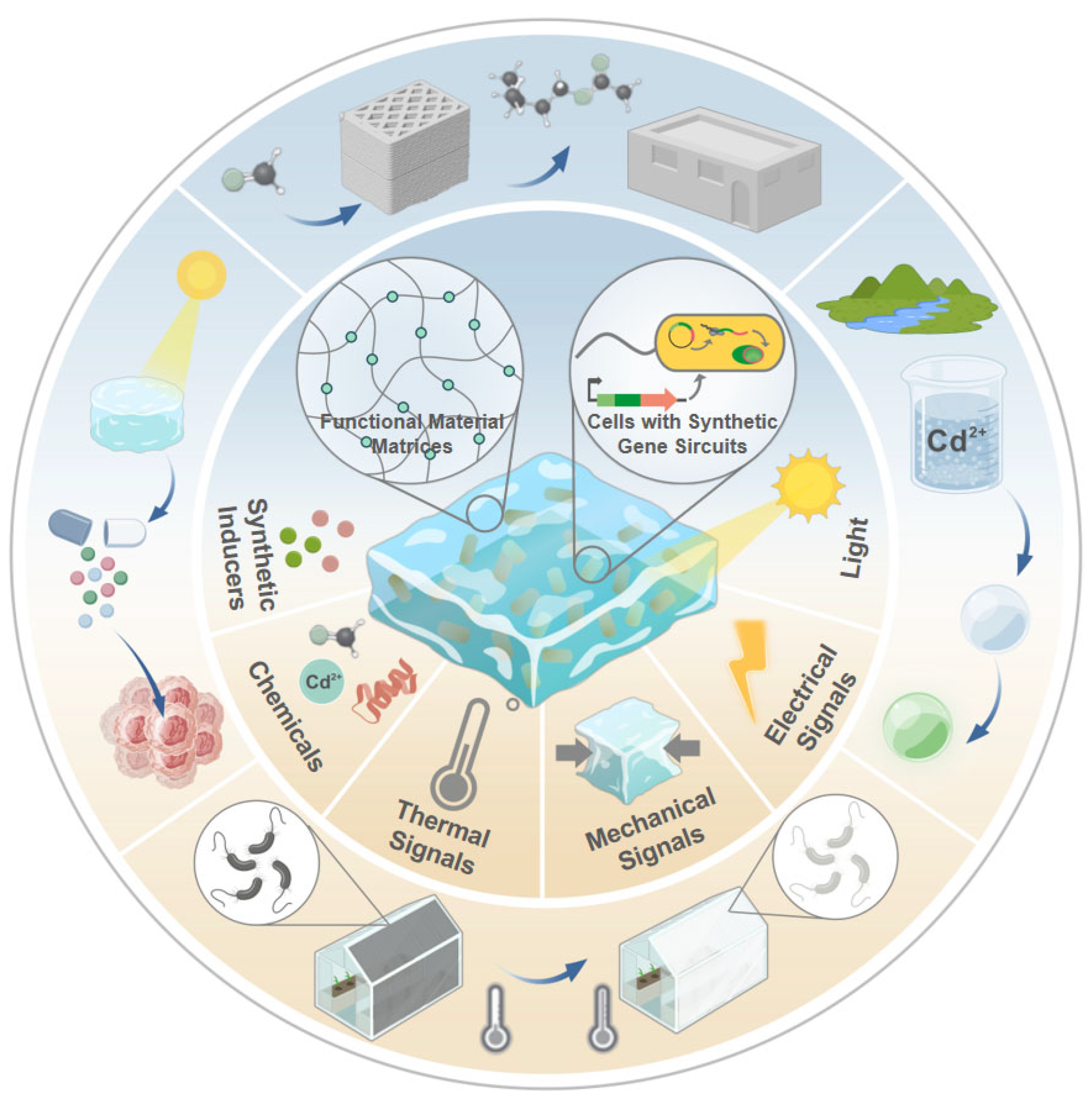
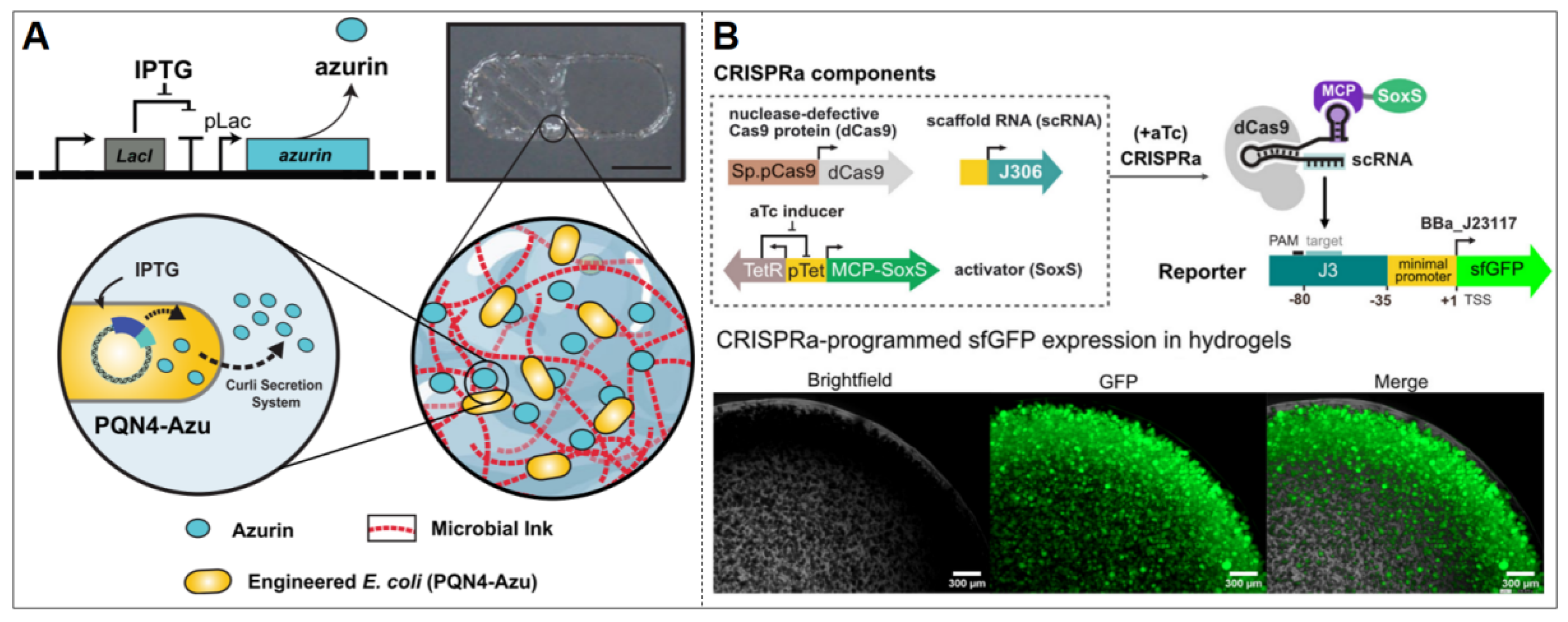

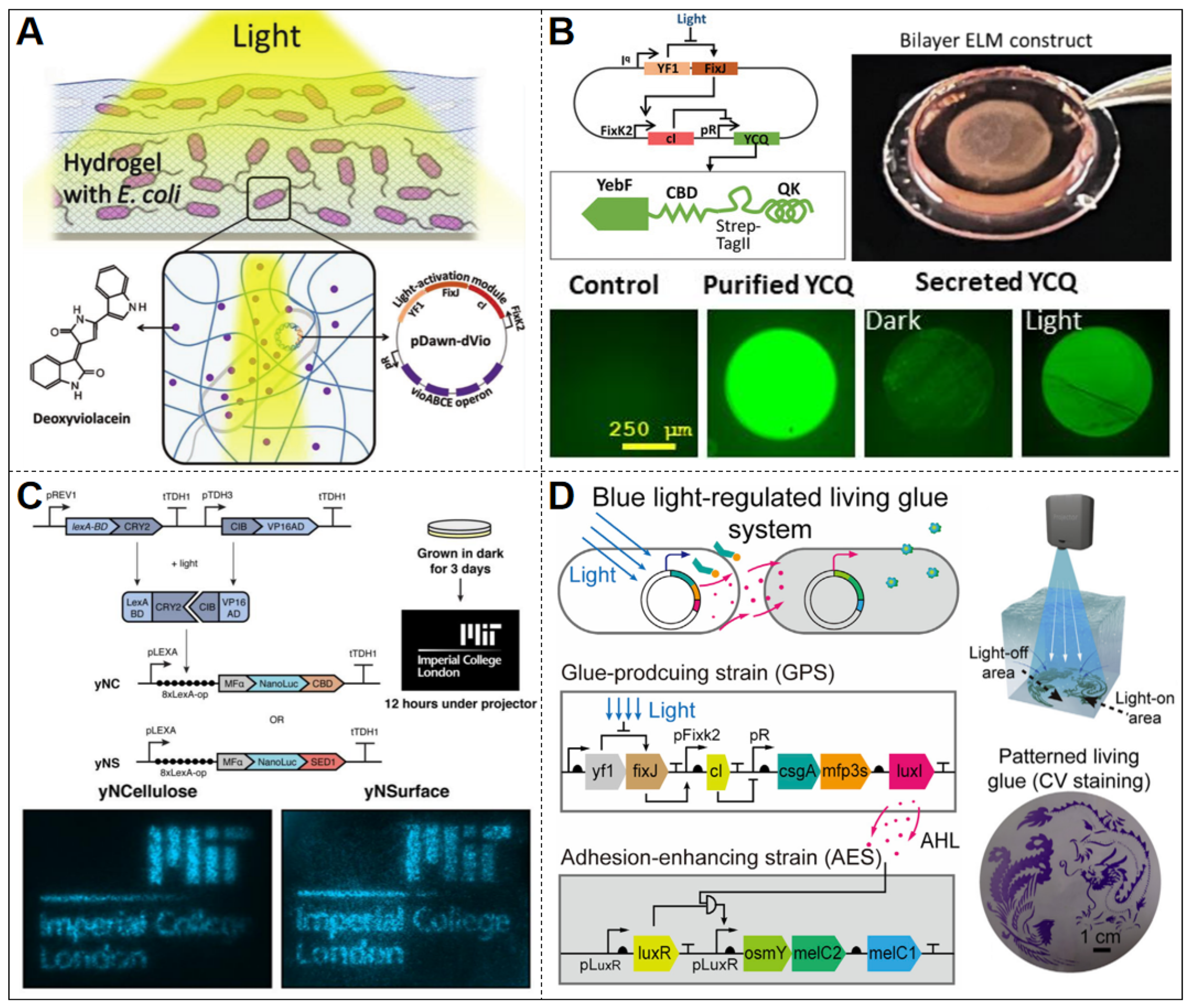
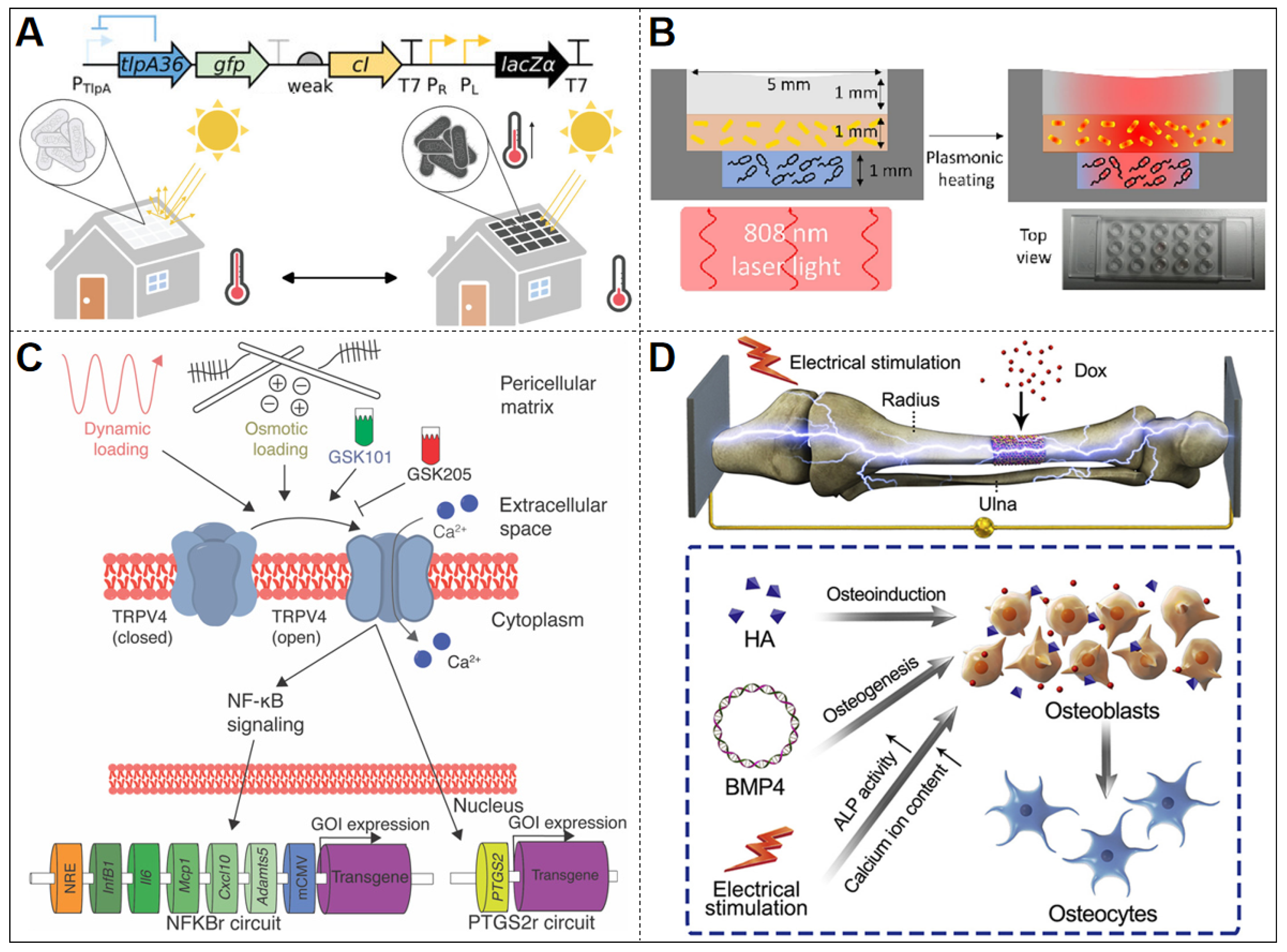
| Stimulus Type | Input Signal | Output Signal | Promoter | Reporter Gene | Host Organism | Material | Threshold | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Synthetic Inducers | IPTG* | RFP* (fluorescence) | PLac | RFP* | E. coli | hydrogel | 0.1–1 mM | >72 h | [27] |
| aTc* | RFP* (fluorescence) | PTet | RFP* | E. coli | 50–200 ng/mL | ||||
| VAI* | γ-PGA* (biopolymer) | Pspank(V) | γ-PGA* | B. subtilis | 1 mM | ||||
| VAI* | Catechol 2,3-dioxygenase (enzyme) | Pspank(V) | xylE | B. subtilis | 1 mM | ||||
| IPTG* | azurin (protein) | PLac | Azurin, | E. coli | CsgA*-αγ hydrogel | ≥0.1 mM | effective one-time release upon induction | [28] | |
| IPTG* | endoribonuclease | PLac | MazF | ≥0.1 mM | |||||
| Ara* | RFP* (fluorescence) | ParaBAD | mRFP1 | E. coli,
F. Gluconacetobacter hansenii | BC*, gelatin | 0.1% (w/v) | ~5 days expt. | [29] | |
| aTc* | sfGFP* (fluorescence) | PTet | sfGFP* | E. coli | Pluronic F127-BUM hydrogel | 0–100 ng/mL | >48 h | [17] | |
| IPTG* | GFP* (fluorescence) | Pspank(hy) | GFP* | B. subtilis | hydrogel | 1 mM | >6 months | [30] | |
| xylose | GFP* (fluorescence) | Pxyl(s) | GFP* | 1% w/v | |||||
| vanillic acid | GFP* (fluorescence) | Pspank(V) | GFP* | 1 mM | |||||
| Cuminic acid | GFP* (fluorescence) | Pspank(C) | GFP* | 0.5 mM | |||||
| Theophylline | YFP* (fluorescence) | PconII | YFP* | S. elongatus | hydrogel | ~0.5 mM | >7 days | [31] | |
| Chemicals | Pb2+ | mtagBFP* (fluorescence) | Ppbr | mtagBFP* | B. subtilis | biofilm@biochar | 0.1 μg/L | >7 days | [32] |
| Cu2+ | eGFP (fluorescence) | PcopA | eGFP* | 1.0 μg/L | |||||
| Hg2+ | mCherry (fluorescence) | Pmer | mCherry | 0.05 μg/L | |||||
| Cd2+; | GFP* (fluorescence) | PzntR | GFP* | E. coli | CsgA* amyloid fibrils | 0.1–10 μM | not explicitly quantified | [33] | |
| Cd2+ | GFP* (fluorescence) | PzntA | GFP*, | E. coli | polyacrylamide-alginate hydrogel | 0.01 μM | >5 days | [34] | |
| heme | luminescence | PL(hrto) | luxCDABE | ~10 μM | |||||
| L-lactate | CreiLOV (fluorescent protein) | PlldR | CreiLOV | E. coli | hydrogel | 5–100 mM | >7 days | [35] | |
| heme | luminescence | PL(hrto) | luxCDABE | E. coli | magnetic living hydrogels | ~10 μM | >7 days | [36] | |
| formaldehyde | IAct* (odor) | Pm4 | ATF1 | E. coli | porous ceramics | ~0.12 ppm | >2 months | [37] | |
| Light | light | NanoLuc (luminescence) | PLexA | NanoLuc | S. cerevisiae | BC | 470 nm | >7 days | [38] |
| light | YCQ (pro-angiogenic fusion protein) | PFixK2 | YCQ | E. coli | hydrogel | ~0.5 μmol·m−2·s−1 | >9 days | [39] | |
| light | RFP*(fluorescence) | PFixK2 | RFP* | E. coli | agarose hydrogel | ~5 μmol·m−2·s−1 | >4 days | [40] | |
| light | CsgA*-Mfp3s (adhesive protein) | PFixK2 | CsgA*-Mfp3s | E. coli | Curli amyloid fibrils | ~50 μmol·m−2·s−1 | not explicitly quantified | [41] | |
| light | deoxyviolacein (anticancer) | PFixK2 | vioABCE | E. coli | hydrogel | ~1 μmol·m−2·s−1 | >14 days | [42] | |
| Other Stimulus | heat | mCherry (fluorescence) | PtlpA39 | mCherry | E. coli | GNC hydrogel | >39 °C | not explicitly quantified | [43] |
| heat | β-galactosidase (enzyme for pigment synthesis) | PtlpA36 | LacZ | E. coli | polycarbonate membranes | ~42 °C (Repression threshold) | not explicitly quantified | [44] | |
| mechanical loading | IL-1Ra* (anti-inflammatory protein) | PTGS2r | IL1Ra* | chondrocytes | agarose hydrogels | 15% compressive strain | ≥3 days | [45] | |
| electricity | hBMP-4* (osteogenic protein) | PTRE | hBMP-4* | rabbit osteoblasts | PLGA/HA*/PLA-AP/phBMP-4* scaffold | 200 mV/cm | ≥14 days | [46] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Wang, Y.; Hu, S. Synthetic Gene Circuits Enable Sensing in Engineered Living Materials. Biosensors 2025, 15, 556. https://doi.org/10.3390/bios15090556
Cai Y, Wang Y, Hu S. Synthetic Gene Circuits Enable Sensing in Engineered Living Materials. Biosensors. 2025; 15(9):556. https://doi.org/10.3390/bios15090556
Chicago/Turabian StyleCai, Yaxuan, Yujie Wang, and Shengbiao Hu. 2025. "Synthetic Gene Circuits Enable Sensing in Engineered Living Materials" Biosensors 15, no. 9: 556. https://doi.org/10.3390/bios15090556
APA StyleCai, Y., Wang, Y., & Hu, S. (2025). Synthetic Gene Circuits Enable Sensing in Engineered Living Materials. Biosensors, 15(9), 556. https://doi.org/10.3390/bios15090556






