Cutting-Edge Sensor Technologies for Exosome Detection: Reviewing Role of Antibodies and Aptamers
Abstract
1. Introduction
2. Techniques for the Separation of Exosomes as Well as Challenges in Exosome Separation from Body Fluids and Different Functionalization Methods
2.1. Traditional and Advanced Techniques for Exosome Isolation from Body Fluids
2.2. Innovations and Challenges in Exosome Separation: Immunoaffinity and Microfluidic Approaches
2.3. Different Functionalization Methods
2.4. Aptamer
2.5. Antibody
3. Advanced Sensing Methods for Exosome Detection Using Aptamers and Antibodies
| Sensing Methods | Mechanism | Target | LOD | Reference | Strengths | Possible Limitations |
|---|---|---|---|---|---|---|
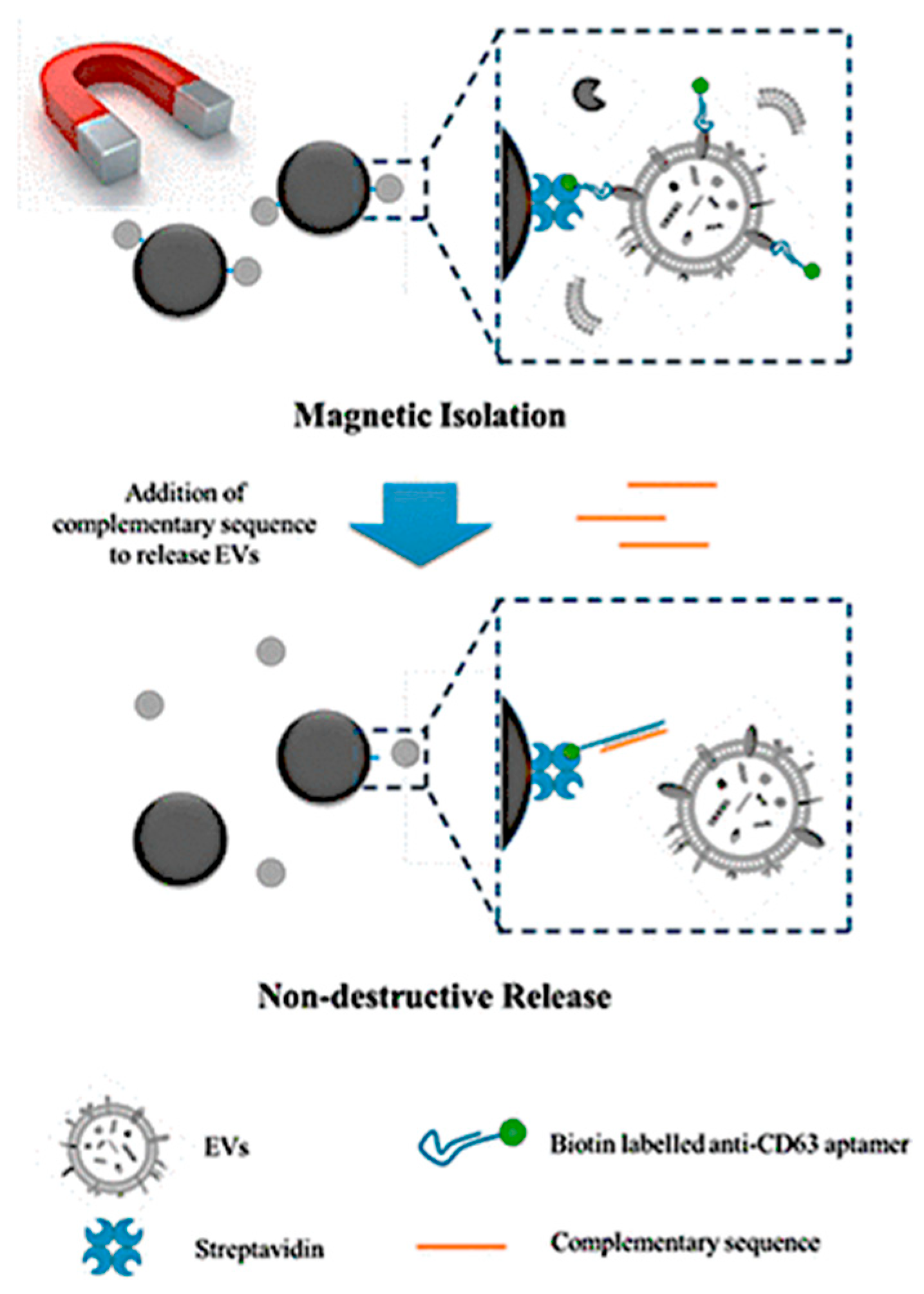
| Magnetic | Magnetic separation approach | Cell-exosome uptake and wound-healing using CD63 proteins | - | [66] |
|
|
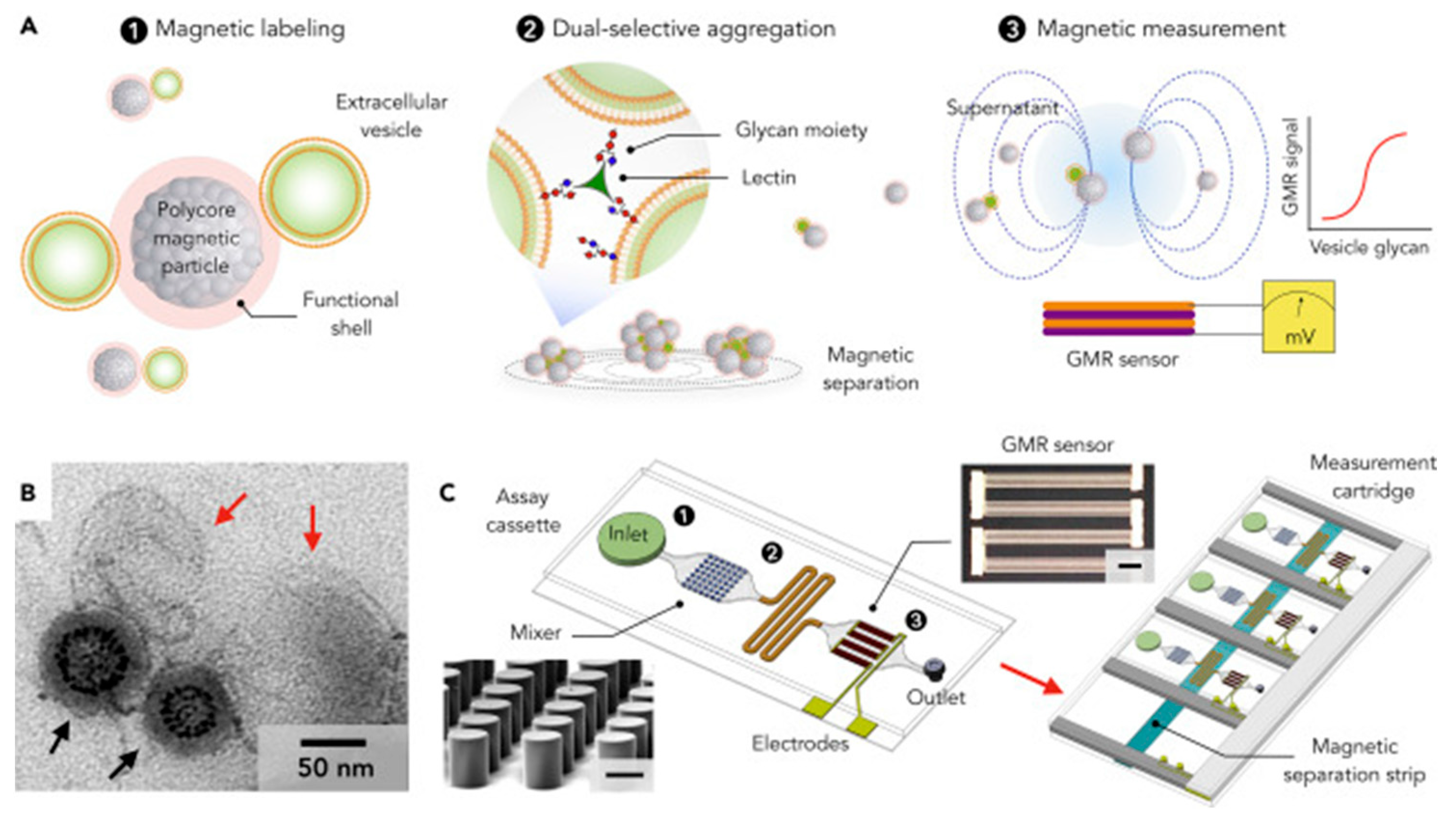
| Labeled integrated magnetic analysis of glycans in extracellular vesicles | EV glycan role in natural biofluids against CD63 proteins | ~104 vesicles | [67] | |||
| SPR | Dual AuNP-assisted amplification of the signal | MCF-7 breast cancer cells using CD63 proteins | 5 × 103 particles/µL | [68] |
|
|
| Polydopamine-functionalized gold nanoparticle (Au@PDA NP)–assisted signal amplification | human hepatic carcinoma (SMMC-7721) cell line offered SMMC-7721 exosomes in liquid biopsy and they were detected using CD63 proteins | 5.6 × 105 particles mL−1 | [69] | |||
| The laser beam is split into two beams by an optical splitter | Non-small cell lung cancer diagnostics using CD63 proteins | 2 × 1010 exosomes/mL | [70] | |||
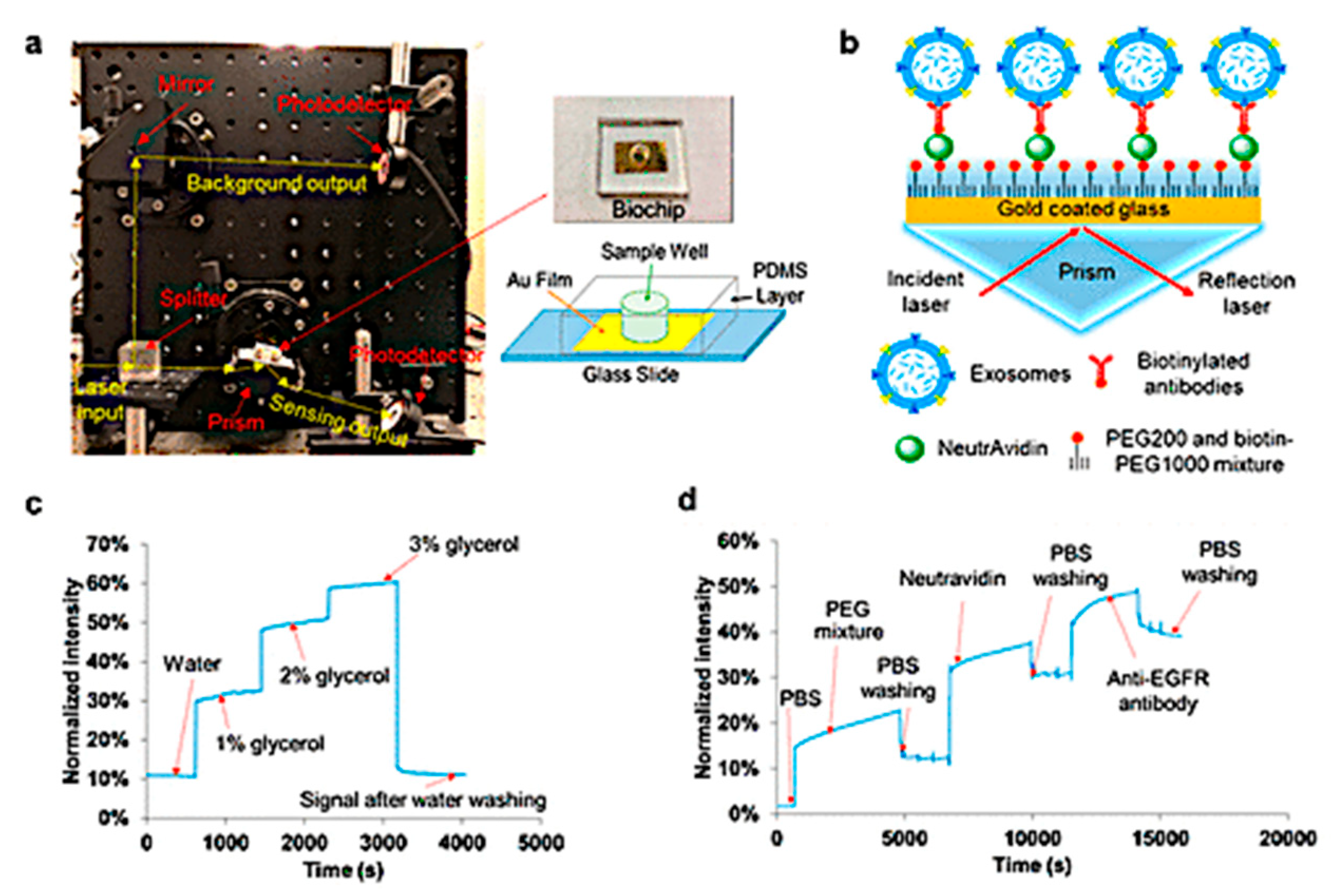
| Intravesicular nanoplasmonic system platform used nanohole-based SPR for molecular detection | Transmembrane (CD63, EpCAM, EGFR) proteins, were measured in ovarian cancer cell lines | 104 EVs | [71] | |||
| Sandwich approach for the detection of clinically relevant exosomes | Isolate bulk exosomes and detect ≥10% HER2(+) exosomes in mixed samples; 14–35% HER2(+) exosomes in patient samples using CD9 and CD63 proteins | 2070 exosomes/μL | [72] | |||
| Plasmonic biosensor for the analysis of EVs based on GC-SPR with wavelength interrogation | mesenchymal stem cell EVs using CD81 protein | 670 aM | [73] | |||
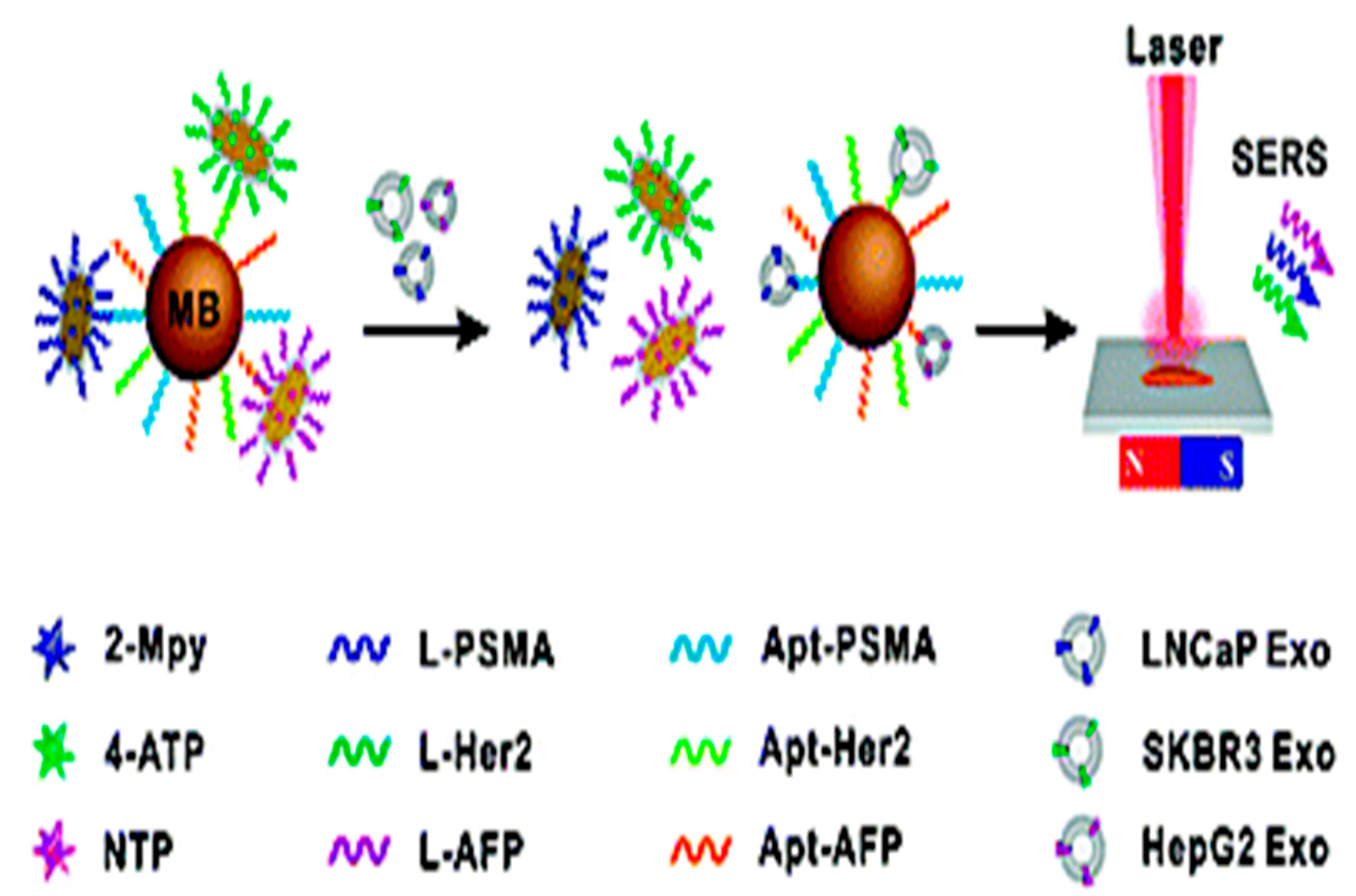
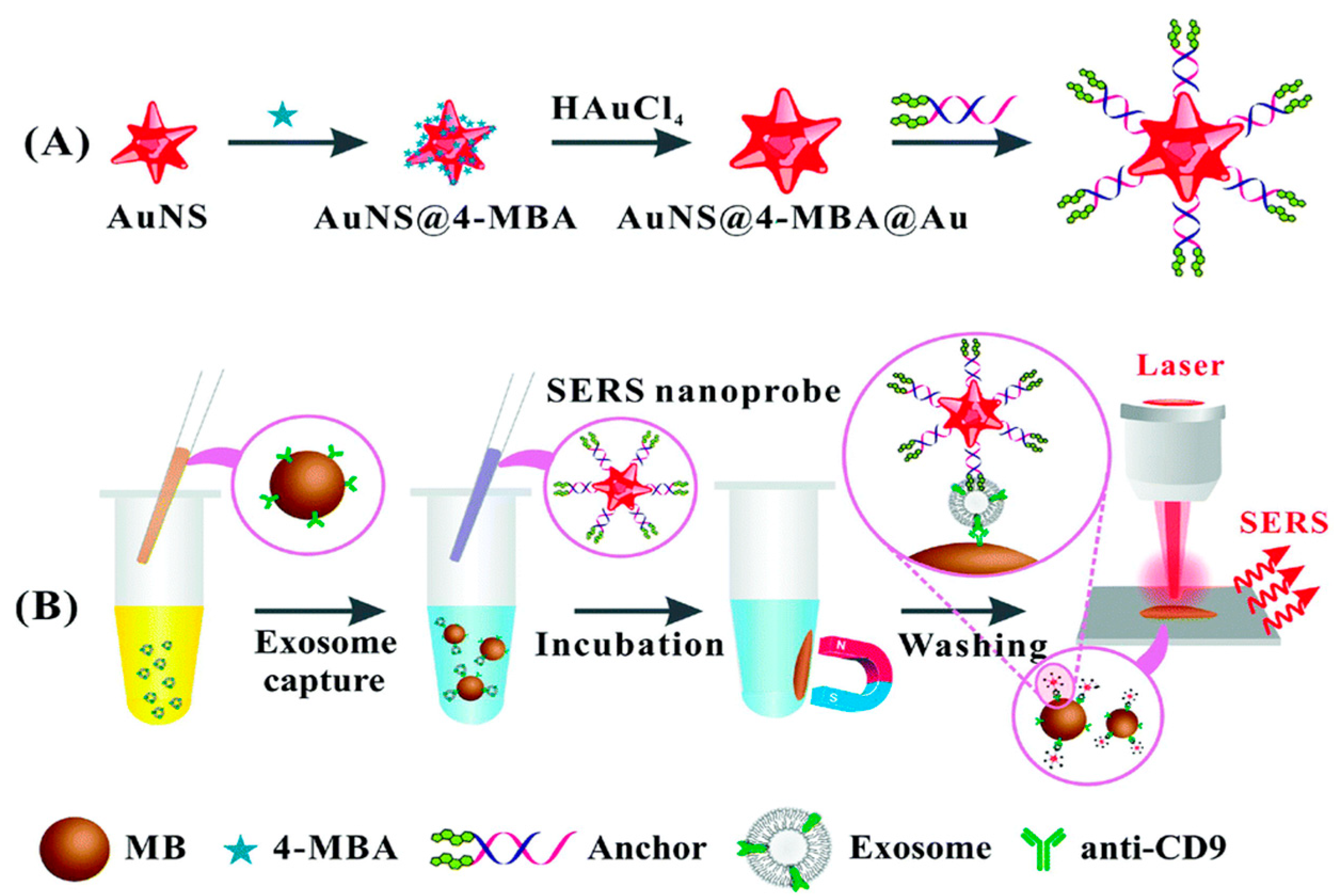
| SERS | AuNPs for targeting exosomes and a Raman reporter for signal readout source | Breast, colorectal, and prostate cancers detection using CD63 proteins | 32, 73, 203 particles/µL | [74] |
|
|
| Gold–silver bimetallic SERS-active nanotags nanotrepangs and a Raman reporter | Various exosome species for cancer | 26, 72, 35 particles/μL | [75] | |||
| Sandwich-type immunocomplex | CD9 protein on the surfaces of exosomes using CD9 proteins | 27 particles/µL | [76] | |||
| H-SERS substrate and a rapid enrichment strategy MEDP | LRG1-Exosomes and GPC1-Exosomes were selected to discriminate against pancreatic cancer using CD63 proteins | 15 particles µL−1 | [77] | |||
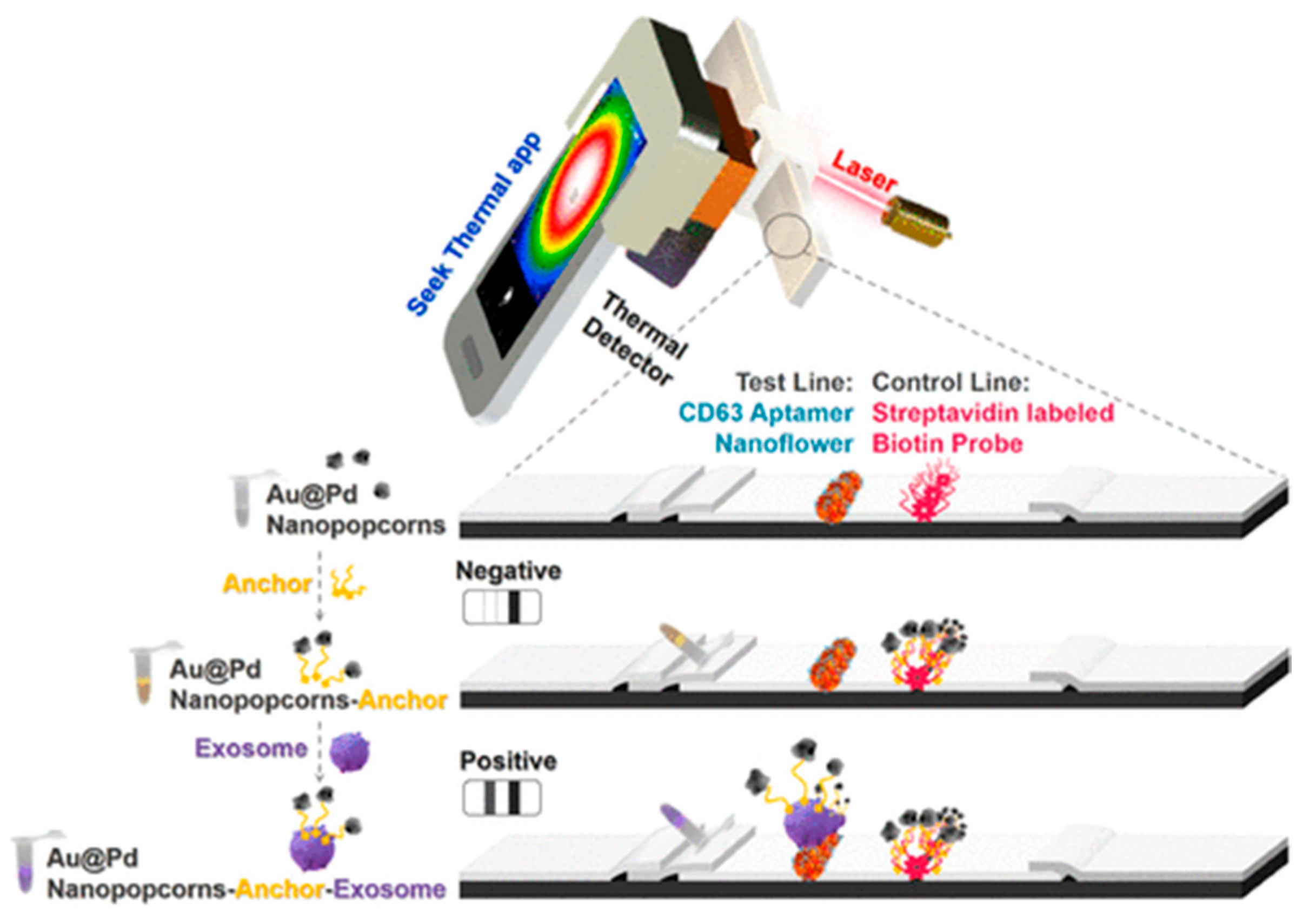
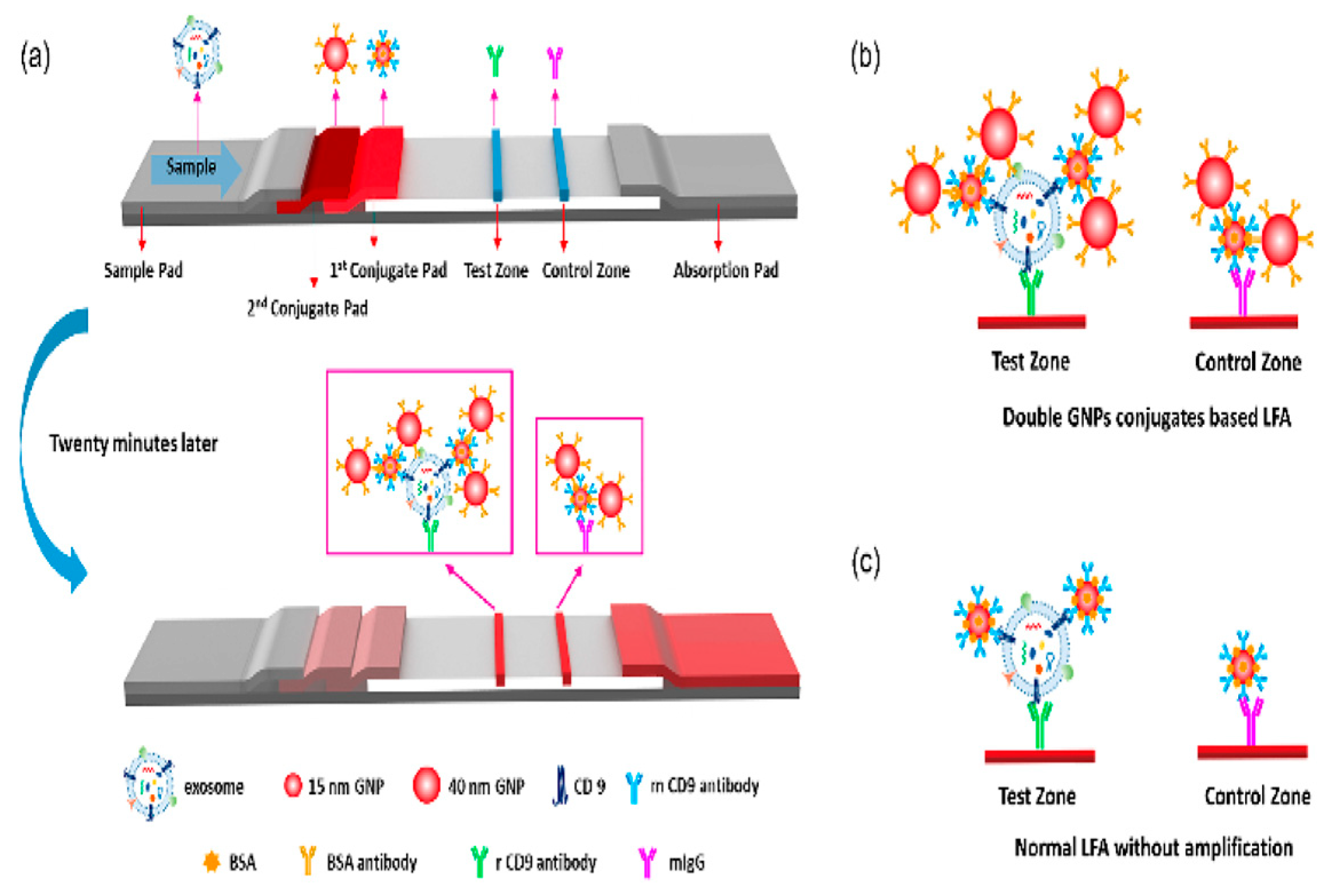
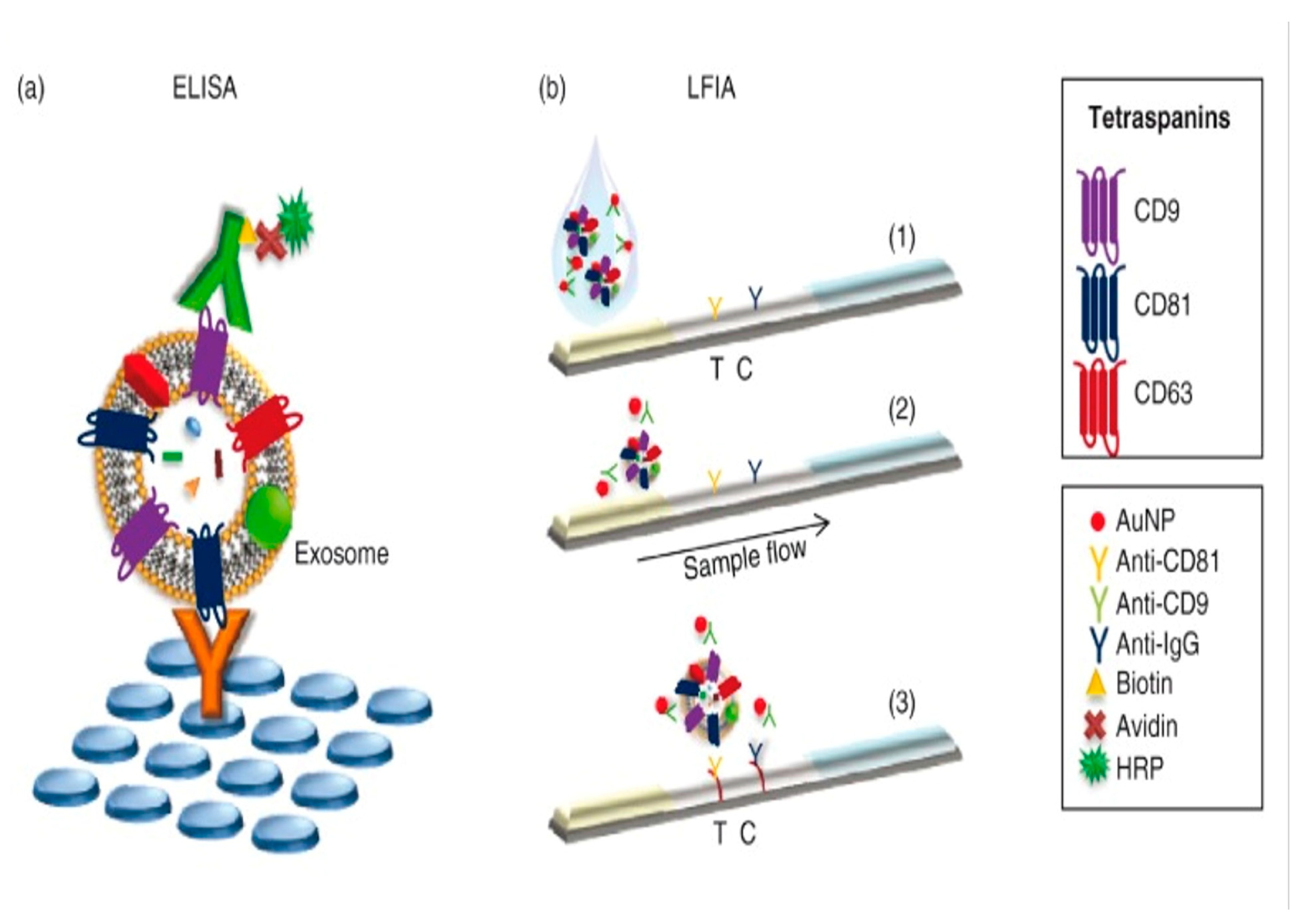
| Lateral Flow Strip | Sandwich strip assay | CD63 protein antigen against different clinical samples | 1.4 × 107 particle/μL | [78] |
|
|
| Competitive strip assay | CD63 protein on exosome from human lung carcinoma cells | 6.4 × 109 particles/mL | [79] | |||
| Sandwich strip assay | Exosomes from fetal bovine serum using CD9 proteins | 1.3 × 103 particles/μL | [80] | |||
| Non-competitive strip assay | Malignant melanoma cell line using CD9, CD 63 and CD81 mixed proteins | 8.54 × 105 exosomes/mL | [81] | |||
| Luminescence | Electrochemiluminescence emitters of Ru(dcbpy)32+ along with sandwich format | Exosomes-related disease diagnosis using CD63 proteins | 37 particle/μL | [82] |
|
|
| UCNPs as a donor and TAMRA as an acceptor | Exosomes derived from breast cancer cells using CD63 proteins | 80 particles/μL | [83] | |||
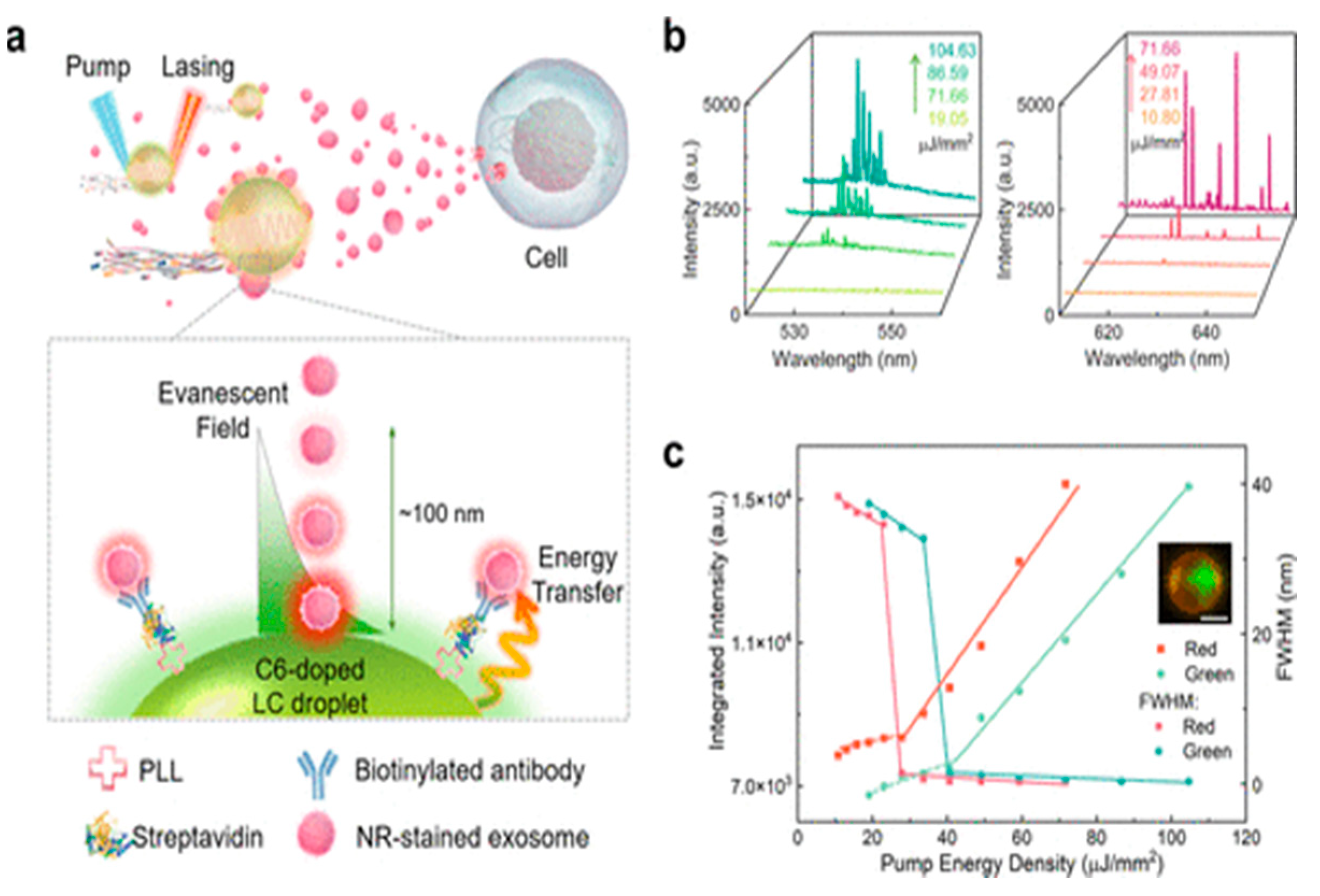
| Autonomous microlaser formation with liquid crystal microdroplets undertaking micellar solubilization in a surfactant | Extracellular biomarkers in circulating biological fluids using CD63 proteins | 1.1 × 104 particles/μL | [84] | |||
| Colorimetric | Dual signal amplification, enzyme-induced silver deposition on Au NRs, and HCR process | Colon cancer LoVo cells and breast cancer exosomes from serum samples using CD63 proteins | 1.6 × 102 particles/µL (UV visible spectroscopy) and 9 × 103 particles/µL (naked eyes) | [85] |
|
|
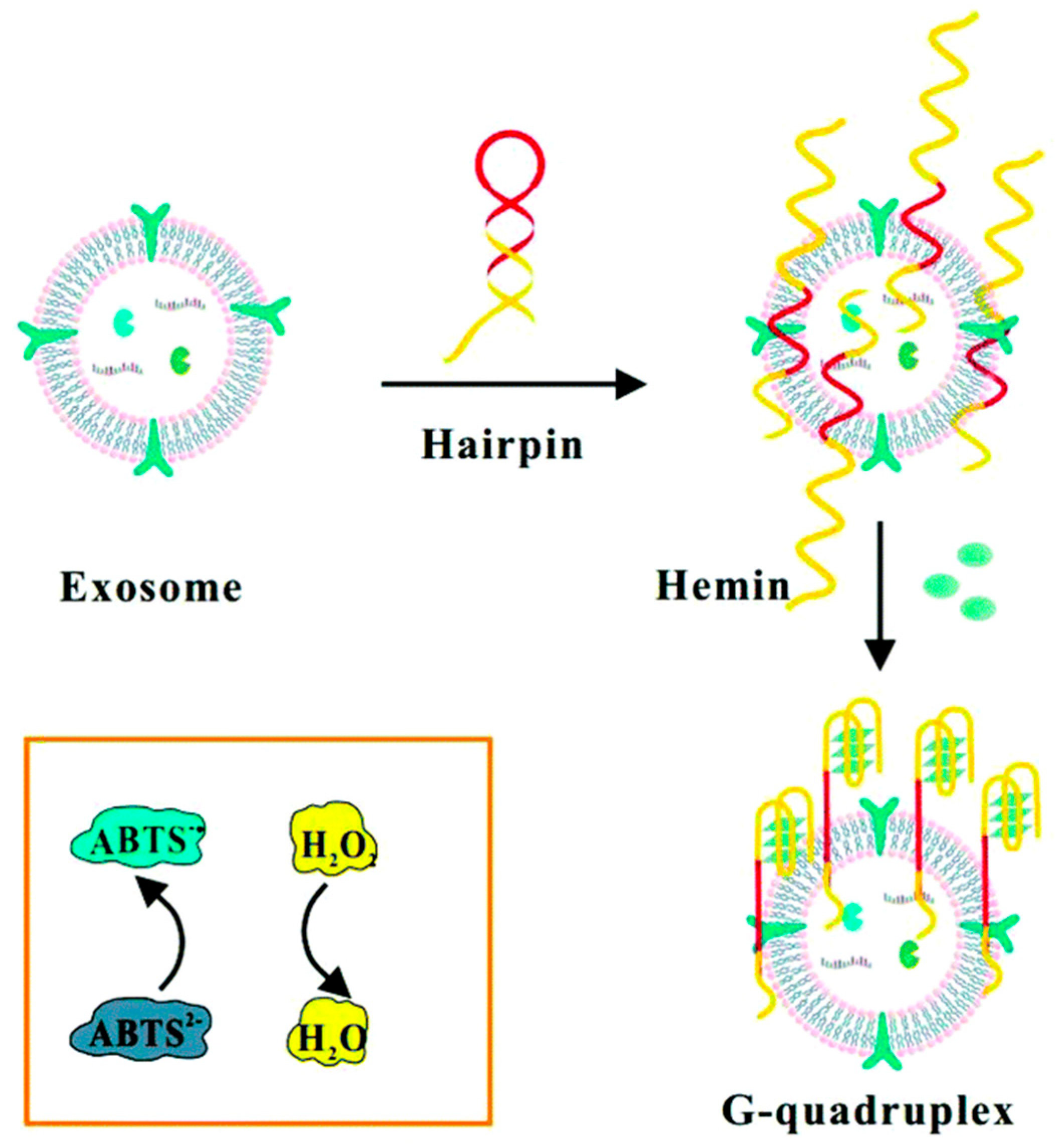
| HRP-mimicking DNAzyme, the hemin/G-quadruplex toward H2O2 reduction | Breast cancer exosomes using CD63 proteins | 3.94 × 105 particles/mL | [86] | |||
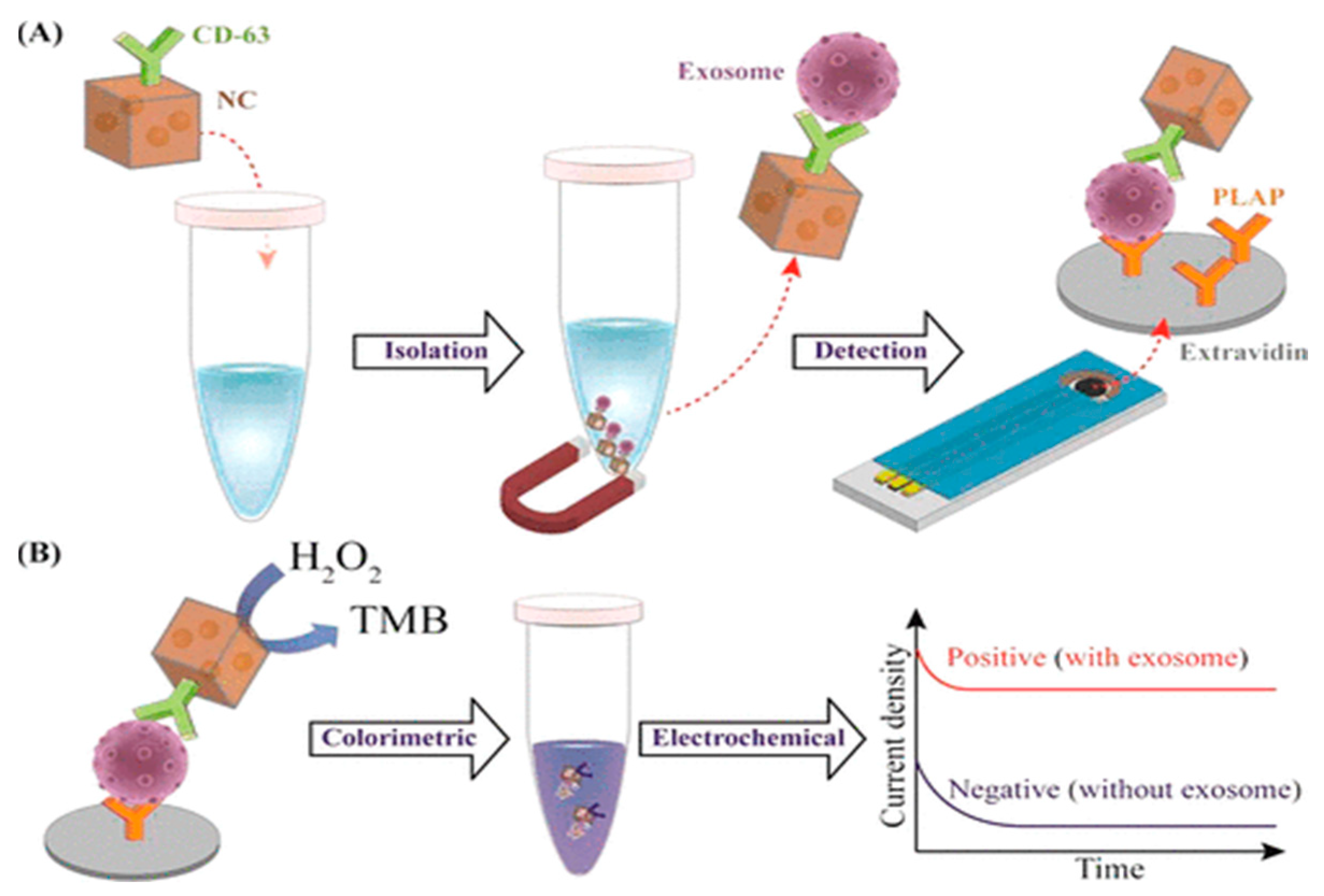
| Au-NPFe2O3NC functionalized with a generic tetraspanin antibody-modified, screen-printed electrode with PLAP | Placental cell-derived exosomes release using CD63 proteins | 103 exosomes/mL | [87] | |||
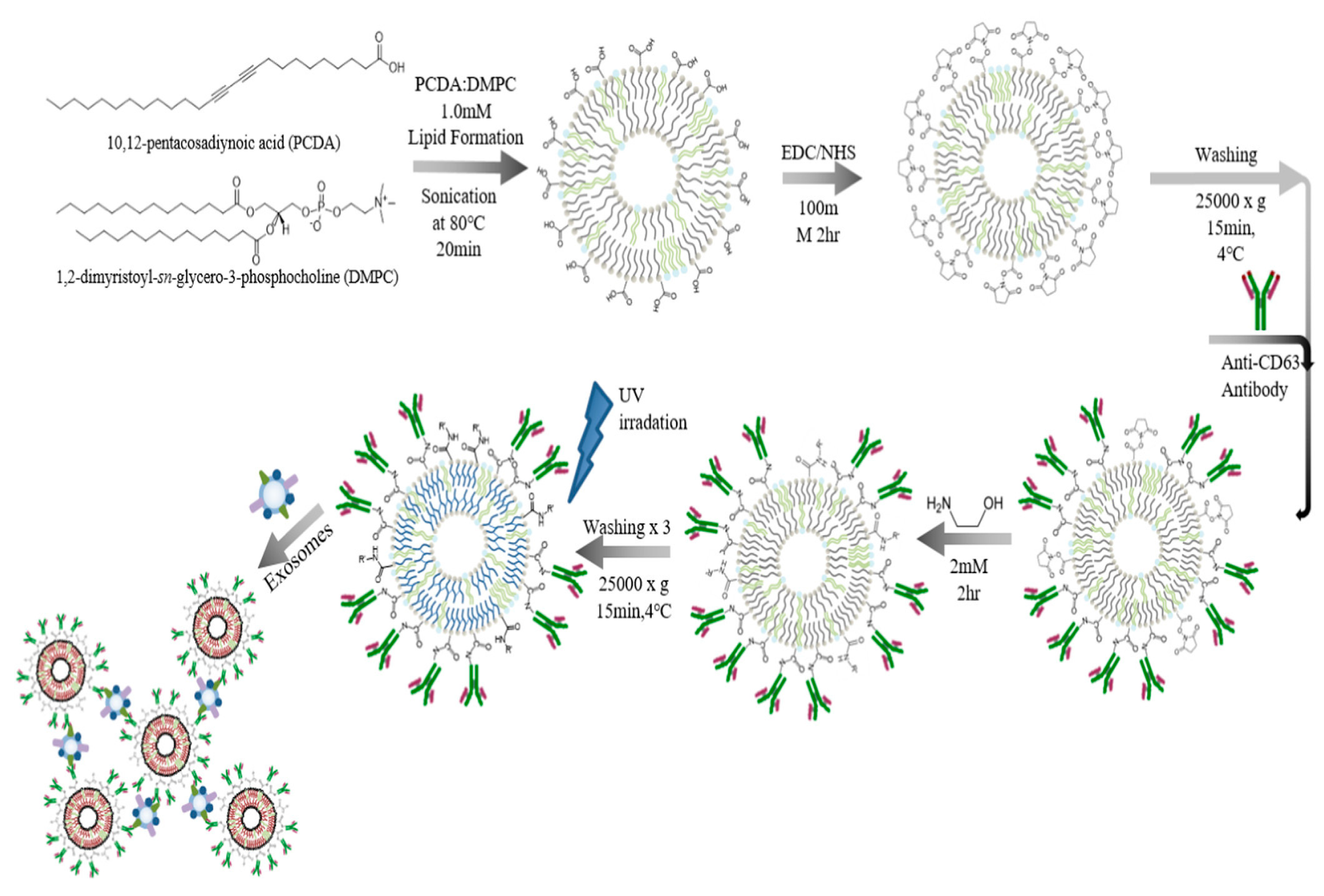
| PDA conjugate polymer with distinctive optical characteristics | Exosomes isolated from the human plasma using CD63 proteins | 3 × 108 vesicles/mL | [88] | |||
| Electrochemical | Immobilization-free dual-aptamer identification sensing method and hyperbranched DNA superstructure signal amplification | Isolation and quantification of tumor exosomes using CD63 proteins | 3.0 × 104 particles mL−1 | [89] |
|
|
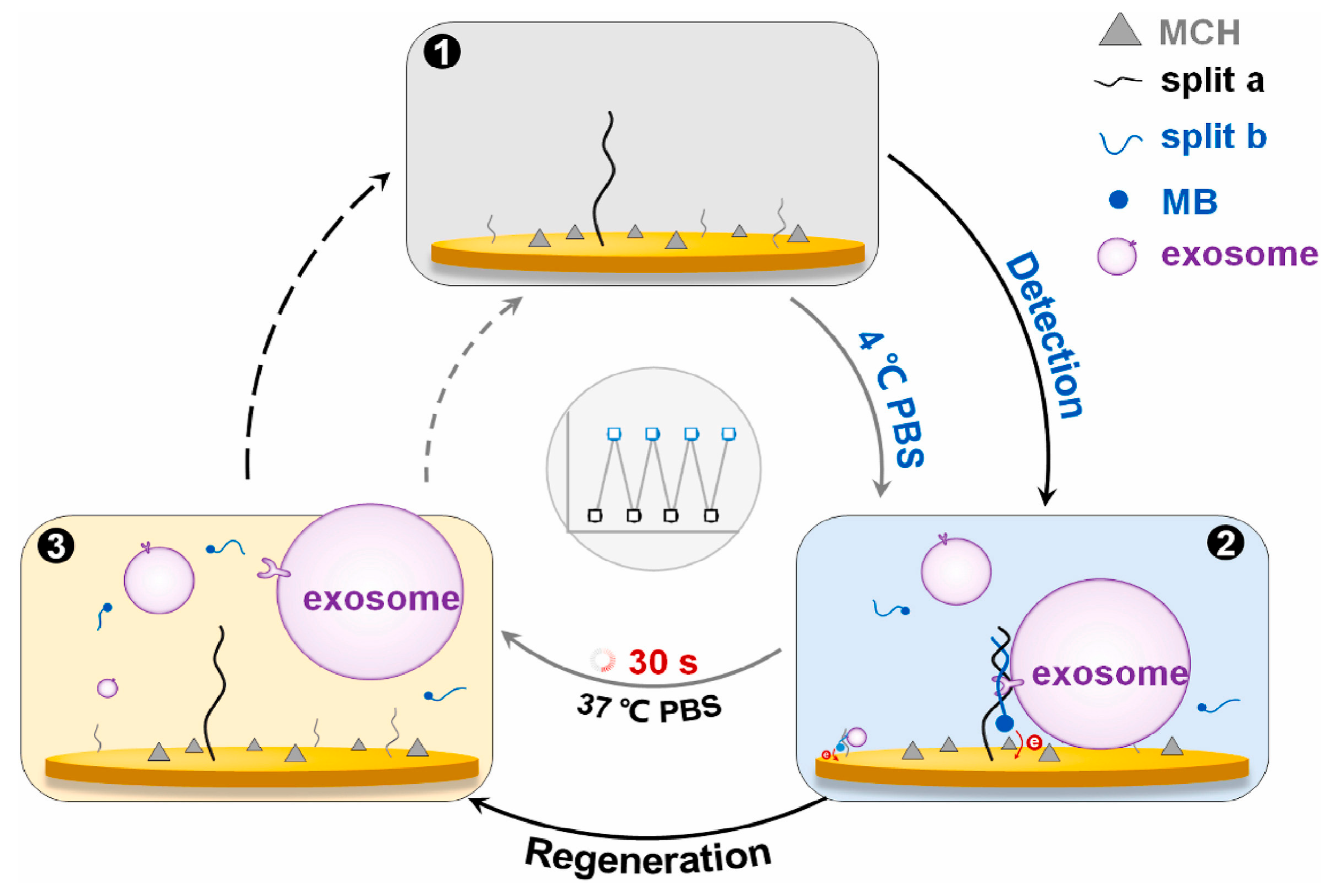
| SMRT biosensor comprised two split-a and split-b fragments | Tumor exosomes from SMMC-7721 (human hepatocellular carcinoma cell line) | 1.5 × 106 particles/mL | [90] | |||
| Sandwich-type reduction of HRP-oxidized TMB electrochemical immunoassay | Distinguishes between exosomes and other EVs using CD9 proteins | 200 particles/µL | [91] | |||
| An electrochemical paper-based analytical instrument having electrode-bound antibodies | Exosomes captured with ovarian cancer-specific CA125 antibodies using CD9 proteins | 9.3 × 107 exosomes/mL | [92] | |||
| Fluorescent | TdT mediated forming ultra-long poly T as the activator | NPC-derived exosomes using CD63 proteins | 100 particles mL−1 | [93] |
|
|
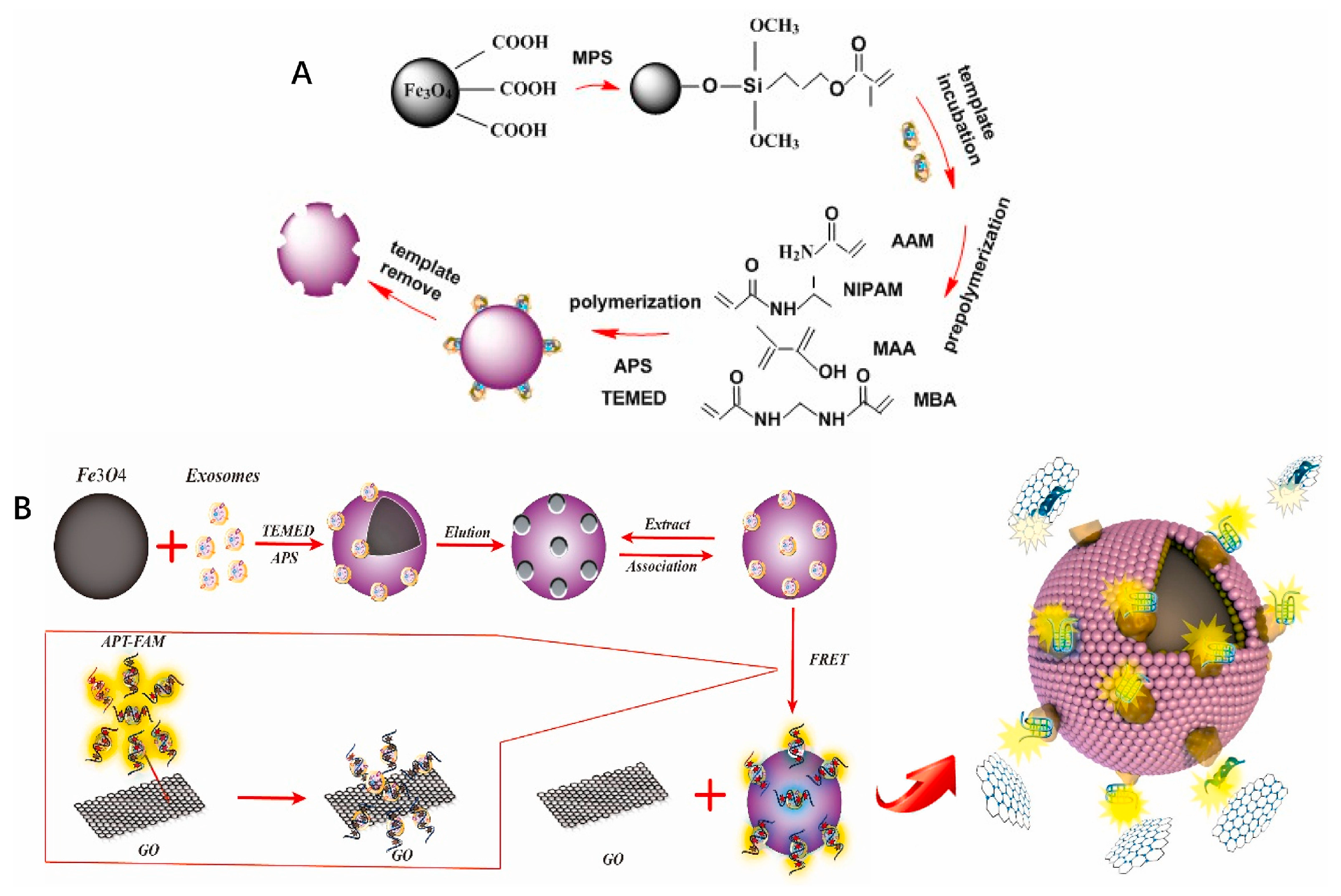
| MIP and aptamer/GO-based FRET system-based optical sensing in a sandwich mode | Lyz proteins and exosomes using CD63 proteins | 2.43 × 106 particles/mL | [94] | |||
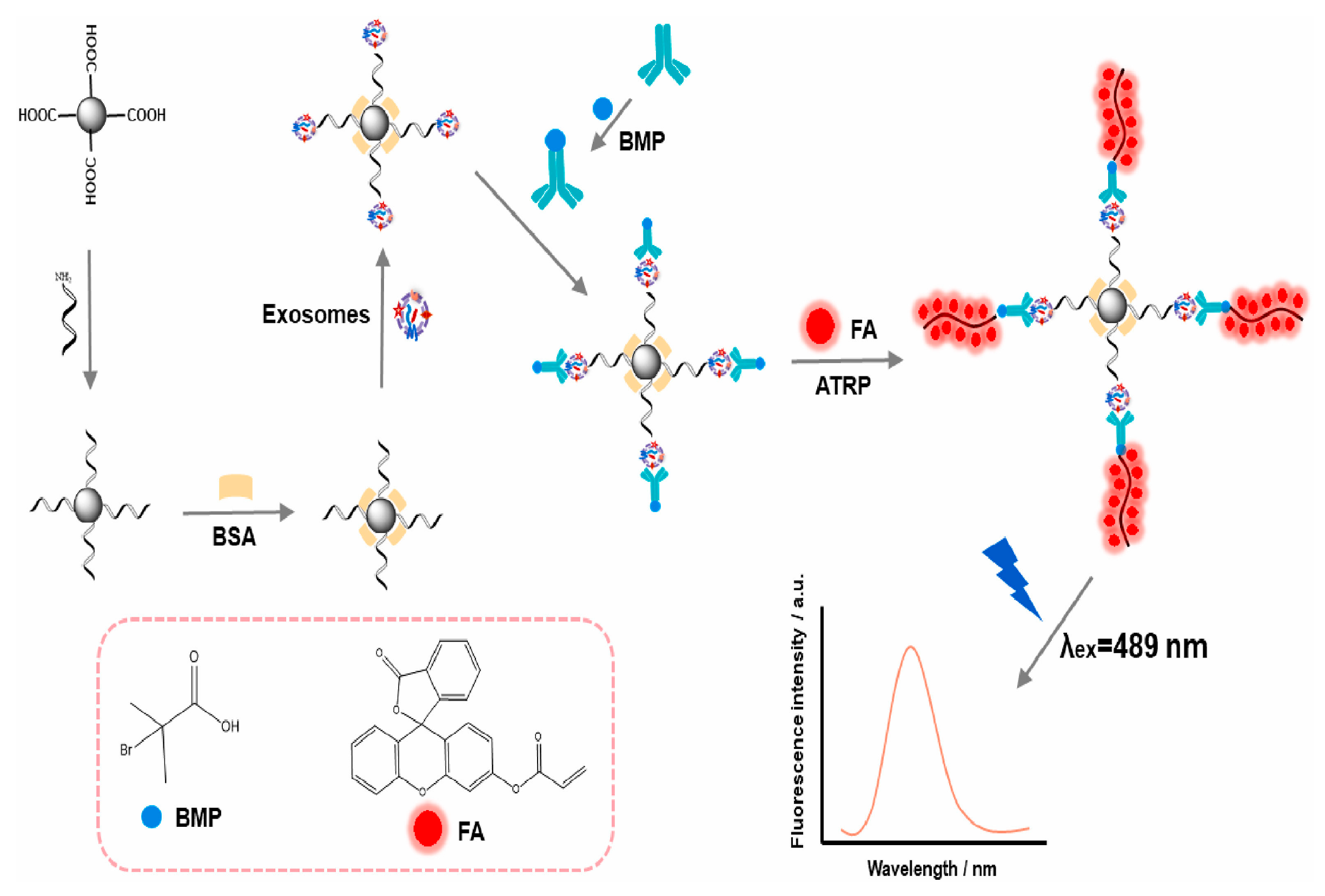
| ARGET ATRP polymerization | The early stage of lung cancer using exosomes using CD63 proteins | 11,610 exosomes/mL | [95] | |||
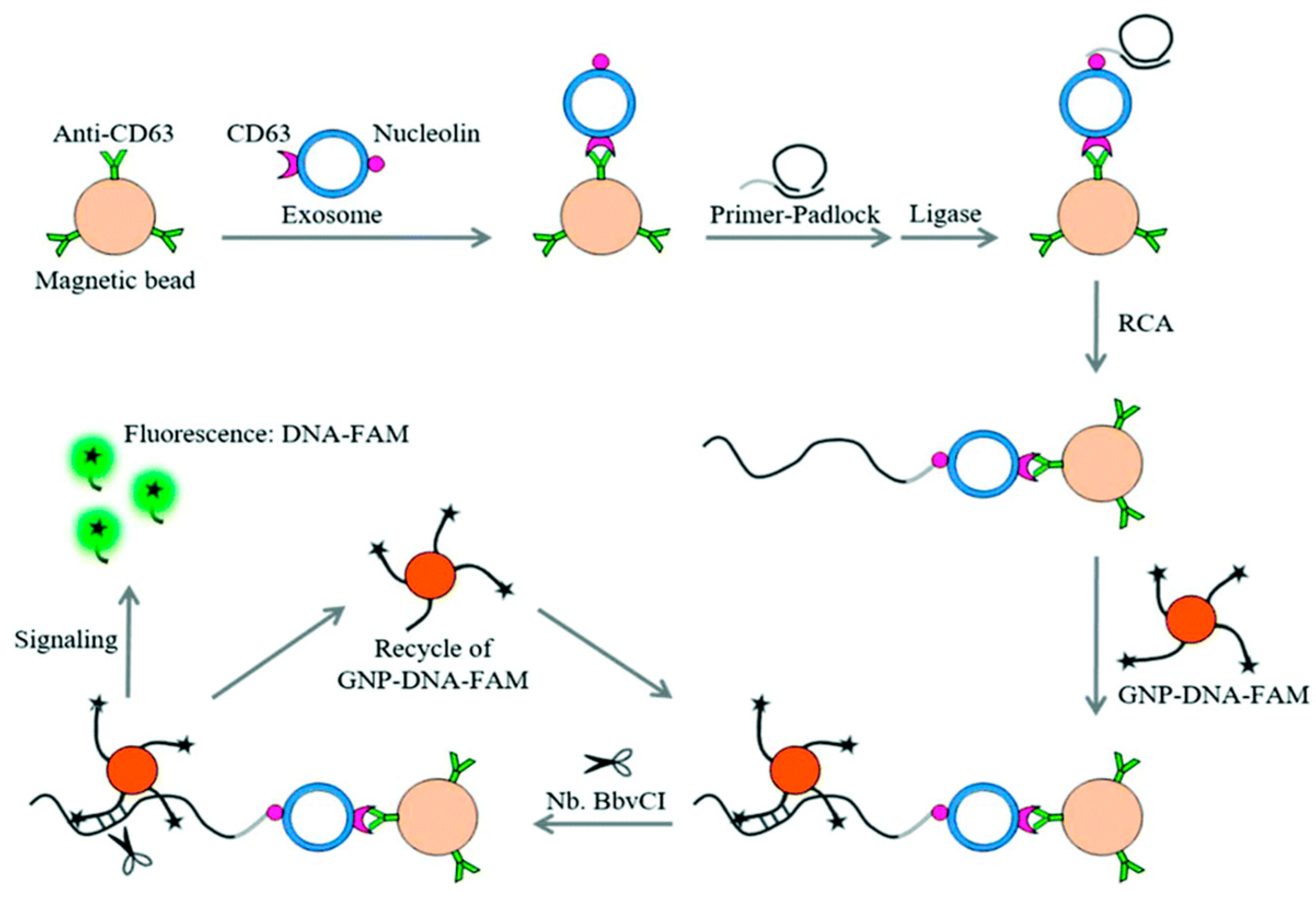
| GNP–DNA–FAM signal amplification technology | Leukemia cell-derived exosomes using CD63 proteins | 1 × 102 particles/L | [96] |
3.1. Magnetic Biosensors
3.2. Surface Plasmon Resonance (SPR) Biosensors
3.3. Surface-Enhanced Raman Scattering (SERS) Biosensors
3.4. Lateral Flow Strip Biosensors
3.5. Luminescence Biosensors
3.5.1. Electrochemiluminescence Biosensors
3.5.2. Luminescence Resonance Energy Transfer Biosensors
3.6. Colorimetric Biosensors
3.7. Electrochemical Biosensors
3.8. Fluorescent Biosensors
4. The Comparison Between Advantages and Disadvantages of Antibody-Based Sensors and Aptamer-Based Sensors
5. Presently Demonstrated and Upcoming Applications
6. Future Trends
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| EV | Extracellular vesicle |
| MVB | Multivesicular body |
| ILVs | Intraluminal vesicles |
| Lyz | Lysosomes |
| SPR | Surface Plasmon Resonance |
| SERS | Surface-Enhanced Raman Scattering |
| CSF | Cerebrospinal fluid |
| SEC | Size-exclusion chromatography |
| FFF | Field-flow fractionation |
| TFF | Tangential flow filtration |
| SELEX | Systematic Evolution of Ligands by EXponential Enrichment |
| H | Heavy |
| L | Light |
| V | Variable |
| C | Constant |
| MB | Magnetic beads |
| iMAGE | Integrated Magnetic Analysis of Glycans in Extracellular Vesicles |
| LOD | Limit of detection |
| AuNP | Gold nanoparticle |
| Au@PDA NP | Polydopamine-functionalized gold nanoparticle |
| DTPs | DNA tetrahedron probes |
| RIU | Refractive index unit |
| EGFR | Exosomal epidermal growth factor receptor |
| PD-L1 | Programmed death-ligand 1 |
| NSCLC | Non-small cell lung cancer |
| iNPS | Intravesicular nanoplasmonic system |
| HER2 | Human epidermal growth factor receptor 2 |
| GC-SPR | Grating-coupled surface plasmon resonance |
| GSSNTs | Gold–silver–silver core-shell shell nanotrepangs |
| GNT | Gold surface nanotrepangs |
| AuNS@4-MBA@Au | Gold nanostar@4-mercaptobenzoic acid@nanoshell structures |
| B-Chol | Bivalent cholesterol |
| H-SERS | Hierarchical-SERS |
| MEDP | Magnetic beads @ exosomes @ SERS detection probe |
| LRG1-Exosomes | LRG1-positive exosomes |
| GPC1-Exosomes | GPC1-positive exosomes |
| AUC | Area under the operating characteristic curve |
| GNPs | Gold nanoparticles |
| D-LFA | Double gold nanoparticles (GNPs) conjugates based lateral flow assay |
| LFIA | Lateral flow immunoassay |
| ECL | Electrochemiluminescence |
| BPQDs | Black phosphorous quantum dots |
| Ru(dcbpy)32+ | Tris (4,4′-dicarboxylicacid-2,2′-bipyridyl) ruthenium(II) dichloride |
| BPQDs | Black phosphorus quantum dots |
| Ru(dcbpy)32+@BPQDs | Tris (4,4′-dicarboxylicacid-2,2′-bipyridyl) ruthenium(II) dichloride @ Black phosphorus quantum dots |
| LRET | Luminescence resonance energy transfer |
| UCNPs | Upconversion nanoparticles |
| TAMRA | Tetramethyl rhodamine |
| EpCAM | Epithelial cell adhesion molecule |
| HCR | Hybridization chain reaction |
| ALP | Alkaline phosphatase |
| Au-NPFe2O3NC | Gold-loaded ferric oxide nanocubes |
| PLAP | Placenta alkaline phosphatase |
| PDA | Polydiacetylene |
| SMRT | Split-aptamer mediated regenerable temperature-sensitive |
| HRP | Horseradish peroxidase |
| NPC | Nasopharyngeal cancer |
| MNPs | Magnetic nanoparticles |
| TdT | Terminal deoxynucleotidyl transferase |
| GO | Graphene oxide |
| FRET | Fluorescence resonance energy transfer |
| ARGET | Activator regenerated by electron transfer |
| ATRP | Atom transfer radical polymerization |
| MB-CD63 | Anti-CD63 antibody-modified MB conjugates |
| RCA | Rolling circle amplification |
| GNP–DNA–FAM | Gold nanoparticle (GNP)-DNA-fluorescent dye (FAM) conjugates |
References
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [CrossRef] [PubMed]
- Sjöstrand, M.; Ekström, K.; Lee, J.J.; Valadi, H.; Bossios, A.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar]
- Waldenström, A.; Gennebäck, N.; Hellman, U.; Ronquist, G. Cardiomyocyte microvesicles contain DNA/RNA and convey biological messages to target cells. PLoS ONE 2012, 7, e34653. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Lim, J.W.; Moritz, R.L.; Mathivanan, S. Exosomes: Proteomic insights and diagnostic potential. Expert Rev. Proteom. 2009, 6, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Sainte-Marie, J.; Philippot, J.R.; Bienvenue, A. Asymmetric distribution of phospholipids in the membrane of vesicles released during in vitro maturation of guinea pig reticulocytes: Evidence precluding a role for “aminophospholipid translocase”. J. Cell Physiol. 1989, 140, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Jordan, S.; Haroon, J. The Role of Exosomes in Regenerative Medicine. In Essentials of Regenerative Medicine in Interventional Pain Management; Springer: Berlin/Heidelberg, Germany, 2024; pp. 195–201. [Google Scholar]
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81. [Google Scholar] [CrossRef]
- Gangoda, L.; Boukouris, S.; Liem, M.; Kalra, H.; Mathivanan, S. Extracellular vesicles including exosomes are mediators of signal transduction: Are they protective or pathogenic? Proteomics 2015, 15, 260–271. [Google Scholar] [CrossRef]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.A.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Camussi, G.; Deregibus, M.C.; Bruno, S.; Cantaluppi, V.; Biancone, L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010, 78, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Nazimek, K.; Bryniarski, K.; Santocki, M.; Ptak, W. Exosomes as mediators of intercellular communication: Clinical implications. Pol. Arch. Med. Wewnętrznej 2015, 125, 370–380. [Google Scholar] [CrossRef]
- Simons, M.; Raposo, G. Exosomes—Vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Y.; Wang, H.; Zhu, Z.; Xiao, Z. Visualizing of the cellular uptake and intracellular trafficking of exosomes by live-cell microscopy. J. Cell Biochem. 2010, 111, 488–496. [Google Scholar] [CrossRef]
- Fu, M.; Gu, J.; Jiang, P.; Qian, H.; Xu, W.; Zhang, X. Exosomes in gastric cancer: Roles, mechanisms, and applications. Mol. Cancer 2019, 18, 41. [Google Scholar] [CrossRef]
- Khalyfa, A.; Gozal, D. Exosomal miRNAs as potential biomarkers of cardiovascular risk in children. J. Transl. Med. 2014, 12, 162. [Google Scholar] [CrossRef]
- Deatherage, B.L.; Cookson, B.T. Membrane vesicle release in bacteria, eukaryotes, and archaea: A conserved yet underappreciated aspect of microbial life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Zhao, Y.; Banks, W.A.; Bullock, K.M.; Haney, M.; Batrakova, E.; Kabanov, A.V. Macrophage exosomes as natural nanocarriers for protein delivery to inflamed brain. Biomaterials 2017, 142, 1–12. [Google Scholar] [CrossRef]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of Exosome Composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef]
- Zhou, Y.; Tian, T.; Zhu, Y.; Ali, D.J.; Hu, F.; Qi, Y.; Sun, B.; Xiao, Z. Exosomes transfer among different species cells and mediating miRNAs delivery. J. Cell Biochem. 2017, 118, 4267–4274. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Xiao, K.; Xiang, S.; Li, Z.; Weng, X. Emerging role of exosomes in the joint diseases. Cell. Physiol. Biochem. 2018, 47, 2008–2017. [Google Scholar] [CrossRef]
- Minciacchi, V.R.; Freeman, M.R.; Di Vizio, D. Extracellular Vesicles in Cancer: Exosomes, Microvesicles and the Emerging Role of Large Oncosomes. Semin. Cell Dev. Biol. 2015, 40, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.; Kaushik, S.; Clement, C.C.; Cannizzo, E.S.; Scharf, B.; Follenzi, A.; Potolicchio, I.; Nieves, E.; Cuervo, A.M.; Santambrogio, L. Microautophagy of Cytosolic Proteins by Late Endosomes. Dev. Cell 2011, 20, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Record, M. Intercellular communication by exosomes in placenta: A possible role in cell fusion? Placenta 2014, 35, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.-D.; Abdel-Bakky, M.; Gutwein, P.; Altevogt, P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007, 72, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Madison, M.N.; Jones, P.H.; Okeoma, C.M. Exosomes in human semen restrict HIV-1 transmission by vaginal cells and block intravaginal replication of LP-BM5 murine AIDS virus complex. Virology 2015, 482, 189–201. [Google Scholar] [CrossRef]
- Qin, W.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in human breast milk promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef]
- Machida, T.; Tomofuji, T.; Ekuni, D.; Maruyama, T.; Yoneda, T.; Kawabata, Y.; Mizuno, H.; Miyai, H.; Kunitomo, M.; Morita, M. MicroRNAs in salivary exosome as potential biomarkers of aging. Int. J. Mol. Sci. 2015, 16, 21294–21309. [Google Scholar] [CrossRef]
- Street, J.M.; Koritzinsky, E.H.; Glispie, D.M.; Star, R.A.; Yuen, P.S.T. Chapter Three—Urine Exosomes: An Emerging Trove of Biomarkers. Adv. Clin. Chem. 2017, 78, 103–122. [Google Scholar] [CrossRef]
- Peng, P.; Yan, Y.; Keng, S. Exosomes in the ascites of ovarian cancer patients: Origin and effects on anti-tumor immunity. Oncol. Rep. 2011, 25, 749–762. [Google Scholar] [CrossRef]
- Yagi, Y.; Ohkubo, T.; Kawaji, H.; Machida, A.; Miyata, H.; Goda, S.; Roy, S.; Hayashizaki, Y.; Suzuki, H.; Yokota, T. Next-generation sequencing-based small RNA profiling of cerebrospinal fluid exosomes. Neurosci. Lett. 2017, 636, 48–57. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Harding, C.V. Extracellular vesicles and infectious diseases: New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.H.; Xiang, D.; Tran, T.T.; Yin, W.; Zhang, Y.; Kong, L.; Chen, K.; Sun, M.; Li, Y.; Hou, Y.; et al. Exosomes and nanoengineering: A match made for precision therapeutics. Adv. Mater. 2020, 32, 1904040. [Google Scholar] [CrossRef]
- Alenquer, M.; Amorim, M.J. Exosome biogenesis, regulation, and function in viral infection. Viruses 2015, 7, 5066–5083. [Google Scholar] [CrossRef]
- Sivalingam, A.M.; Sureshkumar, D.D. Exosomes in Regulating miRNAs for Biomarkers of Neurodegenerative Disorders. Mol. Neurobiol. 2025, 62, 7576–7596. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.C.; Choi, D.; Lee, D.; Oh, J.S. Engineering Exosomes for CNS Disorders: Advances, Challenges, and Therapeutic Potential. Int. J. Mol. Sci. 2025, 26, 3137. [Google Scholar] [CrossRef]
- Ting, Y.; Dong, X.; Wang, X.; Liu, X.; Fu, L.; Li, L. Engineering exosomes for mRNA delivery: A review. Int. J. Biol. Macromol. 2025, 316, 144662. [Google Scholar] [CrossRef]
- Sharma, D.; Srivastava, S.; Babu, M.R. Precision exosome engineering for neurological therapeutics: Molecular mechanisms and targeted strategies. Mol. Biol. Rep. 2025, 52, 518. [Google Scholar] [CrossRef]
- Akter, A.; Kamal, T.; Akter, S.; Auwal, A.; Islam, F. Exosomes: A potential tool in the diagnosis, prognosis and treatment of patients with colorectal cancer. Future Oncol. 2025, 21, 2347–2365. [Google Scholar] [CrossRef]
- Sharma, H.; Tyagi, S.J.; Chandra, P.; Verma, A.; Kumar, P.; Ashique, S.; Hussain, A. Role of Exosomes in Parkinson’s and Alzheimer’s Diseases. In Exosomes Based Drug Delivery Strategies for Brain Disorders; Springer: Berlin/Heidelberg, Germany, 2024; pp. 147–182. [Google Scholar]
- Edgar, J.R. Q&A: What are exosomes, exactly? BMC Biol. 2016, 14, 46. [Google Scholar] [CrossRef]
- Zheng, L.; Li, J.; Li, Y.; Sun, W.; Ma, L.; Qu, F.; Tan, W. Empowering exosomes with aptamers for Precision Theranostics. Small Methods 2024, 9, e2400551. [Google Scholar] [CrossRef]
- Hu, S.; Zhang, L.; Su, Y.; Liang, X.; Yang, J.; Luo, Q.; Luo, H. Sensitive detection of multiple blood biomarkers via immunomagnetic exosomal PCR for the diagnosis of Alzheimer’s disease. Sci. Adv. 2024, 10, eabm3088. [Google Scholar] [CrossRef]
- Martins, T.S.; Vaz, M.; Henriques, A.G. A review on comparative studies addressing exosome isolation methods from body fluids. Anal. Bioanal. Chem. 2023, 415, 1239–1263. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Li, A.; Hu, J.; Feng, L.; Liu, L.; Shen, Z. Recent developments in isolating methods for exosomes. Front. Bioeng. Biotechnol. 2023, 10, 1100892. [Google Scholar] [CrossRef] [PubMed]
- Mukerjee, N.; Bhattacharya, A.; Maitra, S.; Kaur, M.; Ganesan, S.; Mishra, S.; Ashraf, A.; Rizwan, M.; Kesari, K.K.; Tabish, T.A.; et al. Exosome isolation and characterization for advanced diagnostic and therapeutic applications. Mater. Today Bio 2025, 31, 101613. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Herrera, A.; Rossi, J.J.; Zhou, J. Current advances in aptamers for cancer diagnosis and therapy. Cancers 2018, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Chiu, T.; Huang, C. Aptamer-functionalized nano-biosensors. Sensors 2009, 9, 10356–10388. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Sharma, T.K.; Bruno, J.G.; Dhiman, A. ABCs of DNA aptamer and related assay development. Biotechnol. Adv. 2017, 35, 275–301. [Google Scholar] [CrossRef]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.B.; Tang, T. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef]
- Ku, T.; Zhang, T.; Luo, H.; Yen, T.M.; Chen, P.-W.; Han, Y.; Lo, Y.-H. Nucleic acid aptamers: An emerging tool for biotechnology and biomedical sensing. Sensors 2015, 15, 16281–16313. [Google Scholar] [CrossRef]
- Davies, D.R.; Chacko, S. Antibody structure. Acc. Chem. Res. 1993, 26, 421–427. [Google Scholar] [CrossRef]
- Marquart, M.; Deisenhofer, J. The 3-dimensional structure of antibodies. Immunol. Today 1982, 3, 160–166. [Google Scholar] [CrossRef]
- Alzari, P.M.; Lascombe, M.B.; Poljak, R.J. Three-Dimensional Structure of Antibodies. Annu. Rev. Immunol. 1988, 6, 555–580. [Google Scholar] [CrossRef]
- Chothia, C. Antigen recognition: Current opinion in structural biology 1991, 1:53–59. Curr. Opin. Struct. Biol. 1991, 1, 53–59. [Google Scholar] [CrossRef]
- Colman, P.M. Antigen-antigen receptor interactions: Current Opinion in Structural Biology 1991, 1:232–236. Curr. Opin. Struct. Biol. 1991, 1, 232–236. [Google Scholar] [CrossRef]
- Mian, I.S.; Bradwell, A.R.; Olson, A.J. Structure, function and properties of antibody binding sites. J. Mol. Biol. 1991, 217, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.R.; Padlan, E.A.; Sheriff, S. Antibody-Antigen Complexes. Annu. Rev. Biochem. 1990, 59, 439–473. [Google Scholar] [CrossRef]
- Davies, D.R.; Padlan, E.A. Twisting into shape. Curr. Biol. 1992, 2, 254–256. [Google Scholar] [CrossRef]
- Sheriff, S. Antibody-Protein Complexes. ImmunoMethods 1993, 3, 222–227. [Google Scholar] [CrossRef]
- Paul, W.E. Fundamental Immunology, 2nd ed.; Raven Press: Dublin, Ireland, 1989. [Google Scholar]
- Zhang, K.; Yue, Y.; Wu, S.; Liu, W.; Shi, J.; Zhang, Z. Rapid capture and nondestructive release of extracellular vesicles using aptamer-based magnetic isolation. ACS Sens. 2019, 4, 1245–1251. [Google Scholar] [CrossRef]
- Wang, Z.; Sun, X.; Natalia, A.; Tang, C.S.L.; Ang, C.B.T.; Ong, C.-A.J.; Teo, M.C.C.; So, J.B.Y.; Shao, H. Dual-Selective Magnetic Analysis of Extracellular Vesicle Glycans. Matter 2020, 2, 150–166. [Google Scholar] [CrossRef]
- Wang, Q.; Zou, L.; Yang, X.; Liu, X.; Nie, W.; Zheng, Y.; Cheng, Q.; Wang, K. Direct quantification of cancerous exosomes via surface plasmon resonance with dual gold nanoparticle-assisted signal amplification. Biosens. Bioelectron. 2019, 135, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Liu, X.; Yang, X.; Wang, Q.; Geng, X.; Zou, L.; Liu, Y.; Li, S.; Zheng, Y.; Wang, K. Surface plasmon resonance assay for exosomes based on aptamer recognition and polydopamine-functionalized gold nanoparticles for signal amplification. Microchim. Acta 2020, 187, 251. [Google Scholar] [CrossRef]
- Liu, C.; Zeng, X.; An, Z.; Yang, Y.; Eisenbaum, M.; Gu, X.; Jornet, J.M.; Dy, G.K.; Reid, M.E.; Gan, Q.; et al. Sensitive Detection of Exosomal Proteins via a Compact Surface Plasmon Resonance Biosensor for Cancer Diagnosis. ACS Sens. 2018, 3, 1471–1479. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Im, H.; Hong, S.; Castro, C.M.; Weissleder, R.; Lee, H. Analyses of intravesicular exosomal proteins using a nano-plasmonic system. ACS Photonics 2018, 5, 487–494. [Google Scholar] [CrossRef]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef]
- Reiner, A.T.; Ferrer, N.; Venugopalan, P.; Lai, R.C.; Lim, S.K.; Dostálek, J. Magnetic nanoparticle-enhanced surface plasmon resonance biosensor for extracellular vesicle analysis. Analyst 2017, 142, 3913–3921. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef] [PubMed]
- Ning, C.; Wang, L.; Tian, Y.; Yin, B.; Ye, B. Multiple and sensitive SERS detection of cancer-related exosomes based on gold–silver bimetallic nanotrepangs. Analyst 2020, 145, 2795–2804. [Google Scholar] [CrossRef]
- Tian, Y.; Ning, C.; He, F.; Yin, B.; Ye, B. Highly sensitive detection of exosomes by SERS using gold nanostar@Raman reporter@nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst 2018, 143, 4915–4922. [Google Scholar] [CrossRef]
- Li, J.; Li, Y.; Chen, S.; Duan, W.; Kong, X.; Wang, Y.; Zhou, L.; Li, P.; Zhang, C.; Du, L.; et al. Highly Sensitive Exosome Detection for Early Diagnosis of Pancreatic Cancer Using Immunoassay Based on Hierarchical Surface-Enhanced Raman Scattering Substrate. Small Methods 2022, 6, e2200154. [Google Scholar] [CrossRef]
- Cheng, N.; Song, Y.; Shi, Q.; Du, D.; Liu, D.; Luo, Y.; Xu, W.; Lin, Y. Au@ Pd nanopopcorn and aptamer nanoflower assisted lateral flow strip for thermal detection of exosomes. Anal. Chem. 2019, 91, 13986–13993. [Google Scholar] [CrossRef]
- Yu, Q.; Zhao, Q.; Wang, S.; Zhao, S.; Zhang, S.; Yin, Y.; Dong, Y. Development of a lateral flow aptamer assay strip for facile identification of theranostic exosomes isolated from human lung carcinoma cells. Anal. Biochem. 2020, 594, 113591. [Google Scholar] [CrossRef]
- Wu, T.; Yang, Y.; Cao, Y.; Huang, Y.; Xu, L.-P.; Zhang, X.; Wang, S. Enhanced lateral flow assay with double conjugates for the detection of exosomes. Sci. China Chem. 2018, 61, 1423–1429. [Google Scholar] [CrossRef]
- Oliveira-Rodríguez, M.; López-Cobo, S.; Reyburn, H.T.; Costa-García, A.; López-Martín, S.; Yáñez-Mó, M.; Cernuda-Morollón, E.; Paschen, A.; Valés-Gómez, M.; Blanco-López, M.C. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell. Vesicles 2016, 5, 31803. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Zhao, D.; Zhang, S.; Huang, Y.; Dai, H.; Lin, Y. Black phosphorus quantum dots functionalized MXenes as the enhanced dual-mode probe for exosomes sensing. Sens. Actuators B Chem. 2020, 305, 127544. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, D.; Fang, Y.; Wu, W.; Wang, Y.; Xia, Y.; Wu, F.; Li, C.; Lan, J.; Chen, J. An aptasensor based on upconversion nanoparticles as LRET donors for the detection of exosomes. Sens. Actuators B Chem. 2019, 298, 126900. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, G.; Gao, Z.; Liao, Y.; Gong, C.; Kim, M.; Chang, G.-E.; Feng, S.; Xu, T.; Liu, T.; et al. Autonomous Microlasers for Profiling Extracellular Vesicles from Cancer Spheroids. Nano Lett. 2023. [CrossRef] [PubMed]
- Zhang, Y.; Wang, D.; Yue, S.; Lu, Y.-B.; Yang, C.; Fang, J.; Xu, Z.-R. Sensitive multicolor visual detection of exosomes via dual signal amplification strategy of enzyme-catalyzed metallization of Au nanorods and hybridization chain reaction. ACS Sens. 2019, 4, 3210–3218. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Wang, H.; Ye, B. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst 2020, 145, 107–114. [Google Scholar] [CrossRef]
- Boriachek, K.; Masud, M.K.; Palma, C.; Phan, H.-P.; Yamauchi, Y.; Hossain, S.A.; Nguyen, N.-T.; Salomon, C.; Shiddiky, M.J.A. Avoiding Pre-Isolation Step in Exosome Analysis: Direct Isolation and Sensitive Detection of Exosomes Using Gold-Loaded Nanoporous Ferric Oxide Nanozymes. Anal. Chem. 2019, 91, 3827–3834. [Google Scholar] [CrossRef]
- Kim, C.; Lee, K. Polydiacetylene (PDA) Liposome-Based Immunosensor for the Detection of Exosomes. Biomacromolecules 2019, 20, 3392–3398. [Google Scholar] [CrossRef]
- Yang, L.; Yin, X.; An, B.; Li, F. Precise Capture and Direct Quantification of Tumor Exosomes via a Highly Efficient Dual-Aptamer Recognition-Assisted Ratiometric Immobilization-Free Electrochemical Strategy. Anal. Chem. 2021, 93, 1709–1716. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tang, J.; Xu, H.; Yuan, K.; Aryee, A.A.; Zhang, C.; Meng, H.; Qu, L.; Li, Z. Split-aptamer mediated regenerable temperature-sensitive electrochemical biosensor for the detection of tumour exosomes. Anal. Chim. Acta 2022, 1219, 340027. [Google Scholar] [CrossRef] [PubMed]
- Doldán, X.; Fagúndez, P.; Cayota, A.; Laíz, J.; Tosar, J.P. Electrochemical Sandwich Immunosensor for Determination of Exosomes Based on Surface Marker-Mediated Signal Amplification. Anal. Chem. 2016, 88, 10466–10473. [Google Scholar] [CrossRef] [PubMed]
- Kasetsirikul, S.; Tran, K.T.; Clack, K.; Soda, N.; Shiddiky, M.J.A.; Nguyen, N. Low-cost electrochemical paper-based device for exosome detection. Analyst 2022, 147, 3732–3740. [Google Scholar] [CrossRef] [PubMed]
- Yi, P.; Luo, D.; Gao, Z.; Chen, Q.; Zhou, Y. Fluorescent aptasensor based on the MNPs-CRISPR/Cas12a-TdT for the determination of nasopharyngeal carcinoma-derived exosomes. Microchim. Acta 2023, 190, 74. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Ren, M.; Miao, Y.; Liao, Z.; Zhang, T.; Chen, S.; Ye, K.; Zhang, P.; Ma, X.; Ni, J.; et al. Dual selective sensor for exosomes in serum using magnetic imprinted polymer isolation sandwiched with aptamer/graphene oxide based FRET fluorescent ignition. Biosens. Bioelectron. 2022, 207, 114112. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Jin, Z.; Cui, Z.; Guo, L.; Kong, J. A specific sensor system based on in-situ synthesis fluorescent polymers by ARGET ATRP achieving sensitive exosome detection. Talanta 2023, 253, 124059. [Google Scholar] [CrossRef]
- Huang, L.; Wang, D.; Singh, N.; Yang, F.; Gu, N.; Zhang, X. A dual-signal amplification platform for sensitive fluorescence biosensing of leukemia-derived exosomes. Nanoscale 2018, 10, 20289–20295. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zou, S.; Huang, Z.; Sun, C.; Liu, G. Isolation and quantification of L1CAM-positive extracellular vesicles on a chip as a potential biomarker for Parkinson’s disease. J. Extracell. Vesicles 2024, 13, e12467. [Google Scholar] [CrossRef] [PubMed]












| Features | Aptamer-Based Sensor | Antibody-Based Sensor |
|---|---|---|
| Recognition Element | Aptamer (ssDNA or RNA) | Antibody |
| Production | Chemical synthesis, fast, cost effective | Biological (animals/cells), slow, costly |
| Specificity/Affinity | High, can be tuned | High, well characterized |
| Stability | More stable, resists harsh conditions | Sensitive to heat, pH, organic solvents |
| Modifiability | Flexible chemical modification | Limited modification options |
| Batch Variability | High reproducibility | Can vary between batches |
| Shelf Life | Long (especially if modified) | Short to moderate |
| Size | Small (≈8–25 kDa) | Large (≈150 kDa) |
| Reusability | High (can be regenerated) | Limited (may denature) |
| Nuclease Sensitivity | Sensitive (but can be chemically protected) | Not applicable |
| Clinical Acceptance | Gaining acceptance | Well-accepted, validated |
| Cost per assay | Low to moderate | Moderate to high |
| Common Sensor Types | All (SPR, SERS, LFA, etc.) | All (SPR, SERS, LFA, etc.) |
| Limitations | Nuclease sensitivity, sometimes lower affinity | Costly, stability issues |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prabhu, S.N.; Liu, G. Cutting-Edge Sensor Technologies for Exosome Detection: Reviewing Role of Antibodies and Aptamers. Biosensors 2025, 15, 511. https://doi.org/10.3390/bios15080511
Prabhu SN, Liu G. Cutting-Edge Sensor Technologies for Exosome Detection: Reviewing Role of Antibodies and Aptamers. Biosensors. 2025; 15(8):511. https://doi.org/10.3390/bios15080511
Chicago/Turabian StylePrabhu, Sumedha Nitin, and Guozhen Liu. 2025. "Cutting-Edge Sensor Technologies for Exosome Detection: Reviewing Role of Antibodies and Aptamers" Biosensors 15, no. 8: 511. https://doi.org/10.3390/bios15080511
APA StylePrabhu, S. N., & Liu, G. (2025). Cutting-Edge Sensor Technologies for Exosome Detection: Reviewing Role of Antibodies and Aptamers. Biosensors, 15(8), 511. https://doi.org/10.3390/bios15080511






