Enhancing ELISA Sensitivity: From Surface Engineering to Synthetic Biology
Abstract
1. Introduction
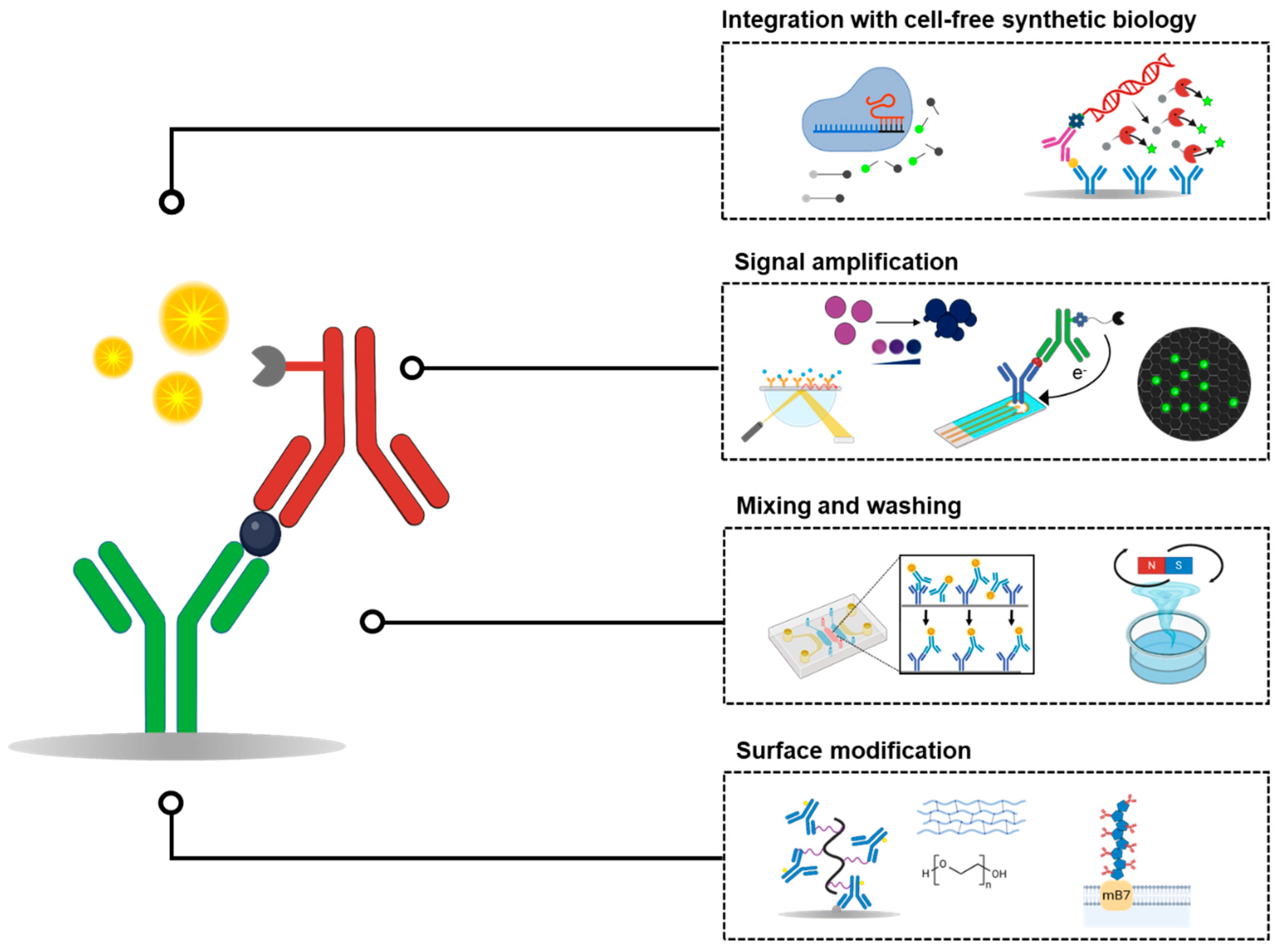
2. Surface Modification Strategies for Enhanced Antibody Coating
2.1. Use of Blocking Agents to Reduce Non-Specific Binding
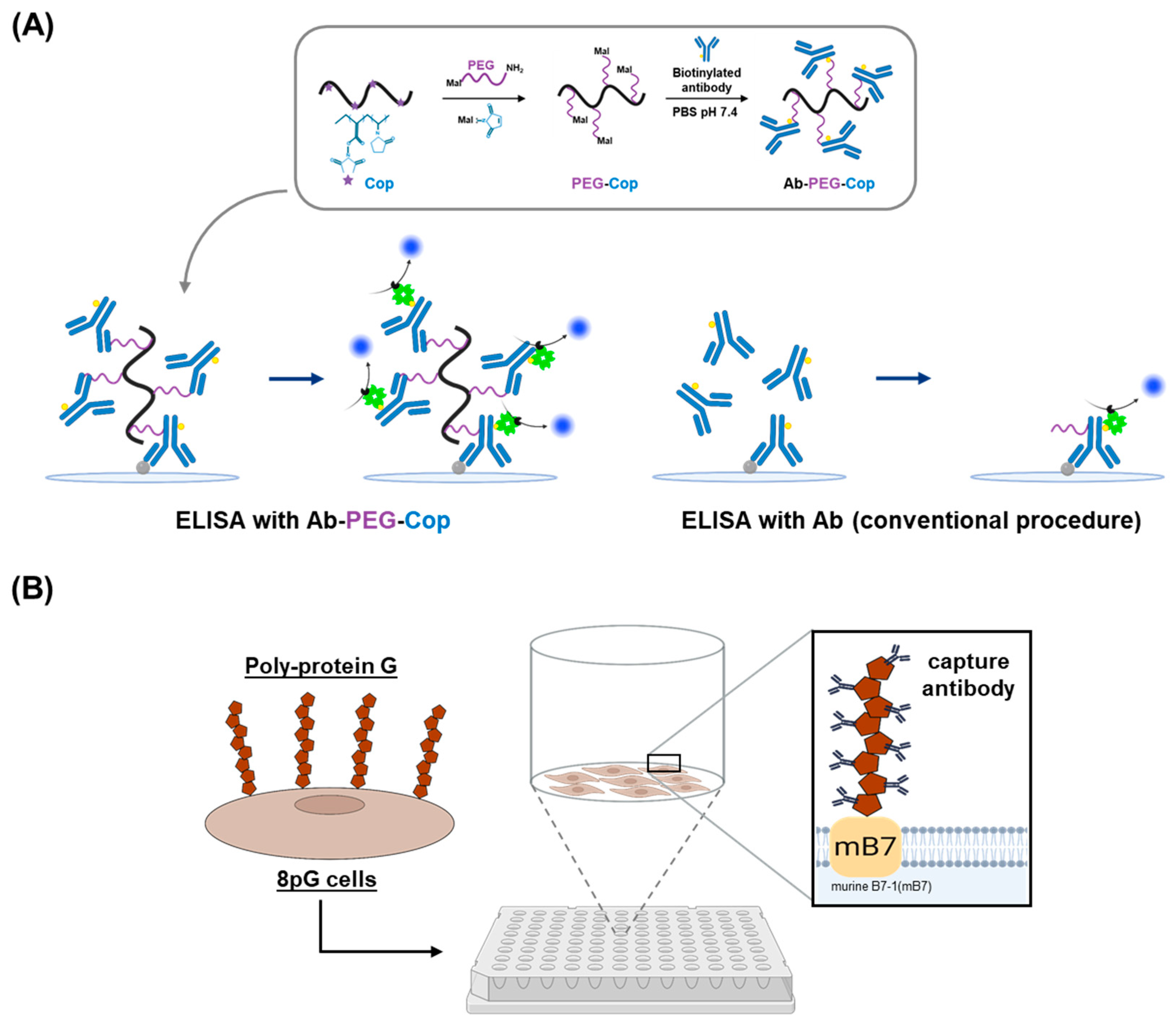
2.2. Nonfouling Surface Modifications Using Synthetic Polymers and Polysaccharides
2.3. Antibody Orientation Strategies for Enhanced Binding Efficiency
3. Mixing and Washing Efficiency in ELISA
3.1. Implementing Microfluidic Systems
3.2. Implementing Micro-Stirring Mechanisms
3.3. Applying Acoustic Waves
4. Enhancing ELISA Sensitivity Through Alternative Signal-Generation Mechanisms
4.1. Nanozyme ELISA
4.2. Plasmonic ELISA

4.3. Electrochemical ELISA
4.4. SERS-ELISA
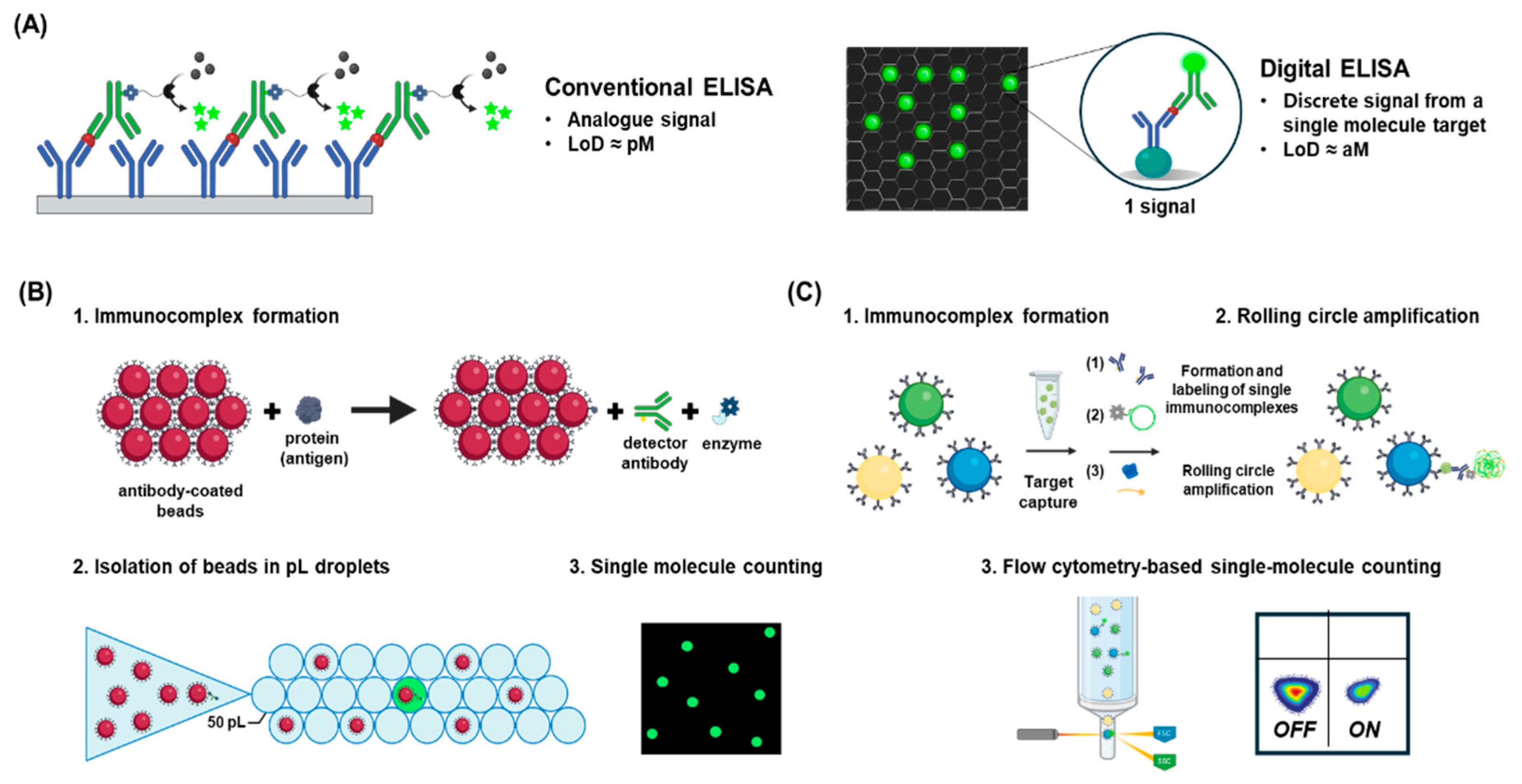
4.5. Digital ELISA
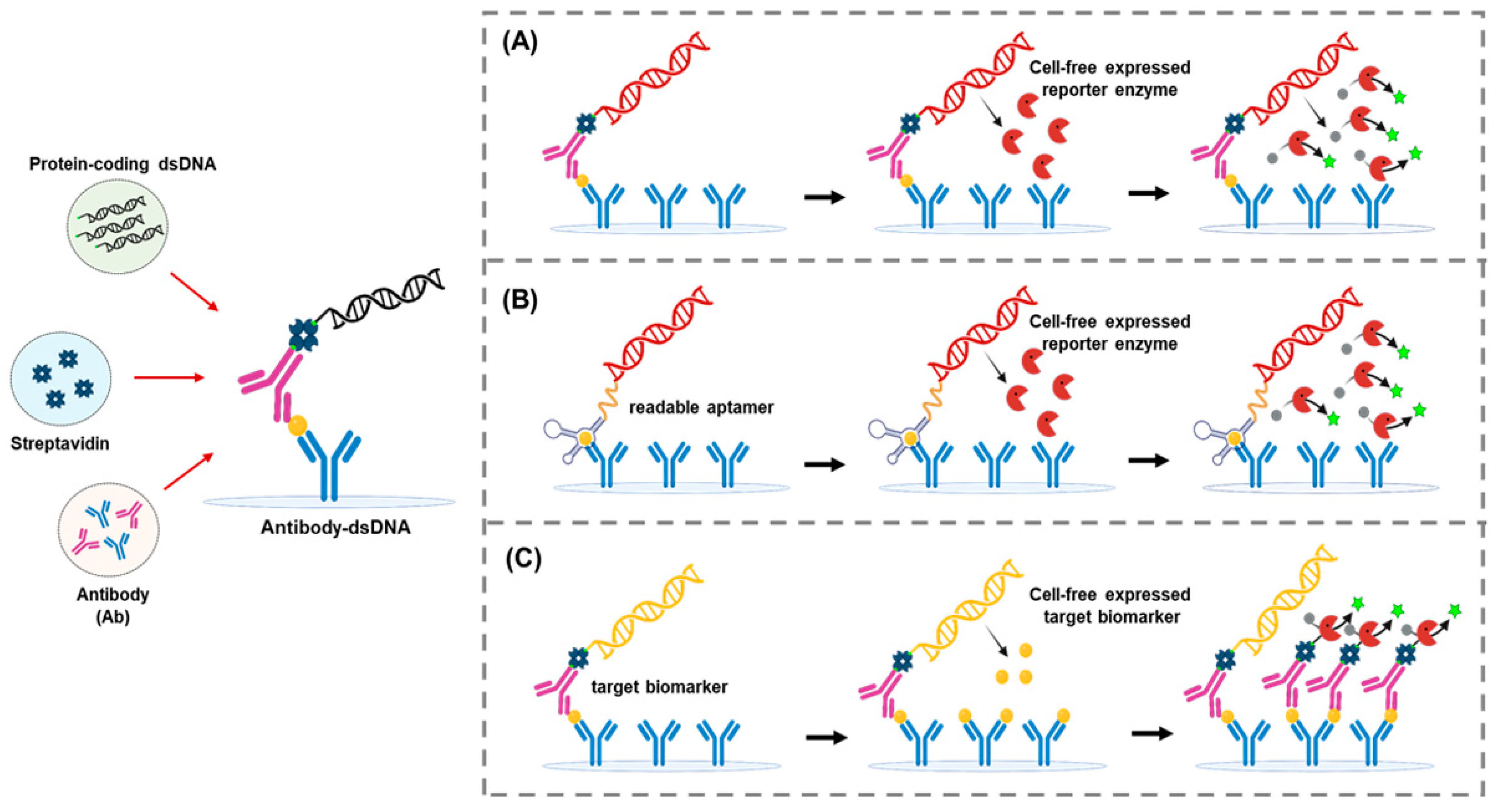
5. Implementing Cell-Free Synthetic Biology to Improve the Sensitivity and Flexibility of ELISA
5.1. Immuno-PCR
5.2. CRISPR-Linked Immunosorbent Assay (CLISA)
5.3. Expression Immunoassay
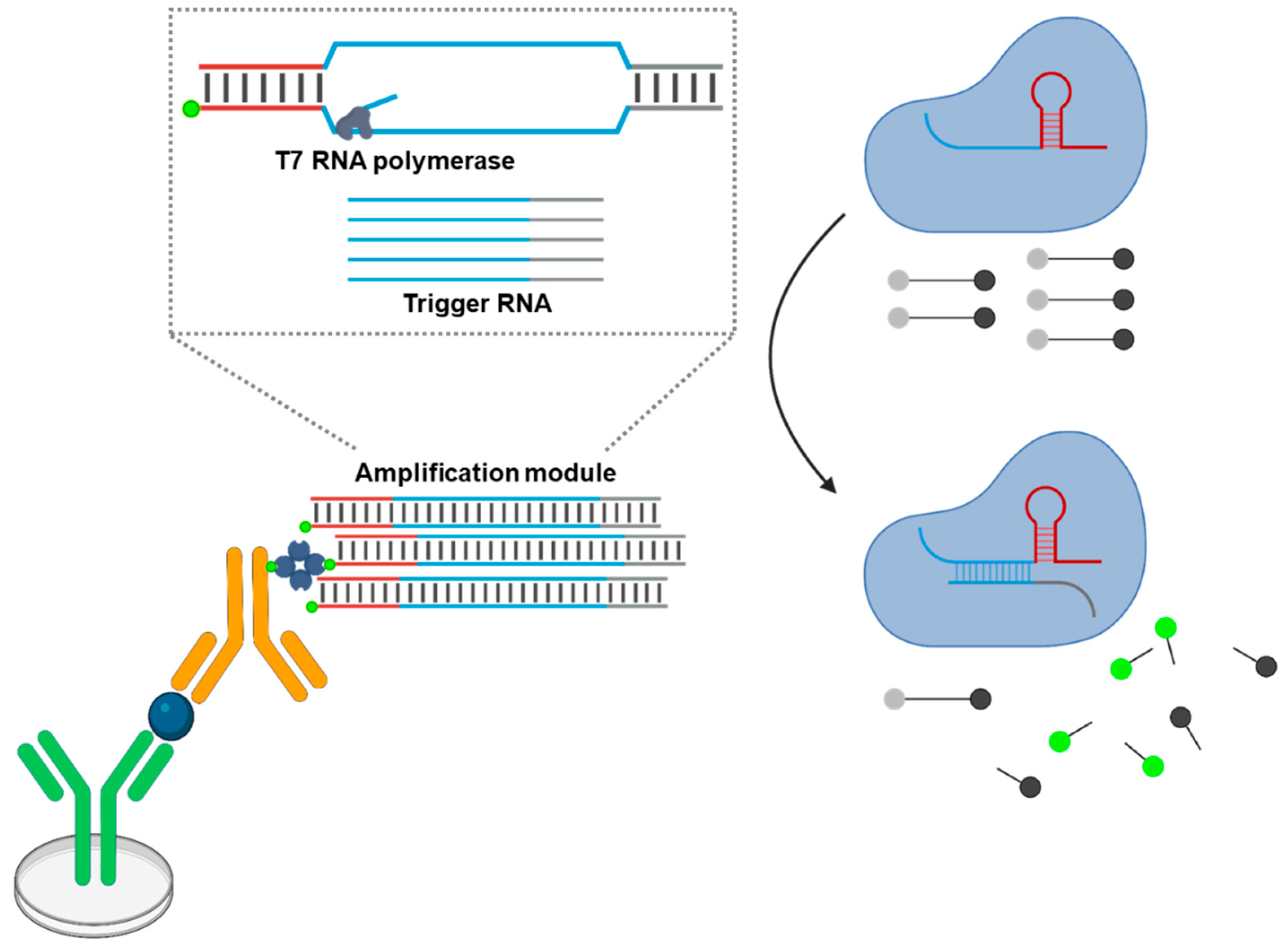
5.4. TLISA
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Liu, Q.; Jin, X.; Cheng, J.; Zhou, H.; Zhang, Y.; Dai, Y. Advances in the application of molecular diagnostic techniques for the detection of infectious disease pathogens (Review). Mol. Med. Rep. 2023, 27, 104. [Google Scholar] [CrossRef] [PubMed]
- Vashist, S.K.; Luppa, P.B.; Yeo, L.Y.; Ozcan, A.; Luong, J.H. Emerging technologies for next-generation point-of-care testing. Trends Biotechnol. 2015, 33, 692–705. [Google Scholar] [CrossRef]
- Habibzadeh, P.; Mofatteh, M.; Silawi, M.; Ghavami, S.; Faghihi, M.A. Molecular diagnostic assays for COVID-19: An overview. Crit. Rev. Clin. Lab. Sci. 2021, 58, 385–398. [Google Scholar] [CrossRef]
- Shao, J.; Wei, L.; Wang, H.; Sun, Y.; Zhang, L.-F.; Li, J.; Dong, J.-Q. Relationship between hepatitis B virus DNA levels and liver histology in patients with chronic hepatitis B. World J. Gastroenterol. 2007, 13, 2104. [Google Scholar] [CrossRef]
- Abdulrahman, A.; Mallah, S.I.; Alqahtani, M. COVID-19 viral load not associated with disease severity: Findings from a retrospective cohort study. BMC Infect. Dis. 2021, 21, 688. [Google Scholar] [CrossRef] [PubMed]
- Egger, M.E.; Alexander, E.; Van Meter, T.; Kong, M.; Maung, A.A.; Valdes, R., Jr.; Hall, M.B.; Linder, M.W. Corresponding ctDNA and tumor burden dynamics in metastatic melanoma patients on systemic treatment. Transl. Oncol. 2024, 42, 101883. [Google Scholar] [CrossRef]
- Panet, F.; Papakonstantinou, A.; Borrell, M.; Vivancos, J.; Vivancos, A.; Oliveira, M. Use of ctDNA in early breast cancer: Analytical validity and clinical potential. NPJ Breast Cancer 2024, 10, 50. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.; Singh, B.; Theofilatos, K.; Sorensen, N.A.; Lehmacher, J.; Hartikainen, T.; Haller, P.M.; Westermann, D.; Zeller, T.; Blankenberg, S.; et al. Serial measurements of protein and microRNA biomarkers to specify myocardial infarction subtypes. J. Mol. Cell. Cardiol. Plus 2022, 1, 100014. [Google Scholar] [CrossRef]
- Tasaki, S.; Xu, J.; Avey, D.R.; Johnson, L.; Petyuk, V.A.; Dawe, R.J.; Bennett, D.A.; Wang, Y.; Gaiteri, C. Inferring protein expression changes from mRNA in Alzheimer’s dementia using deep neural networks. Nat. Commun. 2022, 13, 655. [Google Scholar] [CrossRef]
- Gómez-Bañuelos, E.; Goldman, D.W.; Andrade, V.; Darrah, E.; Petri, M.; Andrade, F. Uncoupling interferons and the interferon signature explains clinical and transcriptional subsets in SLE. Cell Rep. Med. 2024, 5, 101569. [Google Scholar] [CrossRef]
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; de Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.D.; Montagut, C. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- Felekkis, K.; Papaneophytou, C. Challenges in using circulating micro-RNAs as biomarkers for cardiovascular diseases. Int. J. Mol. Sci. 2020, 21, 561. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Consortium, C. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Budd, J.; Miller, B.S.; Weckman, N.E.; Cherkaoui, D.; Huang, D.; Decruz, A.T.; Fongwen, N.; Han, G.-R.; Broto, M.; Estcourt, C.S.; et al. Lateral flow test engineering and lessons learned from COVID-19. Nat. Rev. Bioeng. 2023, 1, 13–31. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Álvarez-Diduk, R.; Parolo, C.; Piper, A.; Merkoçi, A. Toward Next Generation Lateral Flow Assays: Integration of Nanomaterials. Chem. Rev. 2022, 122, 14881–14910. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme immunoassay (EIA)/enzyme-linked immunosorbent assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Liu, C.; Li, Z.; Xue, Z.; Mao, P.; Hu, J.; Xu, F.; Yao, C.; You, M. Emerging ELISA derived technologies for in vitro diagnostics. TrAC Trends Anal. Chem. 2022, 152, 116605. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, J.; Zhou, B.; Ian, H.; Shao, H. Advanced sensitivity amplification strategies for voltammetric immunosensors of tumor marker: State of the art. Biosens. Bioelectron. 2021, 178, 113021. [Google Scholar] [CrossRef]
- Lee, S.J.; Kim, D.-M. Cell-free synthetic biology: Navigating the new frontiers of biomanufacturing and biological engineering. Curr. Opin. Syst. Biol. 2024, 37, 100488. [Google Scholar] [CrossRef]
- Waritani, T.; Chang, J.; McKinney, B.; Terato, K. An ELISA protocol to improve the accuracy and reliability of serological antibody assays. MethodsX 2017, 4, 153–165. [Google Scholar] [CrossRef]
- Vashist, S.K.; Marion Schneider, E.; Lam, E.; Hrapovic, S.; Luong, J.H. One-step antibody immobilization-based rapid and highly-sensitive sandwich ELISA procedure for potential in vitro diagnostics. Sci. Rep. 2014, 4, 4407. [Google Scholar] [CrossRef] [PubMed]
- Alugubelly, N.; Stokes, J.V.; Cross, C.E.; Ross, A.-M.L.; Crawford, A.E.; Fiihr, G.F.; Varela-Stokes, A.S. Beyond the IFA: Revisiting the ELISA as a more sensitive, objective, and Quantitative Evaluation of Spotted Fever Group Rickettsia exposure. Pathogens 2021, 10, 88. [Google Scholar] [CrossRef]
- Jiang, X.; Wu, M.; Albo, J.; Rao, Q. Non-specific binding and cross-reaction of ELISA: A case study of porcine hemoglobin detection. Foods 2021, 10, 1708. [Google Scholar] [CrossRef]
- Trimaille, T.; Verrier, B.; Coiffier, C.; Gigmes, D. Multiple antibody conjugation to PEG grafted copolymer improves sensitivity of immunoassays. Eur. Polym. J. 2024, 216, 113263. [Google Scholar] [CrossRef]
- Lin, W.-W.; Hsieh, Y.-C.; Cheng, Y.-A.; Chuang, K.-H.; Huang, C.-C.; Chuang, C.-H.; Chen, I.-J.; Cheng, K.-W.; Lu, Y.-C.; Cheng, T.-C. Optimization of an anti-poly (ethylene glycol)(anti-PEG) cell-based capture system to quantify PEG and PEGylated molecules. Anal. Chem. 2016, 88, 12371–12379. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-L.; Tseng, Y.-T.; Lai, M.-C.; Chau, L.-K. Ultrasensitive and rapid detection of N-terminal pro-B-type natriuretic peptide (NT-proBNP) using fiber optic nanogold-linked immunosorbent assay. Biosensors 2022, 12, 746. [Google Scholar] [CrossRef]
- Hoang, T.X.; Phan, L.M.T.; Vo, T.A.T.; Cho, S. Advanced signal-amplification strategies for paper-based analytical devices: A comprehensive review. Biomedicines 2021, 9, 540. [Google Scholar] [CrossRef]
- García-Maceira, T.; García-Maceira, F.I.; González-Reyes, J.A.; Paz-Rojas, E. Highly enhanced ELISA sensitivity using acetylated chitosan surfaces. BMC Biotechnol. 2020, 20, 41. [Google Scholar] [CrossRef]
- Cruz, D.F.; Fontes, C.M.; Semeniak, D.; Huang, J.; Hucknall, A.; Chilkoti, A.; Mikkelsen, M.H. Ultrabright fluorescence readout of an inkjet-printed immunoassay using plasmonic nanogap cavities. Nano Lett. 2020, 20, 4330–4336. [Google Scholar] [CrossRef]
- Björck, L.; Kronvall, G. Purification and some properties of streptococcal protein G, a novel IgG-binding reagent. J. Immunol. 1984, 133, 969–974. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Chen, M.; Hsieh, Y.-C.; Su, Y.-C.; Wang, C.-H.; Cheng, C.-M.; Kao, A.-P.; Wang, K.-H.; Cheng, J.-J.; Chuang, K.-H. Development of a highly sensitive enzyme-linked immunosorbent assay (ELISA) through use of poly-protein G-expressing cell-based microplates. Sci. Rep. 2018, 8, 17868. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Chiang, Y.-H.; Chiang, H.-Y. A label-free electrochemical impedimetric immunosensor with biotinylated-antibody for SARS-CoV-2 nucleoprotein detection in saliva. Biosensors 2022, 12, 265. [Google Scholar] [CrossRef] [PubMed]
- Beitello, E.; Osei, K.; Kobulnicky, T.; Breausche, F.; Friesen, J.A.; Driskell, J.D. Oriented Surface Immobilization of Antibodies Using Enzyme-Mediated Site-Specific Biotinylation for Enhanced Antigen-Binding Capacity. Langmuir 2025, 41, 10576–10585. [Google Scholar] [CrossRef]
- Høyer-Hansen, G.; Hamers, M.J.; Pedersen, A.N.; Nielsen, H.J.; Brünner, N.; Danø, K.; Stephens, R.W. Loss of ELISA specificity due to biotinylation of monoclonal antibodies. J. Immunol. Methods 2000, 235, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.-Y.; Chan, Y.-H. The importance of antibody orientation for enhancing sensitivity and selectivity in lateral flow immunoassays. Sens. Diagn. 2024, 3, 1613–1634. [Google Scholar] [CrossRef]
- Welch, N.G.; Scoble, J.A.; Muir, B.W.; Pigram, P.J. Orientation and characterization of immobilized antibodies for improved immunoassays. Biointerphases 2017, 12, 02D301. [Google Scholar] [CrossRef]
- Jung, Y.; Lee, J.M.; Kim, J.-W.; Yoon, J.; Cho, H.; Chung, B.H. Photoactivable antibody binding protein: Site-selective and covalent coupling of antibody. Anal. Chem. 2009, 81, 936–942. [Google Scholar] [CrossRef]
- Ha, Y.; Kim, I. Recent Developments in Innovative Magnetic Nanoparticles-Based Immunoassays: From Improvement of Conventional Immunoassays to Diagnosis of COVID-19. BioChip J. 2022, 16, 351–365. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, J.; Xu, F.; Wen, X.; Li, P.; Zhang, X.; Qiao, S.; Ge, S.; Xia, N.; Qian, S.; et al. A paper-based microfluidic Dot-ELISA system with smartphone for the detection of influenza A. Microfluid. Nanofluid. 2017, 21, 43. [Google Scholar] [CrossRef]
- Martinez, A.W.; Phillips, S.T.; Whitesides, G.M.; Carrilho, E. Diagnostics for the developing world: Microfluidic paper-based analytical devices. Anal. Chem. 2010, 82, 3–10. [Google Scholar] [CrossRef]
- Whitesides, G.M. The origins and the future of microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Nan, L.; Zhang, H.; Weitz, D.A.; Shum, H.C. Development and future of droplet microfluidics. Lab Chip 2024, 24, 1135–1153. [Google Scholar] [CrossRef] [PubMed]
- Safavieh, R.; Juncker, D. Capillarics: Pre-programmed, self-powered microfluidic circuits built from capillary elements. Lab Chip 2013, 13, 4180–4189. [Google Scholar] [CrossRef]
- Olanrewaju, A.; Beaugrand, M.; Yafia, M.; Juncker, D. Capillary microfluidics in microchannels: From microfluidic networks to capillaric circuits. Lab Chip 2018, 18, 2323–2347. [Google Scholar] [CrossRef] [PubMed]
- Kai, J.; Moore, V.; Puntambekar, A.; Sehy, D.; Lakkis, M.; Lee, S.; Han, J.; Ahn, C. Optimiser™ Microplate enables simultaneous multi-analyte quantitation using ultra-low sample volumes in a 2-hour assay: Validation of a Novel 10-Analyte Th17 Cell Panel (124.7). J. Immunol. 2012, 188, 124.7. [Google Scholar] [CrossRef]
- Uddin, M.J.; Bhuiyan, N.H.; Shim, J.S. Fully integrated rapid microfluidic device translated from conventional 96-well ELISA kit. Sci. Rep. 2021, 11, 1986. [Google Scholar] [CrossRef]
- Yafia, M.; Ymbern, O.; Olanrewaju, A.O.; Parandakh, A.; Sohrabi Kashani, A.; Renault, J.; Jin, Z.; Kim, G.; Ng, A.; Juncker, D. Microfluidic chain reaction of structurally programmed capillary flow events. Nature 2022, 605, 464–469. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, X.; Chen, C.; Chen, Y.; Li, Y.; Ye, H.; Wang, B.; Chen, H.; Guo, J.; Ma, X. Magnetic nanorobots as maneuverable immunoassay probes for automated and efficient enzyme linked immunosorbent assay. ACS Nano 2022, 16, 180–191. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, S.; Floer, C.; Kantubuktha, S.A.R.; Velasco, M.J.G.R.; Friend, J. Surface acoustic wave-driven enhancement of enzyme-linked immunosorbent assays: ELISAW. Anal. Chem. 2024, 96, 9676–9683. [Google Scholar] [CrossRef]
- Rashidian, M.; Dozier, J.K.; Distefano, M.D. Enzymatic labeling of proteins: Techniques and approaches. Bioconjugate Chem. 2013, 24, 1277–1294. [Google Scholar] [CrossRef]
- Veitch, N.C. Horseradish peroxidase: A modern view of a classic enzyme. Phytochemistry 2004, 65, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ren, J.; Qu, X. Catalytically active nanomaterials: A promising candidate for artificial enzymes. Acc. Chem. Res. 2014, 47, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Geng, H.; Vilms Pedersen, S.; Ma, Y.; Haghighi, T.; Dai, H.; Howes, P.D.; Stevens, M.M. Noble metal nanoparticle biosensors: From fundamental studies toward point-of-care diagnostics. Acc. Chem. Res. 2022, 55, 593–604. [Google Scholar] [CrossRef]
- Mohamad, A.; Teo, H.; Keasberry, N.A.; Ahmed, M.U. Recent developments in colorimetric immunoassays using nanozymes and plasmonic nanoparticles. Crit. Rev. Biotechnol. 2019, 39, 50–66. [Google Scholar] [CrossRef]
- Hua, Z.; Yu, T.; Liu, D.; Xianyu, Y. Recent advances in gold nanoparticles-based biosensors for food safety detection. Biosens. Bioelectron. 2021, 179, 113076. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wu, Z.; Liang, C.; Lu, J.; Li, J.; Zhang, L.; Li, T.; Zhao, W.; Fu, Y.; Hou, S. Development of a smartphone-based nanozyme-linked immunosorbent assay for quantitative detection of SARS-CoV-2 nucleocapsid phosphoprotein in blood. Front. Microbiol. 2021, 12, 692831. [Google Scholar] [CrossRef]
- Draz, M.S.; Vasan, A.; Muthupandian, A.; Kanakasabapathy, M.K.; Thirumalaraju, P.; Sreeram, A.; Krishnakumar, S.; Yogesh, V.; Lin, W.; Yu, X.G. Virus detection using nanoparticles and deep neural network–enabled smartphone system. Sci. Adv. 2020, 6, eabd5354. [Google Scholar] [CrossRef]
- Howes, P.D.; Chandrawati, R.; Stevens, M.M. Colloidal nanoparticles as advanced biological sensors. Science 2014, 346, 1247390. [Google Scholar] [CrossRef]
- Xuan, Z.; Li, M.; Rong, P.; Wang, W.; Li, Y.; Liu, D. Plasmonic ELISA based on the controlled growth of silver nanoparticles. Nanoscale 2016, 8, 17271–17277. [Google Scholar] [CrossRef]
- Singh, M.M.; Satija, J. Enzyme-assisted metal nanoparticles etching based plasmonic ELISA: Progress and insights. Anal. Biochem. 2022, 654, 114820. [Google Scholar] [CrossRef]
- De La Rica, R.; Stevens, M.M. Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. Nat. Nanotechnol. 2012, 7, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xu, S.; Ma, X.; Qiu, B.; Lin, Z.; Chen, G. Dual-color plasmonic enzyme-linked immunosorbent assay based on enzyme-mediated etching of Au nanoparticles. Sci. Rep. 2016, 6, 32755. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; He, S.; Qiu, B.; Luo, F.; Guo, L.; Lin, Z. Noble metal nanoparticle-based multicolor immunoassays: An approach toward visual quantification of the analytes with the naked eye. ACS Sens. 2019, 4, 782–791. [Google Scholar] [CrossRef]
- Yao, C.; Yu, S.; Li, X.; Wu, Z.; Liang, J.; Fu, Q.; Xiao, W.; Jiang, T.; Tang, Y. A plasmonic ELISA for the naked-eye detection of chromium ions in water samples. Anal. Bioanal. Chem. 2017, 409, 1093–1100. [Google Scholar] [CrossRef]
- Liu, D.; Yang, J.; Wang, H.-F.; Wang, Z.; Huang, X.; Wang, Z.; Niu, G.; Hight Walker, A.; Chen, X. Glucose oxidase-catalyzed growth of gold nanoparticles enables quantitative detection of attomolar cancer biomarkers. Anal. Chem. 2014, 86, 5800–5806. [Google Scholar] [CrossRef]
- Yang, Q.; Cai, R.; Xiao, W.; Wu, Z.; Liu, X.; Xu, Y.; Xu, M.; Zhong, H.; Sun, G.; Liu, Q. Plasmonic ELISA for sensitive detection of disease biomarkers with a smart phone-based reader. Nanoscale Res. Lett. 2018, 13, 397. [Google Scholar] [CrossRef]
- Arya, S.K.; Estrela, P. Electrochemical ELISA-based platform for bladder cancer protein biomarker detection in urine. Biosens. Bioelectron. 2018, 117, 620–627. [Google Scholar] [CrossRef] [PubMed]
- Arya, S.K.; Estrela, P. Recent advances in enhancement strategies for electrochemical ELISA-based immunoassays for cancer biomarker detection. Sensors 2018, 18, 2010. [Google Scholar] [CrossRef]
- Khanmohammadi, A.; Aghaie, A.; Vahedi, E.; Qazvini, A.; Ghanei, M.; Afkhami, A.; Hajian, A.; Bagheri, H. Electrochemical biosensors for the detection of lung cancer biomarkers: A review. Talanta 2020, 206, 120251. [Google Scholar] [CrossRef]
- Manasa, G.; Mascarenhas, R.J.; Malode, S.J.; Shetti, N.P. Graphene-based electrochemical immunosensors for early detection of oncomarker carcinoembryonic antigen. Biosens. Bioelectron. 2022, 11, 100189. [Google Scholar] [CrossRef]
- Peng, H.; Huang, Z.; Wu, W.; Liu, M.; Huang, K.; Yang, Y.; Deng, H.; Xia, X.; Chen, W. Versatile high-performance electrochemiluminescence ELISA platform based on a gold nanocluster probe. ACS Appl. Mater. Interfaces 2019, 11, 24812–24819. [Google Scholar] [CrossRef] [PubMed]
- Qin, D.; Meng, S.; Wu, Y.; Mo, G.; Jiang, X.; Deng, B. Design of a dual-wavelength ratiometric electrochemiluminescence immunosensor for sensitive detection of amyloid-β protein in human serum. ACS Sustain. Chem. Eng. 2021, 9, 7541–7549. [Google Scholar] [CrossRef]
- Zeng, R.; Li, Y.; Li, Y.; Wan, Q.; Huang, Z.; Qiu, Z.; Tang, D. Smartphone-based photoelectrochemical immunoassay with Co9S8@ ZnIn2S4 for point-of-care diagnosis of breast cancer biomarker. Research 2022, 2022, 9831521. [Google Scholar] [CrossRef]
- Wu, J.; Liu, H.; Chen, W.; Ma, B.; Ju, H. Device integration of electrochemical biosensors. Nat. Rev. Bioeng. 2023, 1, 346–360. [Google Scholar] [CrossRef] [PubMed]
- Chanarsa, S.; Jakmunee, J.; Ounnunkad, K. A bifunctional nanosilver-reduced graphene oxide nanocomposite for label-free electrochemical immunosensing. Front. Chem. 2021, 9, 631571. [Google Scholar] [CrossRef]
- Candia, M.L.; Piccinini, E.; Azzaroni, O.; Marmisollé, W.A. Digitalization of Enzyme-Linked Immunosorbent Assay with Graphene Field-Effect Transistors (G-ELISA) for Portable Ferritin Determination. Biosensors 2024, 14, 394. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wu, L.; Zhu, D.; Cui, Y. SERS-activated platforms for immunoassay: Probes, encoding methods, and applications. Chem. Rev. 2017, 117, 7910–7963. [Google Scholar] [CrossRef]
- Lin, L.L.; Alvarez-Puebla, R.; Liz-Marzán, L.M.; Trau, M.; Wang, J.; Fabris, L.; Wang, X.; Liu, G.; Xu, S.; Han, X.X. Surface-Enhanced Raman Spectroscopy for Biomedical Applications: Recent Advances and Future Challenges. ACS Appl. Mater. Interfaces 2025, 17, 16287–16379. [Google Scholar] [CrossRef]
- Wang, X.; Park, S.G.; Ko, J.; Xiao, X.; Giannini, V.; Maier, S.A.; Kim, D.H.; Choo, J. Sensitive and reproducible immunoassay of multiple mycotoxins using surface-enhanced Raman scattering mapping on 3D plasmonic nanopillar arrays. Small 2018, 14, 1801623. [Google Scholar] [CrossRef]
- Cheng, Z.; Choi, N.; Wang, R.; Lee, S.; Moon, K.C.; Yoon, S.-Y.; Chen, L.; Choo, J. Simultaneous detection of dual prostate specific antigens using surface-enhanced Raman scattering-based immunoassay for accurate diagnosis of prostate cancer. Acs Nano 2017, 11, 4926–4933. [Google Scholar] [CrossRef]
- Gong, T.; Hong, Z.-Y.; Chen, C.-H.; Tsai, C.-Y.; Liao, L.-D.; Kong, K.V. Optical interference-free surface-enhanced Raman scattering CO-nanotags for logical multiplex detection of vascular disease-related biomarkers. ACS Nano 2017, 11, 3365–3375. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Kong, K.V.; Goh, D.; Olivo, M.; Yong, K.-T. Sensitive surface enhanced Raman scattering multiplexed detection of matrix metalloproteinase 2 and 7 cancer markers. Biomed. Opt. Express 2015, 6, 2076–2087. [Google Scholar] [CrossRef]
- Lee, H.; Kim, W.; Song, M.Y.; Kim, D.H.; Jung, H.S.; Kim, W.; Choi, S. One-stop plasmonic nanocube-excited SERS immunoassay platform of multiple cardiac biomarkers for rapid screening and progressive tracing of acute myocardial infarction. Small 2024, 20, 2304999. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.; Hsieh, C.-L.; Hsu, K.-C.; Liao, P.-H.; Qiu, S.; Gong, T.; Yong, K.-T.; Feng, S.; Kong, K.V. Geometrically encoded SERS nanobarcodes for the logical detection of nasopharyngeal carcinoma-related progression biomarkers. Nat. Commun. 2021, 12, 3430. [Google Scholar] [CrossRef] [PubMed]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010, 28, 595–599. [Google Scholar] [CrossRef]
- Duffy, D.C. Digital detection of proteins. Lab Chip 2023, 23, 818–847. [Google Scholar] [CrossRef]
- Genco, E.; Modena, F.; Sarcina, L.; Björkström, K.; Brunetti, C.; Caironi, M.; Caputo, M.; Demartis, V.M.; Di Franco, C.; Frusconi, G. A Single-Molecule Bioelectronic Portable Array for Early Diagnosis of Pancreatic Cancer Precursors. Adv. Mater. 2023, 35, 2304102. [Google Scholar] [CrossRef]
- Kim, S.H.; Iwai, S.; Araki, S.; Sakakihara, S.; Iino, R.; Noji, H. Large-scale femtoliter droplet array for digital counting of single biomolecules. Lab Chip 2012, 12, 4986–4991. [Google Scholar] [CrossRef]
- Kan, C.W.; Tobos, C.I.; Rissin, D.M.; Wiener, A.D.; Meyer, R.E.; Svancara, D.M.; Comperchio, A.; Warwick, C.; Millington, R.; Collier, N. Digital enzyme-linked immunosorbent assays with sub-attomolar detection limits based on low numbers of capture beads combined with high efficiency bead analysis. Lab Chip 2020, 20, 2122–2135. [Google Scholar] [CrossRef]
- Wilson, D.H.; Rissin, D.M.; Kan, C.W.; Fournier, D.R.; Piech, T.; Campbell, T.G.; Meyer, R.E.; Fishburn, M.W.; Cabrera, C.; Patel, P.P. The Simoa HD-1 analyzer: A novel fully automated digital immunoassay analyzer with single-molecule sensitivity and multiplexing. J. Lab. Autom. 2016, 21, 533–547. [Google Scholar] [CrossRef]
- Cohen, L.; Cui, N.; Cai, Y.; Garden, P.M.; Li, X.; Weitz, D.A.; Walt, D.R. Single molecule protein detection with attomolar sensitivity using droplet digital enzyme-linked immunosorbent assay. ACS Nano 2020, 14, 9491–9501. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Dougan, T.J.; Walt, D.R. High-throughput, high-multiplex digital protein detection with attomolar sensitivity. ACS Nano 2022, 16, 1025–1035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.J.; Wu, C.; Walt, D.R. A Multiplexed Digital Platform Enables Detection of Attomolar Protein Levels with Minimal Cross-Reactivity. ACS Nano 2024, 18, 29891–29901. [Google Scholar] [CrossRef]
- Zhao, W.; Zhou, Y.; Feng, Y.-Z.; Niu, X.; Zhao, Y.; Zhao, J.; Dong, Y.; Tan, M.; Xianyu, Y.; Chen, Y. Computer vision-based artificial intelligence-mediated encoding-decoding for multiplexed microfluidic digital immunoassay. ACS Nano 2023, 17, 13700–13714. [Google Scholar] [CrossRef]
- Cohen, L.; Walt, D.R. Highly sensitive and multiplexed protein measurements. Chem. Rev. 2018, 119, 293–321. [Google Scholar] [CrossRef] [PubMed]
- Yelleswarapu, V.; Buser, J.R.; Haber, M.; Baron, J.; Inapuri, E.; Issadore, D. Mobile platform for rapid sub–picogram-per-milliliter, multiplexed, digital droplet detection of proteins. Proc. Natl. Acad. Sci. USA 2019, 116, 4489–4495. [Google Scholar] [CrossRef]
- Lee, K.-H.; Kim, D.-M. Recent advances in development of cell-free protein synthesis systems for fast and efficient production of recombinant proteins. FEMS Microbiol. Lett. 2018, 365, fny174. [Google Scholar] [CrossRef]
- Hunt, A.C.; Rasor, B.J.; Seki, K.; Ekas, H.M.; Warfel, K.F.; Karim, A.S.; Jewett, M.C. Cell-free gene expression: Methods and applications. Chem. Rev. 2024, 125, 91–149. [Google Scholar] [CrossRef]
- Yeom, K.; Park, Y.J.; Kim, H.; Song, D.-Y.; Kim, D.-M.; Park, J.-H. Photothermal heating of cell-free reactions for on-site production of recombinant proteins. Biotechnol. Bioprocess Eng. 2024, 29, 255–261. [Google Scholar] [CrossRef]
- Kim, K.J.; Lee, S.-J.; Kim, D.-M. Cell-free systems for a multi-pronged approach to next-generation therapeutic and diagnostics. Biotechnol. Bioprocess Eng. 2024, 29, 233–239. [Google Scholar] [CrossRef]
- Park, Y.-J.; Kim, D.-M. Production of recombinant horseradish peroxidase in an engineered cell-free protein synthesis system. Front. Bioeng. Biotechnol. 2021, 9, 778496. [Google Scholar] [CrossRef]
- Karim, A.S.; Jewett, M.C. Cell-free synthetic biology for pathway prototyping. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 31–57. [Google Scholar]
- Hunt, A.C.; Vögeli, B.; Hassan, A.O.; Guerrero, L.; Kightlinger, W.; Yoesep, D.J.; Krüger, A.; DeWinter, M.; Diamond, M.S.; Karim, A.S. A rapid cell-free expression and screening platform for antibody discovery. Nat. Commun. 2023, 14, 3897. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Choi, S.; Lee, K.W.; Park, S.-Y.; Song, D.-Y.; Yoo, T.H.; Kim, D.-M. A cell-free biosensor for multiplexed and sensitive detection of biological warfare agents. Biosens. Bioelectron. 2024, 257, 116331. [Google Scholar] [CrossRef] [PubMed]
- Byun, J.-Y.; Lee, K.-H.; Shin, Y.-B.; Kim, D.-M. Cascading amplification of immunoassay signal by cell-free expression of firefly luciferase from detection antibody-conjugated DNA in an Escherichia coli extract. ACS Sens. 2018, 4, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.-H.; Hwang, M.-Y.; Lee, K.-H.; Choi, C.-Y.; Kim, D.-M. Use of signal sequences as an in situ removable sequence element to stimulate protein synthesis in cell-free extracts. Nucleic Acids Res. 2007, 35, e21. [Google Scholar] [CrossRef]
- Kang, S.H.; Kim, D.M.; Kim, H.J.; Jun, S.Y.; Lee, K.Y.; Kim, H.J. Cell-free production of aggregation-prone proteins in soluble and active forms. Biotechnol. Prog. 2005, 21, 1412–1419. [Google Scholar] [CrossRef]
- Byun, J.-Y.; Shin, Y.-B.; Li, T.; Park, J.-H.; Kim, D.-M.; Choi, D.-H.; Kim, M.-G. The use of an engineered single chain variable fragment in a localized surface plasmon resonance method for analysis of the C-reactive protein. Chem. Commun. 2013, 49, 9497–9499. [Google Scholar] [CrossRef]
- Chen, Q.; Tian, T.; Xiong, E.; Wang, P.; Zhou, X. CRISPR/Cas13a signal amplification linked immunosorbent assay for femtomolar protein detection. Anal. Chem. 2019, 92, 573–577. [Google Scholar] [CrossRef]
- Park, J.; Park, M.; Kim, J.; Heo, Y.; Han, B.H.; Choi, N.; Park, C.; Lee, R.; Lee, D.-G.; Chung, S. Beads-and oil-free single molecule assay with immuno-rolling circle amplification for detection of SARS-CoV-2 from saliva. Biosens. Bioelectron. 2023, 232, 115316. [Google Scholar] [CrossRef]
- Burrows, J.; Nitsche, A.; Bayly, B.; Walker, E.; Higgins, G.; Kok, T. Detection and subtyping of Herpes simplex virus in clinical samples by LightCycler PCR, enzyme immunoassay and cell culture. BMC Microbiol. 2002, 2, 12. [Google Scholar] [CrossRef]
- Hashimoto, M.; Aoki, M.; Winblad, B.; Tjernberg, L.O. A novel approach for Aβ1–40 quantification using immuno-PCR. J. Neurosci. Methods 2012, 205, 364–367. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Patfield, S.A. Immuno-PCR assay for sensitive detection of proteins in real time. In ELISA: Methods and Protocols; Springer: New York, NY, USA, 2015; pp. 139–148. [Google Scholar]
- Du, J.; He, J.-S.; Wang, R.; Wu, J.; Yu, X. Ultrasensitive reporter DNA sensors built on nucleic acid amplification techniques: Application in the detection of trace amount of protein. Biosens. Bioelectron. 2024, 243, 115761. [Google Scholar] [CrossRef] [PubMed]
- Malou, N.; Raoult, D. Immuno-PCR: A promising ultrasensitive diagnostic method to detect antigens and antibodies. Trends Microbiol. 2011, 19, 295–302. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Recombinase polymerase amplification combined with a magnetic nanoparticle-based immunoassay for fluorometric determination of troponin T. Microchim. Acta 2019, 186, 549. [Google Scholar] [CrossRef]
- Srivastava, P.; Prasad, D. Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 2023, 13, 200. [Google Scholar] [CrossRef] [PubMed]
- Rahmatpour, S.; Khan, A.H.; Kalmarzi, R.N.; Rajabibazl, M.; Tavoosidana, G.; Motevaseli, E.; Zarghami, N.; Sadroddiny, E. Application of immuno-PCR assay for the detection of serum IgE specific to Bermuda allergen. Mol. Cell. Probes 2017, 32, 1–4. [Google Scholar] [CrossRef]
- Qian, S.; Chen, Y.; Xu, X.; Peng, C.; Wang, X.; Wu, H.; Liu, Y.; Zhong, X.; Xu, J.; Wu, J. Advances in amplification-free detection of nucleic acid: CRISPR/Cas system as a powerful tool. Anal. Biochem. 2022, 643, 114593. [Google Scholar] [CrossRef]
- Gong, S.; Zhang, S.; Lu, F.; Pan, W.; Li, N.; Tang, B. CRISPR/Cas-based in vitro diagnostic platforms for cancer biomarker detection. Anal. Chem. 2021, 93, 11899–11909. [Google Scholar] [CrossRef]
- Zhao, X.; Li, S.; Liu, G.; Wang, Z.; Yang, Z.; Zhang, Q.; Liang, M.; Liu, J.; Li, Z.; Tong, Y. A versatile biosensing platform coupling CRISPR–Cas12a and aptamers for detection of diverse analytes. Sci. Bull. 2021, 66, 69–77. [Google Scholar] [CrossRef]
- Li, Y.; Deng, F.; Goldys, E.M. A simple and versatile CRISPR/Cas12a-based immunosensing platform: Towards attomolar level sensitivity for small protein diagnostics. Talanta 2022, 246, 123469. [Google Scholar] [CrossRef]
- Paialunga, E.; Bagheri, N.; Rossetti, M.; Fabiani, L.; Micheli, L.; Chamorro-Garcia, A.; Porchetta, A. Leveraging Synthetic Antibody–DNA Conjugates to Expand the CRISPR-Cas12a Biosensing Toolbox. ACS Synth. Biol. 2025, 14, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Christopoulos, T.K.; Chiu, N.H. Expression immunoassay. Antigen quantitation using antibodies labeled with enzyme-coding DNA fragments. Anal. Chem. 1995, 67, 4290–4294. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.S.; Seo, S.W.; Hwang, S.; Chu, H.S.; Ahn, J.-H.; Kim, T.-W.; Kim, D.-M.; Jung, G.Y. Design of 5′-untranslated region variants for tunable expression in Escherichia coli. Biochem. Biophys. Res. Commun. 2007, 356, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.-S.; Lee, K.-H.; Byun, J.-Y.; Shin, Y.-B.; Kim, D.-M. Aptamer-linked in vitro expression assay for ultrasensitive detection of biomarkers. Anal. Chim. Acta 2021, 1146, 118–123. [Google Scholar] [CrossRef]
- Byun, J.-Y.; Lee, K.-H.; Park, Y.J.; Song, D.-Y.; Min, Y.-H.; Kim, D.-M. Revisiting ELISA with in situ amplification of biomarkers to boost its sensitivity. Sens. Actuators B Chem. 2025, 423, 136780. [Google Scholar] [CrossRef]
- McSweeney, M.A.; Patterson, A.T.; Loeffler, K.; Cuellar Lelo de Larrea, R.; McNerney, M.P.; Kane, R.S.; Styczynski, M.P. A modular cell-free protein biosensor platform using split T7 RNA polymerase. Sci. Adv. 2025, 11, eado6280. [Google Scholar] [CrossRef]
| Method | Analytes | Limit of Detection | Time to Result | Scalability | Cost | Reference |
|---|---|---|---|---|---|---|
| Nanozyme ELISA | HBV, HCV, ZIKV | 250 copies/mL | 50 min | High | Medium | [57] |
| Plasmonic ELISA | PSA, HIV-1 capsid antigen p24 | 10−6 ng/mL | 7 h | Medium | Low | [61] |
| Serum myoglobin | 0.057 ng/mL | 4 h | High | Medium | [66] | |
| Electrochemical ELISA | Aβ42 | 2.6 fg/mL | 3 h | Medium | Medium | [72] |
| Digital ELISA | ≥20 cytokines and neuro markers (e.g., IL-6, Tau, Aβ42) | 1~10 fg/mL | 30 min–2.5 h | High | High | [90] |
| IFN-γ, IL-2 | 0.51 fg/mL, 1.0 fg/mL | 2–3 h | High | Medium | [91] | |
| IL-6, IL-10, IFN-γ, etc. (up to 8-plex) | 0.3~4.8 pg/mL | 3 h | High | High | [92] | |
| CLISA | IL-6, VEGF | 45.8 fg/mL, 32.3 fg/mL | 4 h | Medium | Medium | [109] |
| IFN-γ, EGFR | 1 fg/mL | 3–4 h | High | Medium | [122] | |
| Expression ELISA | thrombin | 0.1 pg/mL | 4 h | Medium | Medium | [126] |
| AFP, IL-6 | 0.414 pg/mL, 0.0126 pg/mL | 4 h | High | Medium | [127] | |
| TLISA | SARS-CoV-2 spike (S) | 21 μg/mL | <1 h | Medium | Medium | [128] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, H.-B.; Song, D.-Y.; Park, Y.J.; Kim, D.-M. Enhancing ELISA Sensitivity: From Surface Engineering to Synthetic Biology. Biosensors 2025, 15, 434. https://doi.org/10.3390/bios15070434
Jeon H-B, Song D-Y, Park YJ, Kim D-M. Enhancing ELISA Sensitivity: From Surface Engineering to Synthetic Biology. Biosensors. 2025; 15(7):434. https://doi.org/10.3390/bios15070434
Chicago/Turabian StyleJeon, Hye-Bin, Dong-Yeon Song, Yu Jin Park, and Dong-Myung Kim. 2025. "Enhancing ELISA Sensitivity: From Surface Engineering to Synthetic Biology" Biosensors 15, no. 7: 434. https://doi.org/10.3390/bios15070434
APA StyleJeon, H.-B., Song, D.-Y., Park, Y. J., & Kim, D.-M. (2025). Enhancing ELISA Sensitivity: From Surface Engineering to Synthetic Biology. Biosensors, 15(7), 434. https://doi.org/10.3390/bios15070434





