Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review
Abstract
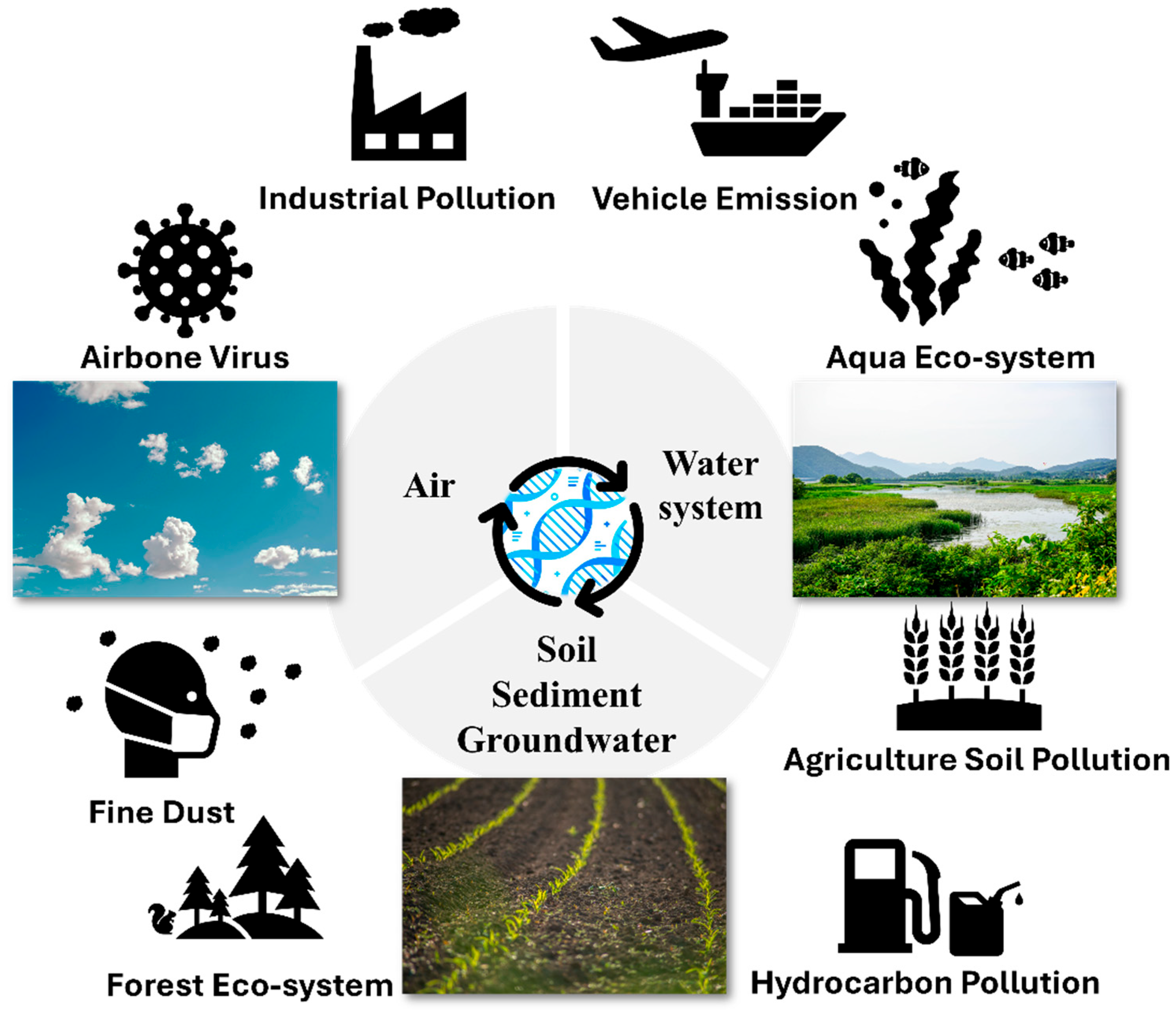
1. Introduction
1.1. Importance of Environmental Monitoring
1.2. Importance of eDNA in Environmental Monitoring
1.3. Trend of eDNA Monitoring
1.4. Technologies in eDNA Monitoring
1.5. Literature Search Strategy
2. Technology and Applications of eDNA Monitoring
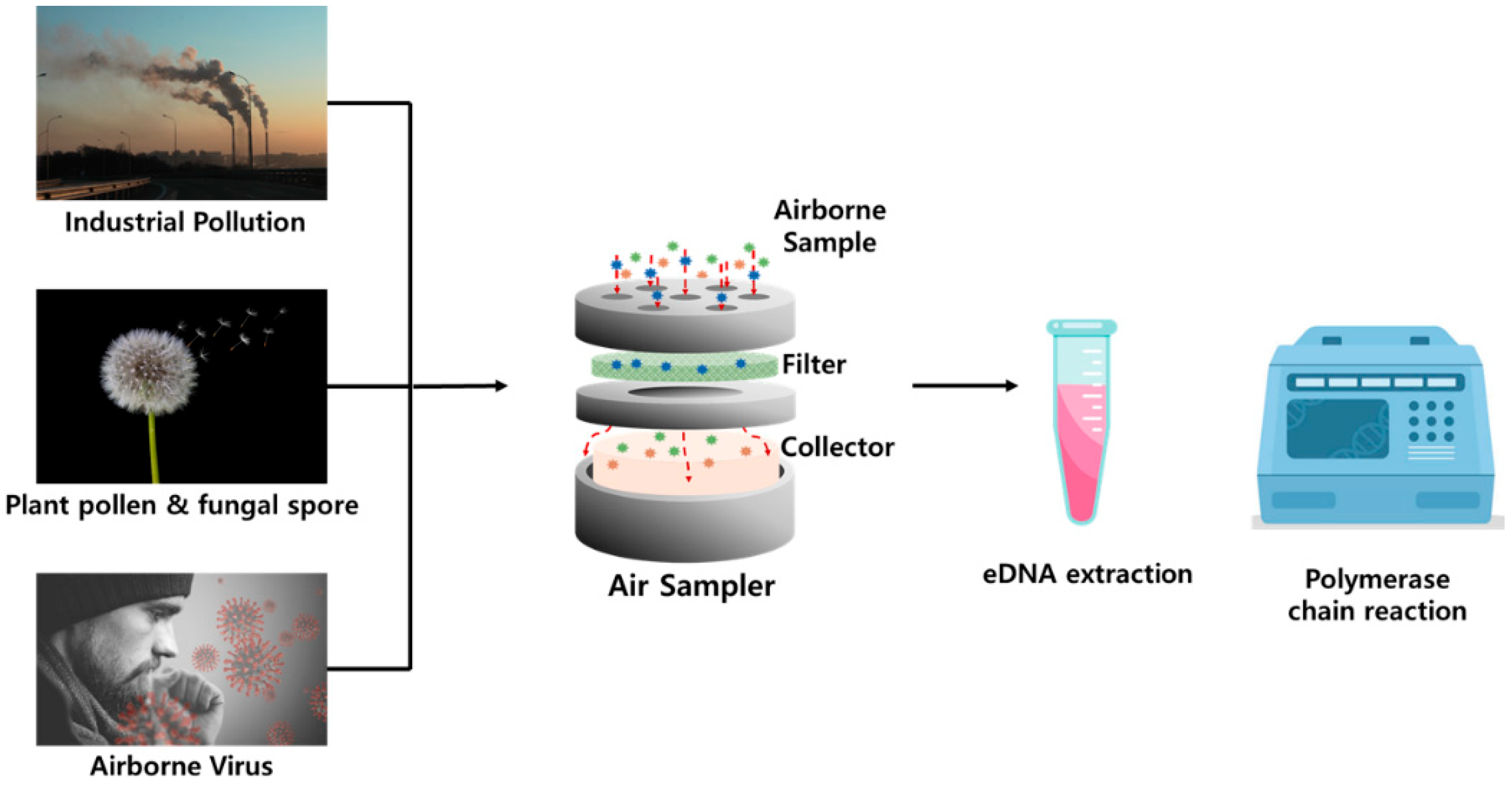
2.1. Methodologies and Environmental Applications of Airborne eDNA
2.1.1. Rationale for Airborne eDNA Analysis
2.1.2. Sampling Methods and Protocols
| Category | Target | Sampling Methods | Ref. |
|---|---|---|---|
| Pollution | Microbial contamination (pathogens, fungi, viruses) | -Filter-based sampling (HEPA, PTFE filters, etc.) -Liquid impinger -Cyclone sampler | [46,47,48] |
| Microbial community shifts due to industrial emissions | -Filter-based sampling -Passive sampling (settling method) | [49,53] | |
| Antibiotic-resistant bacteria monitoring | -Filter-based sampling -Airborne microbial trap | [50,51] | |
| Microbial responses to heavy metals/chemical pollutants | -Filter-based sampling -Passive sampling -Electrostatic precipitation | [49,50,53] | |
| Ecosystem | Airborne insect eDNA (endangered and invasive species) | -Cyclone sampler -Filter-based sampling -Passive sampling | [49,53] |
| Plant pollen and fungal spore distribution | -Filter-based sampling -Cyclone sampler -Passive sampling (settled dust collection) | [49,53] | |
| Seasonal ecological changes | -Passive sampling (long-term monitoring) -Filter-based sampling | [49,51,52,53] | |
| Public Health and Biosecurity | Airborne virus surveillance (SARS-CoV-2, influenza, etc.) | -Liquid impinger -Electrostatic precipitation -Filter-based sampling | [49,50,53] |
| Bioterrorism agent detection (anthrax, viral pathogens, etc.) | -Cyclone sampler -Electrostatic precipitation -Filter-based sampling | [49] | |
| Pathogen transmission monitoring in hospitals and public spaces | -Filter-based sampling -Liquid impinger -Airborne particle collector | [49,53] | |
| Agriculture and Food Safety | Crop pathogen and pest surveillance | -Cyclone sampler -Passive sampling | [49,53] |
| Monitoring pesticide-resistant airborne microbes | -Filter-based sampling -Electrostatic precipitation | [51,52] | |
| Early detection of livestock infectious diseases | -Liquid impinger -Filter-based sampling | [49,53] |
2.1.3. Applications and Case Studies
2.1.4. Challenges and Outlook
2.2. eDNA Monitoring in Soil, Sediment, and Groundwater
2.2.1. The Importance of Terrestrial and Subsurface Environments
2.2.2. Soil, Sediment, and Groundwater eDNA Sampling
2.2.3. Pollution Monitoring and Biodegradation Studies
2.2.4. eDNA Monitoring Case Studies in Soil, Sediment, and Groundwater
| Case Study | Sampling Methods | Region | Analysis Method | Ref. |
|---|---|---|---|---|
| Industrial Brownfields | Surface | France | -PCR amplification (fungal 18S), NGS (Illumina MiSeq), OTU-based analysis | [92] |
| Core | USA | -PCR amplification (16S, 18S, ITS), metabarcoding, ASV inference (DADA2), NGS (MiSeq) | [94] | |
| Surface | Australia | -PCR amplification (16S V4, 18S V7), metabarcoding, OTU clustering (Greenfield pipeline), NGS (MiSeq) | [95] | |
| Core | Italy | -PCR amplification (16S, 18S, COI), metabarcoding, ASV inference (QIIME2), NGS (MiSeq) | [100] | |
| Surface | Australia | -PCR amplification (16S, 18S), metabarcoding, OTU clustering (Greenfield), GS (MiSeq) | [101] | |
| Agricultural Soil | Surface | China | -PCR-based metabarcoding (16S, 18S, ITS, COI), OTU clustering (97%), NGS (MiSeq) | [97] |
| Core | Italy | -PCR-based metabarcoding (COI, 18S), ASV inference (DADA2 in QIIME2), NGS (MiSeq) | [98] | |
| Core | Germany | -PCR-based metabarcoding (COI for arthropods, D2 for fungi), OTU clustering (97%), NGS (MiSeq) | [102] | |
| Core | Denmark | -PCR-based metabarcoding (16S, ITS, 18S), OTU clustering (97%), NGS | [103] | |
| Surface | Germany | -eDNA metabarcoding for AM fungi, decomposers, and protists; ASV/OTU-based analysis; NGS (HiSeq, 454) | [104] | |
| Wetland Restoration | Core | Australia | -PCR amplification (18S, trnL), OTU clustering (97%), NGS (Roche 454) | [105] |
| Surface | USA | -PCR-based metabarcoding (vertebrate 12S), ASV inference, NGS (HiSeq) | [106] | |
| Surface | Canada | -Species-specific qPCR assays (northern leopard frog, boreal chorus frog) | [107] | |
| Surface | China | -PCR amplification (18S V4), NGS (Illumina MiSeq) | [108] |
2.2.5. Key Considerations
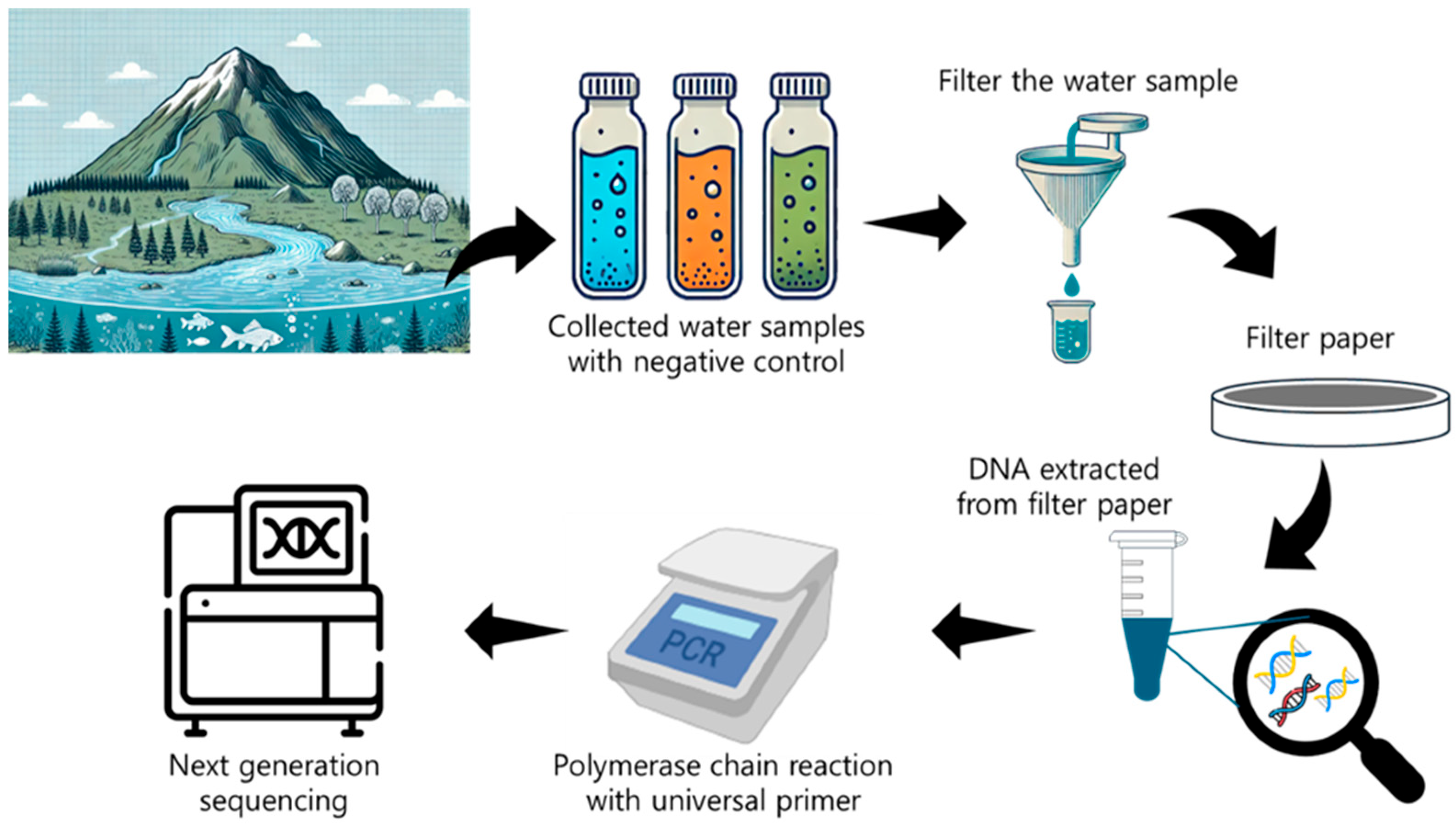
2.3. eDNA Monitoring in Water Systems
2.3.1. Value of Aquatic eDNA
2.3.2. eDNA Sampling Approaches
| Sampling Method | Advantages | Limitations | Typical Applications | Analysis Method | Ref. |
|---|---|---|---|---|---|
| Grab sampling | -Simple and cost-effective -Quick to perform -Minimal equipment required | -Represents only a single time point -May miss temporal variation | -Rapid assessments -Preliminary surveys -Small-scale studies | Two-step PCR, metabarcoding | [122,125] |
| Continuous/ Automated Sampling | -Captures temporal variability -Generates high-resolution datasets -Reduces human error during collection | -Higher cost and maintenance requirements -More complex data management and analysis | -Long-term monitoring programs -Dynamic environments -Detailed temporal studies | [126,127] | |
| Passive Sampling | -Low power requirements -Can be deployed in hard-to-access or remote areas -Minimizes disturbance during sampling | -May have lower control over sampling timing -Potential variability in accumulation rates depending on environmental factors | -Environments with limited human access -Long-term or continuous eDNA accumulation studies | [61,125,128,129,130] | |
| Remote and Autonomous Sampling | -Enables sampling in remote, harsh, or hazardous environments -Real-time data transmission and monitoring -Reduces the need for frequent human intervention | -Requires significant initial investment -Dependence on reliable power sources and communication infrastructure | -Remote locations (e.g., high-altitude streams, polar regions) -Real-time environmental monitoring applications | PCR, qPCR, NGS | [131] |
2.3.3. Wastewater Surveillance
2.3.4. Resource Water Bodies
2.3.5. eDNA Monitoring Applications and Case Studies in Water Systems
3. Future Directions
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviation
| AI | Artificial Intelligence |
| ARGs | Antibiotic Resistance Genes |
| ASV | Amplicon Sequence Variant |
| COI | Cytochrome Oxidase I |
| CRISPR-Cas | Clustered Regularly Interspaced Short Palindromic Repeats–CRISPR-associated proteins |
| dPCR | Digital Polymerase Chain Reaction |
| ddPCR | Droplet Digital Polymerase Chain Reaction |
| eDNA | Environmental DNA |
| GIS | Geographic Information Systems |
| ITS | Internal Transcribed Spacer |
| HEPA | High-Efficiency Particulate Air (filter) |
| HTS | High-Throughput Sequencing |
| ISO | International Organization for Standardization |
| LAMP | Loop-Mediated Isothermal Amplification |
| RPA | Recombinase Polymerase Amplification |
| LSTM | Long Short-Term Memory |
| MIQE | Minimum Information for Publication of Quantitative Real-Time PCR Experiments |
| NGS | Next Generation Sequencing |
| OTU | Operational Taxonomic Unit |
| PCBs | Polychlorinated Biphenyls |
| PCR | Polymerase Chain Reaction |
| PM | Particulate Matter |
| PTFE | Polytetrafluoroethylene |
| qPCR | Quantitative Polymerase Chain Reaction |
| US EPA | United States Environmental Protection Agency |
References
- Li, J.; Pei, Y.; Zhao, S.; Xiao, R.; Sang, X.; Zhang, C. A review of remote sensing for environmental monitoring in China. Remote Sens. 2020, 12, 1130. [Google Scholar] [CrossRef]
- Estoque, R.C.; Murayama, Y. Monitoring surface urban heat island formation in a tropical mountain city using Landsat data (1987–2015). ISPRS-J. Photogramm. 2017, 133, 18–29. [Google Scholar] [CrossRef]
- Ullo, S.L.; Sinha, G.R. Advances in smart environment monitoring systems using IoT and sensors. Sensors 2020, 20, 3113. [Google Scholar] [CrossRef] [PubMed]
- Lindenmayer, D.B.; Likens, G.E. The science and application of ecological monitoring. Biol. Conserv. 2010, 143, 1317–1328. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef] [PubMed]
- Cole, D.N. Experimental trampling of vegetation. I. Relationship between trampling intensity and vegetation response. J. Appl. Ecol. 1995, 32, 203–214. [Google Scholar] [CrossRef]
- Rodriguez-Gil, J.L.; Sauto, J.S.S.; Gonzalez-Alonso, S.; Sanchez, P.S.; Valcarcel, Y.; Catala, M. Development of cost-effective strategies for environmental monitoring of irrigated areas in mediterranean regions: Traditional and new approaches in a changing world. Agric. Ecosyst. Environ. 2013, 181, 41–49. [Google Scholar] [CrossRef]
- Taberlet, P.; Coissac, E.; Hajibabaei, M.; Rieseberg, L.H. Environmental DNA. Mol. Ecol. 2012, 21, 1789–1793. [Google Scholar] [CrossRef]
- Thomsen, P.F.; Willerslev, E. Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 2015, 183, 4–18. [Google Scholar] [CrossRef]
- Ficetola, G.F.; Taberlet, P.; Coissac, E. How to limit false positives in environmental DNA and metabarcoding? Mol. Ecol. Resour. 2016, 16, 604–607. [Google Scholar] [CrossRef]
- Bohara, K.; Yadev, A.K.; Joshi, P. Detection of fish pathogens in freshwater aquaculture using eDNA methods. Diversity 2022, 14, 1015. [Google Scholar] [CrossRef]
- Klymus, K.E.; Richter, C.A.; Chapman, D.C.; Paukert, C. Quantification of eDNA shedding rates from invasive bighead carp Hypophtalmichthys nobilis and silver carp Hypophthalmichthys molitrix. Biol. Conserv. 2015, 183, 77–84. [Google Scholar] [CrossRef]
- Deiner, K.; Fronhofer, E.A.; Machler, E.; Walser, J.C.; Altermatt, F. Environmental DNA reveals that rivers are conveyer belts of biodiversity information. Nat. Commun. 2016, 31, 12544. [Google Scholar] [CrossRef] [PubMed]
- Sahu, A.; Kumar, N.; Singh, C.P.; Singh, M. Environmental DNA (eDNA): Powerful technique for biodiversity conservation. J. Nat. Conserv. 2023, 71, 126325. [Google Scholar] [CrossRef]
- Alfano, N.; Dayaram, A.; Axtner, J.; Tsangaras, K.; Kampmann, M.L.; Mohamed, A.; Wong, S.T.; Gilbert, M.T.; Wilting, A.; Greenwood, A.D. Non-invasive surveys of mammalian viruses using environmental DNA. Methods Ecol. Evol. 2021, 12, 1941–1952. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Melendy, S.A.; Olson, J.R. Identifying Key Environmental Drivers of Reach-Scale Salmonid eDNA Recovery With Random Forest. Environ. DNA 2024, 6, e70001. [Google Scholar] [CrossRef]
- Bauer, D.C.; Wilson, L.O.; Twine, N.A. Artificial Intelligence in Medicine: Applications, Limitations and Future Directions. In Artificial Intelligence in Medicine: Applications, Limitations and Future Directions; Springer: Singapore, 2022; pp. 101–120. [Google Scholar]
- Hinz, S.; Coston-Guarini, J.; Marnane, M.; Guarini, J.M. Evaluating eDNA for use within marine environmental impact assessment. J. Mark. Sci. Eng. 2022, 10, 375. [Google Scholar] [CrossRef]
- Xiong, F.; Shu, L.; Zeng, H.; Gan, X.; He, S.; Peng, Z. Methodology for fish biodiversity monitoring with environmental DNA metabarcoding: The primers, databases and bioinformatic pipelines. Water Biol. Secur. 2022, 1, 100007. [Google Scholar] [CrossRef]
- Mace, B.; Hocde, R.; Marques, V.; Guerin, P.E.; Valentini, A.; Arnal, V.; Pellissier, L.; Manel, S. Evaluating bioinformatics pipelines for population-level inference using environmental DNA. Environ. DNA 2022, 4, 674–686. [Google Scholar] [CrossRef]
- Ding, Y.; Zhang, F.; Zhang, J. Applicability and advantage of mitochondrial metagenomics and metabarcoding in spider biodiversity survey. Diversity 2023, 15, 711. [Google Scholar] [CrossRef]
- O’Donncha, F.; Hu, Y.; Palmes, P.; Burke, M.; Filgueira, R.; Grant, J. A spatio-temporal LSTM model to forecast across multiple temporal and spatial scales. Ecol. Inform. 2022, 69, 101687. [Google Scholar] [CrossRef]
- Park, K.S.; Choi, A.; Kim, H.J.; Park, I.; Eom, M.-S.; Yeo, S.-G.; Son, R.G.; Park, T.-I.; Lee, G.; Soh, H.T. Ultra-sensitive label-free SERS biosensor with high-throughput screened DNA aptamer for universal detection of SARS-CoV-2 variants from clinical samples. Biosens. Bioelectron. 2023, 228, 115202. [Google Scholar] [CrossRef]
- Toshiaki, S.J. Utilizing the state of environmental DNA (eDNA) to incorporate time-scale information into eDNA analysis. Proc. R. Soc. B-Biol. Sci. 2023, 290, 20230979. [Google Scholar]
- Hempel, C.A.; Buchner, D.; Mack, L.; Brasseur, M.V.; Tulpan, D.; Leese, F.; Steinke, D. Predicting environmental stressor levels with machine learning: A comparison between amplicon sequencing, metagenomics, and total RNA sequencing based on taxonomically assigned data. Front. Microbiol. 2023, 14, 1217750. [Google Scholar] [CrossRef]
- Wang, C.; Chen, L.; Li, X.; Gu, J.; Xiang, Y.; Fang, L.; Chen, L.; Li, Y. Development of an all-in-one real-time PCR assay for simultaneous detection of spotted fever group rickettsiae, severe fever with thrombocytopenia syndrome virus and hantaan virus prevalent in central China. PLoS Negl. Trop. Dis. 2024, 18, e0012024. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Liu, H.; Shi, R.; Cai, Y.; Ma, J.; Shao, L.; Rong, V.; Gkitsas, N.; Lei, H.; Highfill, S.L. Application of droplet digital PCR for the detection of vector copy number in clinical CAR/TCR T cell products. J. Transl. Med. 2020, 18, 191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qiao, S.; Zeng, Y.; Gao, D.; Han, N.; Zhou, J. CAE-CNN: Predicting transcription factor binding site with convolutional autoencoder and convolutional neural network. Expert Syst. Appl. 2021, 183, 115404. [Google Scholar] [CrossRef]
- Yan, Z.; Luo, Y.; Chen, X.; Yang, L.; Yao, M. Angling and trolling for eDNA: A novel and effective approach for passive eDNA capture in natural waters. Environ. Int. 2024, 194, 109175. [Google Scholar] [CrossRef]
- Doi, H.; Watanabe, T.; Nishizawa, N.; Saito, T.; Nagata, H.; Kameda, Y.; Maki, N.; Ikeda, K.; Fukuzawa, T. On-site environmental DNA detection of species using ultrarapid mobile PCR. Mol. Ecol. Resour. 2021, 21, 2364–2368. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, H.; Xu, C.; Yin, W.; Luo, L.; Liu, G.; Wang, Y. Smartphone-integrated RPA-CRISPR-Cas12a Detection System with Microneedle Sampling for Point-of-Care Diagnosis of Potato Late Blight in Early Stage. bioRxiv 2025, 2025-06. [Google Scholar] [CrossRef]
- Su, X.; Sutarlie, L.; Loh, X.J. Sensors and Analytical Technologies for Air Quality: Particulate Matters and Bioaerosols. Chem. Asian J. 2020, 15, 4241–4255. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.T.; Chen, H.W.; Chang, E.J.; Kristiani, E.; Nguyen, K.L.P.; Chang, J.S. Current advances and future challenges of AIoT applications in particulate matters (PM) monitoring and control. J. Hazard. Mater. 2021, 419, 126442. [Google Scholar] [CrossRef]
- Liu, X.; Jayaratne, R.; Thai, P.; Kuhn, T.; Zing, I.; Christensen, B.; Lamont, R.; Dunbabin, M.; Zhu, S.; Gao, J.; et al. Low-cost sensors as an alternative for long-term air quality monitoring. Environ. Res. 2020, 185, 109438. [Google Scholar] [CrossRef] [PubMed]
- Petruci, J.F.d.S.; Barreto, D.N.; Dias, M.A.; Felix, E.P.; Cardoso, A.A. Analytical methods applied for ozone gas detection: A review. Trends Anal. Chem. 2022, 149, 116552. [Google Scholar] [CrossRef]
- Yeo, M.J.; Kim, Y.P. Long-term trends of surface ozone in Korea. J. Clean. Prod. 2021, 294, 125352. [Google Scholar] [CrossRef]
- Thangamani, G.J.; Pasha, S.K.K. Titanium dioxide (TiO(2)) nanoparticles reinforced polyvinyl formal (PVF) nanocomposites as chemiresistive gas sensor for sulfur dioxide (SO(2)) monitoring. Chemosphere 2021, 275, 129960. [Google Scholar] [CrossRef]
- Idrees, Z.; Zheng, L. Low cost air pollution monitoring systems: A review of protocols and enabling technologies. J. Ind. Inf. Integr. 2020, 17, 100123. [Google Scholar] [CrossRef]
- Vîrghileanu, M.; Săvulescu, I.; Mihai, B.-A.; Nistor, C.; Dobre, R. Nitrogen Dioxide (NO2) Pollution Monitoring with Sentinel-5P Satellite Imagery over Europe during the Coronavirus Pandemic Outbreak. Remote Sens. 2020, 12, 3575. [Google Scholar] [CrossRef]
- Huang, J.; Wang, D.; Zhu, Y.; Yang, Z.; Yao, M.; Shi, X.; An, T.; Zhang, Q.; Huang, C.; Bi, X.; et al. An overview for monitoring and prediction of pathogenic microorganisms in the atmosphere. Fundam. Res. 2024, 4, 430–441. [Google Scholar] [CrossRef]
- Mahaffee, W.F.; Margairaz, F.; Ulmer, L.; Bailey, B.N.; Stoll, R. Catching Spores: Linking Epidemiology, Pathogen Biology, and Physics to Ground-Based Airborne Inoculum Monitoring. Plant Dis. 2023, 107, 13–33. [Google Scholar] [CrossRef]
- Suanno, C.; Aloisi, I.; Fernandez-Gonzalez, D.; Del Duca, S. Monitoring techniques for pollen allergy risk assessment. Environ. Res. 2021, 197, 111109. [Google Scholar] [CrossRef]
- Khatib, M.; Haick, H. Sensors for Volatile Organic Compounds. ACS Nano 2022, 16, 7080–7115. [Google Scholar] [CrossRef]
- Lynggaard, C.; Bertelsen, M.F.; Jensen, C.V.; Johnson, M.S.; Froslev, T.G.; Olsen, M.T.; Bohmann, K. Airborne environmental DNA for terrestrial vertebrate community monitoring. Curr. Biol. 2022, 32, 701–707. [Google Scholar] [CrossRef]
- Roger, F.; Ghanavi, H.R.; Danielsson, N.; Wahlberg, N.; Löndahl, J.; Pettersson, L.B.; Andersson, G.K.S.; Boke Olén, N.; Clough, Y. Airborne environmental DNA metabarcoding for the monitoring of terrestrial insects—A proof of concept from the field. Environ. DNA 2022, 4, 790–807. [Google Scholar] [CrossRef]
- Sullivan, A.R.; Karlsson, E.; Svensson, D.; Brindefalk, B.; Villegas, J.A.; Mikko, A.; Bellieny, D.; Siddique, A.B.; Johansson, A.-M.; Grahn, H. Airborne eDNA captures three decades of ecosystem biodiversity. bioRxiv 2023. [Google Scholar] [CrossRef]
- Goray, M.; Taylor, D.; Bibbo, E.; Fantinato, C.; Fonnelop, A.E.; Gill, P.; van Oorschot, R.A.H. Emerging use of air eDNA and its application to forensic investigations—A review. Electrophoresis 2024, 45, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Polling, M.; Buij, R.; Laros, I.; de Groot, G.A. Continuous daily sampling of airborne eDNA detects all vertebrate species identified by camera traps. Environ. DNA 2024, 6, edn3.591. [Google Scholar] [CrossRef]
- Yan, S.; Liu, Q.; Liang, B.; Zhang, M.; Chen, W.; Zhang, D.; Wang, C.; Xing, D. Airborne microbes: Sampling, detection, and inactivation. Crit. Rev. Biotechnol. 2025, 45, 556–590. [Google Scholar] [CrossRef]
- Shivaram, K.B.; Bhatt, P.; Verma, M.S.; Clase, K.; Simsek, H. Bacteriophage-based biosensors for detection of pathogenic microbes in wastewater. Sci. Total Environ. 2023, 901, 165859. [Google Scholar] [CrossRef]
- Papaioannou, C.; Geladakis, G.; Kommata, V.; Batargias, C.; Lagoumintzis, G. Insights in Pharmaceutical Pollution: The Prospective Role of eDNA Metabarcoding. Toxics 2023, 11, 903. [Google Scholar] [CrossRef]
- Fronczek, C.F.; Yoon, J.Y. Biosensors for Monitoring Airborne Pathogens. J. Lab. Autom. 2015, 20, 390–410. [Google Scholar] [CrossRef]
- Marselle, M.R.; Hartig, T.; Cox, D.T.; De Bell, S.; Knapp, S.; Lindley, S.; Triguero-Mas, M.; Böhning-Gaese, K.; Braubach, M.; Cook, P.A. Pathways linking biodiversity to human health: A conceptual framework. Environ. Int. 2021, 150, 106420. [Google Scholar] [CrossRef] [PubMed]
- Bass, D.; Christison, K.W.; Stentiford, G.D.; Cook, L.S.J.; Hartikainen, H. Environmental DNA/RNA for pathogen and parasite detection, surveillance, and ecology. Trends Parasitol. 2023, 39, 285–304. [Google Scholar] [CrossRef] [PubMed]
- Rishan, S.T.; Kline, R.J.; Rahman, M.S. Applications of environmental DNA (eDNA) to detect subterranean and aquatic invasive species: A critical review on the challenges and limitations of eDNA metabarcoding. Environ. Adv. 2023, 12, 100370. [Google Scholar] [CrossRef]
- Vasavi, S.; Sripathi, V.; Simma, C.M. Visualization of humpback whale tracking on edge device using space-borne remote sensing data for Indian Ocean. Egypt. J. Remote Sens. Space Sci. 2024, 27, 705–715. [Google Scholar] [CrossRef]
- Sajjad, B.; Hussain, S.; Rasool, K.; Hassan, M.; Almomani, F. Comprehensive insights into advances in ambient bioaerosols sampling, analysis and factors influencing bioaerosols composition. Environ. Pollut. 2023, 336, 122473. [Google Scholar] [CrossRef]
- Ruppert, K.M.; Kline, R.J.; Rahman, M.S. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Glob. Ecol. Conserv. 2019, 17, e00547. [Google Scholar] [CrossRef]
- Yang, J.; Li, C.; Lo, L.S.H.; Zhang, X.; Chen, Z.; Gao, J.; Clara, U.; Dai, Z.; Nakaoka, M.; Yang, H.; et al. Artificial Intelligence-Assisted Environmental DNA Metabarcoding and High-Resolution Underwater Optical Imaging for Noninvasive and Innovative Marine Environmental Monitoring. J. Mar. Sci. Eng. 2024, 12, 1729. [Google Scholar] [CrossRef]
- Valenzuela, E.F.; Menezes, H.C.; Cardeal, Z.L. Passive and grab sampling methods to assess pesticide residues in water. A review. Environ. Chem. Lett. 2020, 18, 1019–1048. [Google Scholar] [CrossRef]
- Pouresmaieli, M.; Ataei, M.; Forouzandeh, P.; Azizollahi, P.; Mahmoudifard, M. Recent progress on sustainable phytoremediation of heavy metals from soil. J. Environ. Chem. Eng. 2022, 10, 108482. [Google Scholar] [CrossRef]
- Rasool, S.; Rasool, T.; Gani, K.M. A review of interactions of pesticides within various interfaces of intrinsic and organic residue amended soil environment. Chem. Eng. J. Adv. 2022, 11, 100301. [Google Scholar] [CrossRef]
- Yahaya, S.M.; Mahmud, A.A.; Abdullahi, M.; Haruna, A. Recent advances in the chemistry of nitrogen, phosphorus and potassium as fertilizers in soil: A review. Pedosphere 2023, 33, 385–406. [Google Scholar] [CrossRef]
- Zhao, S.; Yuan, X.-T.; Wang, X.-H.; Ai, Y.-J.; Li, F.-P. Research Progress and Hotspots in Microbial Remediation for Polluted Soils. Sustainability 2024, 16, 7458. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.; Khalifa, H.O.; Yoon, H.J.; Ki, M.-R.; Pack, S.P. Microbial Immobilized Enzyme Biocatalysts for Multipollutant Mitigation: Harnessing Nature’s Toolkit for Environmental Sustainability. Int. J. Mol. Sci. 2024, 25, 8616. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.; Son, R.G.; Ki, M.-R.; Pack, S.P. Biosilica-coated carbonic anhydrase displayed on Escherichia coli: A novel design approach for efficient and stable biocatalyst for CO2 sequestration. Int. J. Biol. Macromol. 2024, 277, 134058. [Google Scholar] [CrossRef]
- Ariza, M.; Fouks, B.; Mauvisseau, Q.; Halvorsen, R.; Alsos, I.G.; de Boer, H.J. Plant biodiversity assessment through soil eDNA reflects temporal and local diversity. Methods Ecol. Evol. 2023, 14, 415–430. [Google Scholar] [CrossRef]
- Hiiesalu, I.; Oepik, M.; Metsis, M.; Lilje, L.; Davison, J.; Vasar, M.; Moora, M.; Zobel, M.; Wilson, S.D.; Paertel, M. Plant species richness belowground: Higher richness and new patterns revealed by next-generation sequencing. Mol. Ecol. 2012, 21, 2004–2016. [Google Scholar] [CrossRef]
- Wendt, J.; Hauser, S. An equivalent soil mass procedure for monitoring soil organic carbon in multiple soil layers. Eur. J. Soil Sci. 2013, 64, 58–65. [Google Scholar] [CrossRef]
- Fu, Y.; Xue, M.; Cai, R.; Kangasluoma, J.; Jiang, J. Theoretical and experimental analysis of the core sampling method: Reducing diffusional losses in aerosol sampling line. Aerosol. Sci. Technol. 2019, 53, 793–801. [Google Scholar] [CrossRef]
- Hou, D.; O’Connor, D.; Nathanail, P.; Tian, L.; Ma, Y. Integrated GIS and multivariate statistical analysis for regional scale assessment of heavy metal soil contamination: A critical review. Environ. Pollut. 2017, 231, 1188–1200. [Google Scholar] [CrossRef]
- Young, J.M.; Rawlence, N.J.; Weyrich, L.S.; Cooper, A. Limitations and recommendations for successful DNA extraction from forensic soil samples: A review. Sci. Justice 2014, 54, 238–244. [Google Scholar] [CrossRef]
- Roberts, J.; Cozzolino, D. Wet or dry? The effect of sample characteristics on the determination of soil properties by near infrared spectroscopy. Trends Anal. Chem. 2016, 83, 25–30. [Google Scholar] [CrossRef]
- Tuit, C.; Wait, A. A review of marine sediment sampling methods. Environ. Forensics 2020, 21, 291–309. [Google Scholar] [CrossRef]
- He, S.; Peng, Y.; Jin, Y.; Wan, B.; Liu, G. Review and analysis of key techniques in marine sediment sampling. Chin. J. Mech. Eng. 2020, 33, 66. [Google Scholar] [CrossRef]
- Corinaldesi, C.; Barucca, M.; Luna, G.M.; Dell’anno, A. Preservation, origin and genetic imprint of extracellular DNA in permanently anoxic deep-sea sediments. Mol. Ecol. 2011, 20, 642–654. [Google Scholar] [CrossRef]
- Kestel, J.H.; Field, D.L.; Bateman, P.W.; White, N.E.; Allentoft, M.E.; Hopkins, A.J.; Gibberd, M.; Nevill, P. Applications of environmental DNA (eDNA) in agricultural systems: Current uses, limitations and future prospects. Sci. Total Environ. 2022, 847, 157556. [Google Scholar] [CrossRef]
- Sorensen, J.P.; Maurice, L.; Edwards, F.K.; Lapworth, D.J.; Read, D.S.; Allen, D.; Butcher, A.S.; Newbold, L.K.; Townsend, B.R.; Williams, P.J. Using boreholes as windows into groundwater ecosystems. PLoS ONE 2013, 8, e70264. [Google Scholar] [CrossRef]
- Korbel, K.; Chariton, A.; Stephenson, S.; Greenfield, P.; Hose, G.C. Wells provide a distorted view of life in the aquifer: Implications for sampling, monitoring and assessment of groundwater ecosystems. Sci. Rep. 2017, 7, 40702. [Google Scholar] [CrossRef]
- Britt, S.L.; Parker, B.L.; Cherry, J.A. A downhole passive sampling system to avoid bias and error from groundwater sample handling. Environ. Sci. Technol. 2010, 44, 4917–4923. [Google Scholar] [CrossRef]
- Harter, T.; Watanabe, N.; Li, X.; Atwill, E.R.; Samuels, W. Microbial groundwater sampling protocol for fecal-rich environments. Groundwater 2014, 52, 126–136. [Google Scholar] [CrossRef]
- Gomo, M.; Vermeulen, D.; Lourens, P. Groundwater sampling: Flow-through bailer passive method versus conventional purge method. Nat. Resour. Res. 2018, 27, 51–65. [Google Scholar] [CrossRef]
- van der Heyde, M.; Alexander, J.; Nevill, P.; Austin, A.D.; Stevens, N.; Jones, M.; Guzik, M.T. Rapid detection of subterranean fauna from passive sampling of groundwater eDNA. Environ. DNA 2023, 5, 1706–1719. [Google Scholar] [CrossRef]
- Couton, M.; Hürlemann, S.; Studer, A.; Alther, R.; Altermatt, F. Groundwater environmental DNA metabarcoding reveals hidden diversity and reflects land-use and geology. Mol. Ecol. 2023, 32, 3497–3512. [Google Scholar] [CrossRef]
- Wu, P.; Feng, J.; Ju, M.; Wu, S.; Han, W.; Wang, M.; Liao, J.; Zhao, L.; Gao, Y.; Zheng, J. Water filter: A rapid water environmental DNA collector in the field. Front. Environ. Sci. 2024, 12, 1415338. [Google Scholar] [CrossRef]
- Davis, J.; Garcia, E.A.; Gibb, K.S.; Kennard, M.J.; Rose, A.; Stromsoe, N.; Wedd, D. The importance of groundwater for riverine fish faunas in a region of shale gas development in northern Australia. Front. Environ. Sci. 2023, 11, 1106862. [Google Scholar] [CrossRef]
- Hossain, M.S.; Iken, B.; Iyer, R. Whole genome analysis of 26 bacterial strains reveals aromatic and hydrocarbon degrading enzymes from diverse environmental soil samples. Sci. Rep. 2024, 14, 30685. [Google Scholar] [CrossRef] [PubMed]
- Ji, L.; Chang, X.; Wang, L.; Fu, X.; Lai, W.; Zheng, L.; Li, Q.; Xing, Y.; Yang, Z.; Guan, Y. The Mechanism Insight into Bacterial Degradation of Pentachlorobiphenyl. bioRxiv 2024. [Google Scholar] [CrossRef]
- Chunyan, X.; Qaria, M.A.; Qi, X.; Daochen, Z. The role of microorganisms in petroleum degradation: Current development and prospects. Sci. Total Environ. 2023, 865, 161112. [Google Scholar] [CrossRef]
- Guerrero Ramírez, J.R.; Ibarra Muñoz, L.A.; Balagurusamy, N.; Frías Ramírez, J.E.; Alfaro Hernández, L.; Carrillo Campos, J. Microbiology and biochemistry of pesticides biodegradation. Int. J. Mol. Sci. 2023, 24, 15969. [Google Scholar] [CrossRef]
- Lemmel, F.; Maunoury-Danger, F.; Leyval, C.; Cébron, A. Altered fungal communities in contaminated soils from French industrial brownfields. J. Hazard. Mater. 2021, 406, 124296. [Google Scholar] [CrossRef]
- Zhang, R.-D.; Gao, F.-Z.; Shi, Y.-J.; Zhao, J.-L.; Liu, Y.-S.; He, L.-Y.; Ying, G.-G. Metagenomic investigation of antibiotic resistance genes and resistant bacteria contamination in pharmaceutical plant sites in China. Environ. Pollut. 2024, 357, 124482. [Google Scholar] [CrossRef] [PubMed]
- Mejia, M.P.; Rojas, C.A.; Curd, E.; Renshaw, M.A.; Edalati, K.; Shih, B.; Vincent, N.; Lin, M.; Nguyen, P.H.; Wayne, R. Soil microbial community composition and tolerance to contaminants in an urban brownfield site. Microb. Ecol. 2023, 85, 998–1012. [Google Scholar] [CrossRef] [PubMed]
- Kavehei, A.; Hose, G.C.; Chariton, A.A.; Gore, D.B. Application of environmental DNA for assessment of contamination downstream of a legacy base metal mine. J. Hazard. Mater. 2021, 416, 125794. [Google Scholar] [CrossRef] [PubMed]
- Alves Senabio, J.; Correia da Silva, R.; Guariz Pinheiro, D.; Gomes de Vasconcelos, L.; Soares, M.A. The pesticides carbofuran and picloram alter the diversity and abundance of soil microbial communities. PLoS ONE 2024, 19, e0314492. [Google Scholar] [CrossRef]
- Xing, K.; Lu, W.; Huang, Q.; Wu, J.; Shang, H.; Wang, Q.; Guo, F.; Du, Q.; Yin, Z.; Zhang, Y. Soil eDNA biomonitoring reveals changes in multitrophic biodiversity and ecological health of agroecosystems. Environ. Res. 2024, 262, 119931. [Google Scholar] [CrossRef]
- Brunetti, M.; Magoga, G.; Cussigh, A.; Alali, S.; Pizzi, F.; Cremonesi, P.; Di Lelio, I.; Becchimanzi, A.; Comolli, R.; Gallina, P.M. Soil invertebrate biodiversity and functionality within the intensively farmed areas of the Po Valley. Appl. Soil Ecol. 2024, 197, 105326. [Google Scholar] [CrossRef]
- Ruppert, O.M.; Homola, J.J.; Kanefsky, J.; Swinehart, A.; Scribner, K.T.; Robinson, J.D. Optimization of Wetland Environmental DNA Metabarcoding Protocols for Great Lakes Region Herpetofauna. Environ. DNA 2025, 7, e70047. [Google Scholar] [CrossRef]
- Angeles, I.B.; Romero-Martínez, M.L.; Cavaliere, M.; Varrella, S.; Francescangeli, F.; Piredda, R.; Mazzocchi, M.G.; Montresor, M.; Schirone, A.; Delbono, I. Encapsulated in sediments: eDNA deciphers the ecosystem history of one of the most polluted European marine sites. Environ. Int. 2023, 172, 107738. [Google Scholar] [CrossRef]
- Kavehei, A.; Gore, D.B.; Chariton, A.A.; Hose, G.C. Impact assessment of ephemeral discharge of contamination downstream of two legacy base metal mines using environmental DNA. J. Hazard. Mater. 2021, 419, 126483. [Google Scholar] [CrossRef]
- Agerbo Rasmussen, J.; Nielsen, M.; Mak, S.S.; Döring, J.; Klincke, F.; Gopalakrishnan, S.; Dunn, R.R.; Kauer, R.; Gilbert, M.T.P. eDNA-based biomonitoring at an experimental German vineyard to characterize how management regimes shape ecosystem diversity. Environ. DNA 2021, 3, 70–82. [Google Scholar] [CrossRef]
- Frøslev, T.G.; Nielsen, I.B.; Santos, S.S.; Barnes, C.J.; Bruun, H.H.; Ejrnæs, R. The biodiversity effect of reduced tillage on soil microbiota. Ambio 2022, 51, 1022–1033. [Google Scholar] [CrossRef]
- Le Provost, G.; Thiele, J.; Westphal, C.; Penone, C.; Allan, E.; Neyret, M.; Van Der Plas, F.; Ayasse, M.; Bardgett, R.D.; Birkhofer, K. Contrasting responses of above-and belowground diversity to multiple components of land-use intensity. Nat. Commun. 2021, 12, 3918. [Google Scholar] [CrossRef]
- Shackleton, M.; Rees, G.N.; Watson, G.; Campbell, C.; Nielsen, D. Environmental DNA reveals landscape mosaic of wetland plant communities. Glob. Ecol. Conserv. 2019, 19, e00689. [Google Scholar] [CrossRef]
- Tetzlaff, S.J.; Katz, A.D.; Wolff, P.J.; Kleitch, M.E. Comparison of soil eDNA to camera traps for assessing mammal and bird community composition and site use. Ecol. Evol. 2024, 14, e70022. [Google Scholar] [CrossRef] [PubMed]
- Randall, L.A.; Goldberg, C.S.; Moehenschlager, A. Environmental DNA surveys can underestimate amphibian occupancy and overestimate detection probability: Implications for practice. J. Wildl. Manag. 2023, 87, e22463. [Google Scholar] [CrossRef]
- Xie, G.; Lan, J.; Liang, J.; Wang, Q.; Cao, X.; Wang, Y.; Ren, C.; Liu, H.; Zhang, J. Biodiversity and distribution of zoobenthos in the ecological water replenishment area of the Yellow River estuary coastal wetland revealed by eDNA metabarcoding. PLoS ONE 2024, 19, e0315346. [Google Scholar] [CrossRef] [PubMed]
- Pedreira-Segade, U.; Hao, J.; Razafitianamaharavo, A.; Pelletier, M.; Marry, V.; Le Crom, S.; Michot, L.J.; Daniel, I. How do nucleotides adsorb onto clays? Life 2018, 8, 59. [Google Scholar] [CrossRef]
- Abdelhamid, M.A.; Ki, M.-R.; Pack, S.P. Biominerals and Bioinspired materials in Biosensing: Recent advancements and applications. Int. J. Mol. Sci. 2024, 25, 4678. [Google Scholar] [CrossRef]
- Tedetti, M.; Sempéré, R. Penetration of ultraviolet radiation in the marine environment. A review. Photochem. Photobiol. 2006, 82, 389–397. [Google Scholar] [CrossRef]
- Guthrie, A.M.; Cooper, C.E.; Bateman, P.W.; van der Heyde, M.; Allentoft, M.E.; Nevill, P. A quantitative analysis of vertebrate environmental DNA degradation in soil in response to time, UV light, and temperature. Environ. DNA 2024, 6, e581. [Google Scholar] [CrossRef]
- Sirois, S.H.; Buckley, D.H. Factors governing extracellular DNA degradation dynamics in soil. Environ. Microbiol. Rep. 2019, 11, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Barnes, M.A.; Turner, C.R.; Jerde, C.L.; Renshaw, M.A.; Chadderton, W.L.; Lodge, D.M. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 2014, 48, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.C.; Maddison, B.C.; Middleitch, D.J.; Patmore, J.R.M.; Gough, K.C. REVIEW: The detection of aquatic animal species using environmental DNA—a review of eDNA as a survey tool in ecology. J. Appl. Ecol. 2014, 51, 1450–1459. [Google Scholar] [CrossRef]
- Farrell, J.A.; Whitmore, L.; Duffy, D.J. The promise and pitfalls of environmental DNA and RNA approaches for the monitoring of human and animal pathogens from aquatic sources. BioScience 2021, 71, 609–625. [Google Scholar] [CrossRef]
- Paruch, A.M.; Paruch, L. Current status of microbial source tracking applications in constructed wetlands serving as nature-based solutions for water management and wastewater treatment. Environ. Pollut. 2024, 351, 124076. [Google Scholar] [CrossRef]
- Sivalingam, P.; Sabatino, R.; Sbaffi, T.; Corno, G.; Fontaneto, D.; Borgomaneiro, G.; Rogora, M.; Crotti, E.; Mapelli, F.; Borin, S.; et al. Cesare, A.D. Anthropogenic pollution may enhance natural transformation in water, favouring the spread of antibiotic resistance genes. J. Hazard. Mater. 2024, 475, 134885. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.; He, N.; Du, M.; You, P. Exploring riverine aquatic animal diversity and establishing aquatic ecological monitoring approaches tailored to the Qinling region via eDNA technology. Intergr. Zool. 2024; online ahead of print. [Google Scholar] [CrossRef]
- Suren, A.M.; Burdon, F.J.; Wilkinson, S.P. eDNA is a useful environmental monitoring tool for assessing stream ecological health. Environ. DNA 2024, 6, e596. [Google Scholar] [CrossRef]
- Cornman, R.S.; Mckenna, J.E.; Fike, J.; Oyler-McCance, S.J.; Johnson, R. An experimental comparison of composite and grab sampling of stream water for metagenetic analysis of environmental DNA. PeerJ 2018, 6, e5871. [Google Scholar] [CrossRef]
- Schwesig, K.; Zizka, V.; Scherver, C.; Holzel, N. Comparing eDNA and transect methos for aquatic biodiversity assessment in lakes and ponds. Mol. Ecol. Resour. 2024, 25, e14060. [Google Scholar] [CrossRef]
- Govindarajan, A.F.; McCartin, L.; Adams, A.; Allan, E.; Belani, A.; Francoline, R.; Fujii, J.; Gomez-Ibanez, D.; Kukulya, A.; Marin, F.; et al. Improved biodiversity detection using a large-volume environmental DNA sampler with in situ filtration and implications for marine eDNA sampling strategies. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2022, 189, 103871. [Google Scholar] [CrossRef]
- Tadic, D.; Manasfi, R.; Bertrand, M.; Sauvetre, A.; Chiron, S. Use of passive and grab sampling and high-resolution mass spectrometry for non-targeted analysis of emerging contaminants and their semi-quantification in water. Molecules 2022, 27, 3167. [Google Scholar] [CrossRef]
- Kotlash, A.R.; Chessman, B.C. Effects of water sample preservation and storage on nitrogen and phosphorus determinations: Implications for the use of automated sampling equipment. Water. Res. 1998, 32, 3731–3737. [Google Scholar] [CrossRef]
- Coes, A.L.; Paretti, N.V.; Foreman, W.T.; Iverson, J.; Alvarez, D.A. Sampling trace organic compounds in water: A comparison of a continuous active sampler to continuous passive and discrete sampling methods. Sci. Total Environ. 2014, 473–474, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Kot, A.; Zabiegała, B.; Namieśnik, J. Passive sampling for long-term monitoring of organic pollutants in water. Trac-Trends Anal. Chem. 2000, 19, 446–459. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, H.; Jones, K.C. A novel passive water sampler for in situ sampling of antibiotics. J. Environ. Monit. 2012, 14, 1523–1530. [Google Scholar] [CrossRef]
- Chen, C.; Zhang, H.; Ying, G.G.; Jones, K.C. Evidence and recommendations to support the use of a novel passive water sampler to quantify antibiotics in wastewaters. Environ. Sci. Technol. 2013, 47, 13587–13593. [Google Scholar] [CrossRef]
- Schwarzbach, M.; Laiacker, M.; Pazmany, M.M.; Kondak, K. Remote water sampling using flying robots. In Proceedings of the 2014 International Conference on Unmanned Aircraft Systems (ICUAS), Orlando, FL, USA, 27–30 May 2014; pp. 72–76. [Google Scholar]
- Boger, N.; Ozer, M. Monitoring sewer systems to detect the eDNA of missing persons and persons of interest. Forensic Sci. Int. 2023, 349, 111744. [Google Scholar] [CrossRef]
- Choi, P.M.; Tscharke, B.J.; Donner, E.; O’Brien, J.W.; Grant, S.C.; Kaserzon, S.L.; Mackie, R.; O’Malley, E.; Crosible, N.D.; Thomas, K.V.; et al. Wastewater-based epidemiology biomarkers: Past, present, and future. Trac-Trends Anal. Chem. 2018, 105, 453–469. [Google Scholar] [CrossRef]
- Nguyen, A.Q.; Vu, H.P.; Nguyen, L.N.; Wang, Q.; Djordjevic, S.P.; Donner, E.; Yin, H.; Nghiem, L.D. Monitoring antibiotic resistance genes in wastewater treatment: Current strategies and future challenges. Sci. Total Environ. 2021, 783, 146964. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Honda, R. Potential sensitivity of wastewater monitoring for SARS-CoV-2: Comparison with norovirus cases. Environ. Sci. Technol. 2020, 54, 6451–6452. [Google Scholar] [CrossRef] [PubMed]
- Lajoie, A.S.; Holm, R.H.; Anderson, L.B.; Ness, H.D.; Smith, T. Nationwide public perceptions regarding the acceptance of using wastewater for community health monitoring in the United States. PLoS ONE 2022, 17, e0275075. [Google Scholar] [CrossRef] [PubMed]
- Osunmakinde, C.O.; Selvarajan, R.; Mamba, B.B.; Msagati, A.M. Profiling bacterial diversity and potential pathogens in wastewater treatment plats using high throughput sequencing analysis. Microorganisms 2019, 7, 506. [Google Scholar] [CrossRef]
- Oladi, M.; Leontidou, K.; Stoeck, T.; Shokri, M.R. Environmental DNA-based profiling of benthic bacterial and eukaryote communities along a crude oil spill gradient in a coral reef in the Persian Gulf. Mar. Pollut. Bull. 2022, 184, 114143. [Google Scholar] [CrossRef]
- Ki, M.-R.; Kim, S.H.; Park, T.I.; Pack, S.P. Self-entrapment of antimicrobial peptides in silica particles for stable and effective antimicrobial peptide delivery system. Int. J. Mol. Sci. 2023, 24, 16423. [Google Scholar] [CrossRef]
- Park, K.S.; Choi, A.; Park, T.-I.; Pack, S.P. Fluorometric and Colorimetric Method for SARS-CoV-2 Detection Using Designed Aptamer Display Particles. Biosensors 2024, 14, 113. [Google Scholar] [CrossRef]
- Min, K.H.; Kim, K.H.; Ki, M.-R.; Pack, S.P. Antimicrobial peptides and their biomedical applications: A review. Antibiotics 2024, 13, 794. [Google Scholar] [CrossRef]
- Rishan, S.T.; Kline, R.J.; Rahman, M.S. Exploitation of environmental DNA (eDNA) for ecotoxicological research: A critical review on eDNA metabarcoding in assessing marine pollution. Chemosphere 2024, 351, 141238. [Google Scholar] [CrossRef]
- Hernandez-Alomia, F.; Ballesteros, I.; Castillejo, P. Bioremediation potential of glyphosate-degrading microorganisms in eutrophicated Ecuadorian water bodies. Saudi J. Biol. Sci. 2022, 29, 1550–1558. [Google Scholar] [CrossRef]
- Song, T.; Zi, F.; Huang, Y.; Fang, L.; Zhang, Y.; Liu, Y.; Chang, J.; Li, J. Assessment of aquatic ecosystem health in the Irtysh river basin using eDNA metabarcoding. Water 2025, 17, 246. [Google Scholar] [CrossRef]
- Levy, N.; Simon-Blecher, N.; Ben-Ezra, S.; Yuval, M.; Doniger, T.; Leray, M.; Karako-Lampert, S.; Tarazi, E.; Levy, O. Evaluating biodiversity for coral reef reformation and monitoring on complex 3D structures using environmental DNA (eDNA) metabarcoding. Sci. Total Environ. 2023, 865, 159051. [Google Scholar] [CrossRef]
- Manaff, A.H.N.A.; Hil, K.S.; Luo, Z.; Liu, M.; Law, I.K.; Teng, S.T.; Akhir, M.F.; Gu, H.; Leaw, C.P.; Lim, P.T. Mapping harmful microalgal species by eDNA monitoring: A large-scale survey across the southwestern South China Sea. Harmful Algae 2023, 129, 102515. [Google Scholar] [CrossRef] [PubMed]
- Johnson, G.; Nour, A.A.; Nolan, T.; Huggett, J.; Bustin, S. Minimum information necessary for quantitative real-time PCR experiments. In Quantitative Real-Time PCR: Methods and Protocols; Humana Press: New York, NY, USA, 2014; pp. 5–17. [Google Scholar]
- Cunningham, S.W.; Tessler, M.; Johnson-Rosemond, J.; Whittaker, I.S.; Brugler, M.R. Environmental DNA Isolation, Validation, and Preservation Methods. In DNA Barcoding: Methods and Protocols; Humana Press: New York, NY, USA, 2024; pp. 171–180. [Google Scholar]
- Bruno, F.; Marinella, M.; Santamaria, M. e-DNA meta-barcoding: From NGS raw data to taxonomic profiling. In RNA Bioinformatics; Humana Press: New York, NY, USA, 2014; pp. 257–278. [Google Scholar]
- Christensen, H.; Olsen, J.E. Full Shotgun DNA Metagenomics. In Introduction to Bioinformatics in Microbiology; Springer: Cham, Switzerland, 2023; pp. 183–200. [Google Scholar]
- Wood, S.A.; Pochon, X.; Laroche, O.; von Ammon, U.; Adamson, J.; Zaiko, A. A comparison of droplet digital polymerase chain reaction (PCR), quantitative PCR and metabarcoding for species-specific detection in environmental DNA. Mol. Ecol. Resour. 2019, 19, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Sruoga, V.; Stunzenas, V.; Paulaviciute, B. COI Gene as a Molecular Marker of Elachista Species (Lepidoptera: Elachistidae: Elachistinae) from Different Lithuanian Populations; Latvian Academy of Sciences: Rīga, Latvia, 2009; p. 21. [Google Scholar]
- Panicker, V.P.; Haridas, P.C.; Narayanan, A.; Mohammed, S.; Babu, B.C. Mitochondrial 12S rRNA gene sequence analysis, a tool for species identification. J. Wildl. Biodivers. 2019, 3, 29–35. [Google Scholar]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Wang, P.; Yan, Z.; Yang, S.; Wang, S.; Zheng, X.; Fan, J.; Zhang, T. Environmental DNA: An emerging tool in ecological assessment. Bull. Environ. Contam. Toxicol. 2019, 103, 651–656. [Google Scholar] [CrossRef]
- Deiner, K.; Bik, H.M.; Mächler, E.; Seymour, M.; Lacoursière-Roussel, A.; Altermatt, F.; Creer, S.; Bista, I.; Lodge, D.M.; De Vere, N. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Mol. Ecol. 2017, 26, 5872–5895. [Google Scholar] [CrossRef]
- Olawade, D.B.; Wada, O.Z.; Ige, A.O.; Egbewole, B.I.; Olojo, A.; Oladapo, B.I. Artificial intelligence in environmental monitoring: Advancements, challenges, and future directions. Hyg. Environ. Health Adv. 2024, 12, 100114. [Google Scholar] [CrossRef]
- Ahuja, A.; Al-Zogbi, L.; Krieger, A. Application of noise-reduction techniques to machine learning algorithms for breast cancer tumor identification. Comput. Biol. Med. 2021, 135, 104576. [Google Scholar] [CrossRef]
- Eraslan, G.; Avsec, Ž.; Gagneur, J.; Theis, F.J. Deep learning: New computational modelling techniques for genomics. Nat. Rev. Genet. 2019, 20, 389–403. [Google Scholar] [CrossRef]
- Frontalini, F.; Greco, M.; Semprucci, F.; Cermakova, K.; Merzi, T.; Pawlowski, J. Developing and testing a new Ecological Quality Status index based on marine nematode metabarcoding: A proof of concept. Chemosphere 2025, 370, 143992. [Google Scholar] [CrossRef]
- Whitmore, L.; McCauley, M.; Farrell, J.A.; Stammnitz, M.R.; Koda, S.A.; Mashkour, N.; Summers, V.; Osborne, T.; Whilde, J.; Duffy, D.J. Inadvertent human genomic bycatch and intentional capture raise beneficial applications and ethical concerns with environmental DNA. Nat. Ecol. Evol. 2023, 7, 873–888. [Google Scholar] [CrossRef]

| Category | Methods | Advantages | Disadvantages |
|---|---|---|---|
| Soil | Core sampling | -Maintains stratigraphic continuity, allowing analysis of vertical pollutant distribution -Suitable for studying long-term environmental changes -Can collect deep layers, making it useful for geological and sedimentological research | -Requires complex and expensive equipment -Time-consuming sample collection process -Possible structural deformation in soft layers |
| Grab sampling | -Fast and easy to perform -Cost-effective with simple equipment -Enables rapid sampling from multiple locations | -Limited to surface-level information -Cannot analyze subsurface contamination or historical changes -May does not represent deeper soil conditions accurately | |
| Sediment | Core | -Provides information on vertical variations in soil composition and contaminants -Allows historical analysis of soil conditions -Essential for studying subsurface pollution migration | -Requires specialized equipment and can be time-consuming -More expensive than surface sampling -May disturb deeper soil layers during extraction |
| Surface | -Quick and easy to perform -Requires minimal equipment and lower cost -Suitable for large-scale surface contamination assessment | -Limited to surface-level information -Cannot analyze subsurface contamination or historical changes -May does not represent deeper soil conditions accurately | |
| Groundwater | Filtration | -Enhances eDNA analysis by concentrating DNA and microorganisms -Remove suspended solids, improving water quality for chemical analysis -Immediate field filtration minimizes sample degradation -Allows selection of various filter materials and pore sizes for optimized results | -Clog easily, especially with high particulate loads -DNA recovery efficiency may vary depending on filter type -Requires field equipment and careful contamination control -Time-consuming when processing large water volumes |
| Stakeholder | Key Recommendations |
|---|---|
| Researchers |
|
| Regulators and Policymakers |
|
| Conservationists |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoon, H.J.; Seo, J.H.; Shin, S.H.; Abdelhamid, M.A.A.; Pack, S.P. Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review. Biosensors 2025, 15, 494. https://doi.org/10.3390/bios15080494
Yoon HJ, Seo JH, Shin SH, Abdelhamid MAA, Pack SP. Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review. Biosensors. 2025; 15(8):494. https://doi.org/10.3390/bios15080494
Chicago/Turabian StyleYoon, Hyo Jik, Joo Hyeong Seo, Seung Hoon Shin, Mohamed A. A. Abdelhamid, and Seung Pil Pack. 2025. "Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review" Biosensors 15, no. 8: 494. https://doi.org/10.3390/bios15080494
APA StyleYoon, H. J., Seo, J. H., Shin, S. H., Abdelhamid, M. A. A., & Pack, S. P. (2025). Bioinformation and Monitoring Technology for Environmental DNA Analysis: A Review. Biosensors, 15(8), 494. https://doi.org/10.3390/bios15080494






