Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis
Abstract
1. Introduction
2. Limitations of Traditional Detection Methods and the Need for Technological Advancements
3. The Rise in Isothermal Nucleic Acid Amplification Technology and Innovative Strategies for Its Use
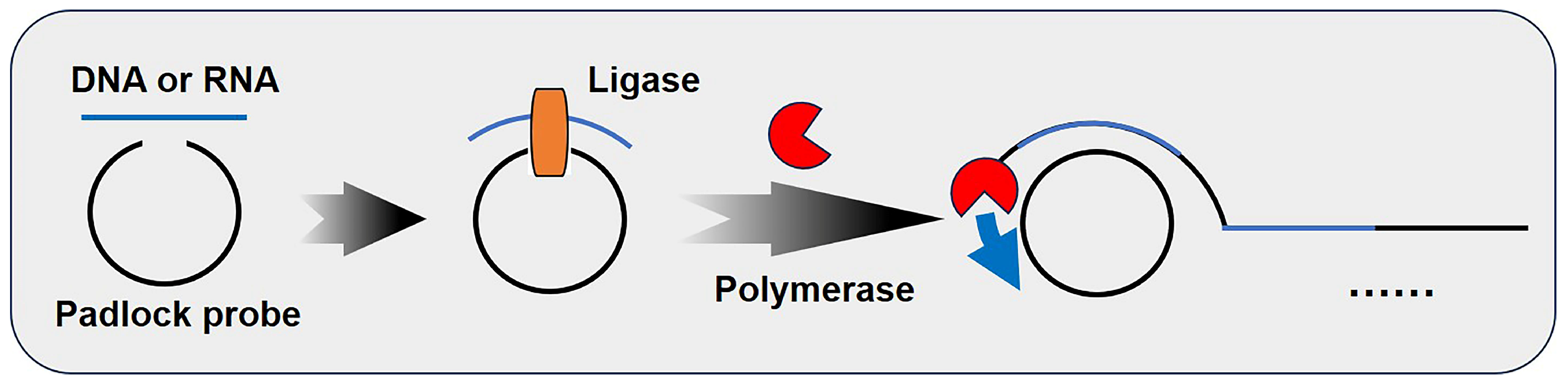
3.1. Rolling Circle Amplification (RCA)
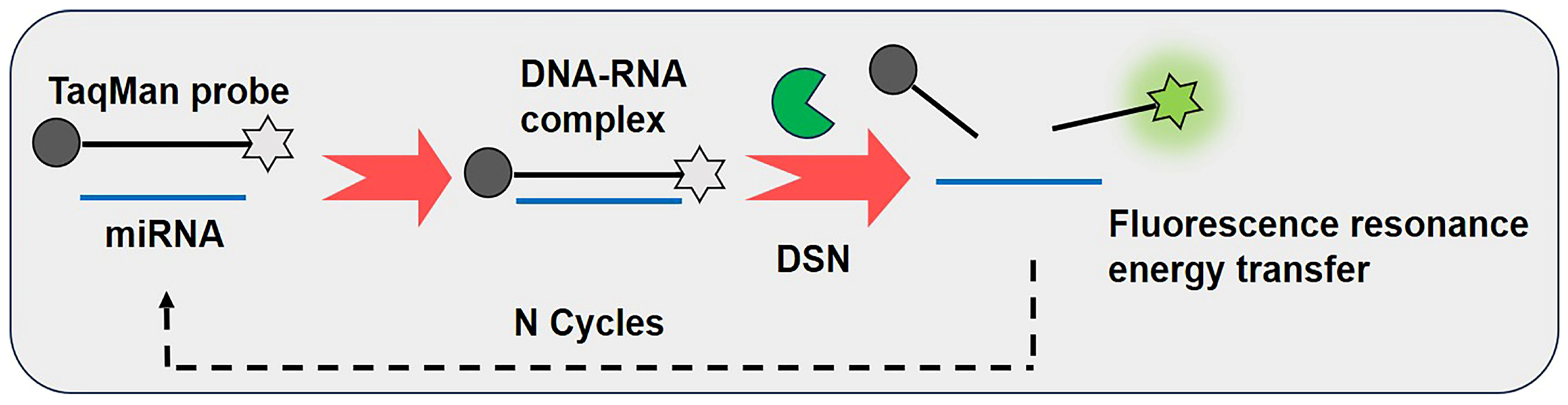
3.2. Duplex-Specific Nuclease-Assisted Signal Amplification (DSNSA)
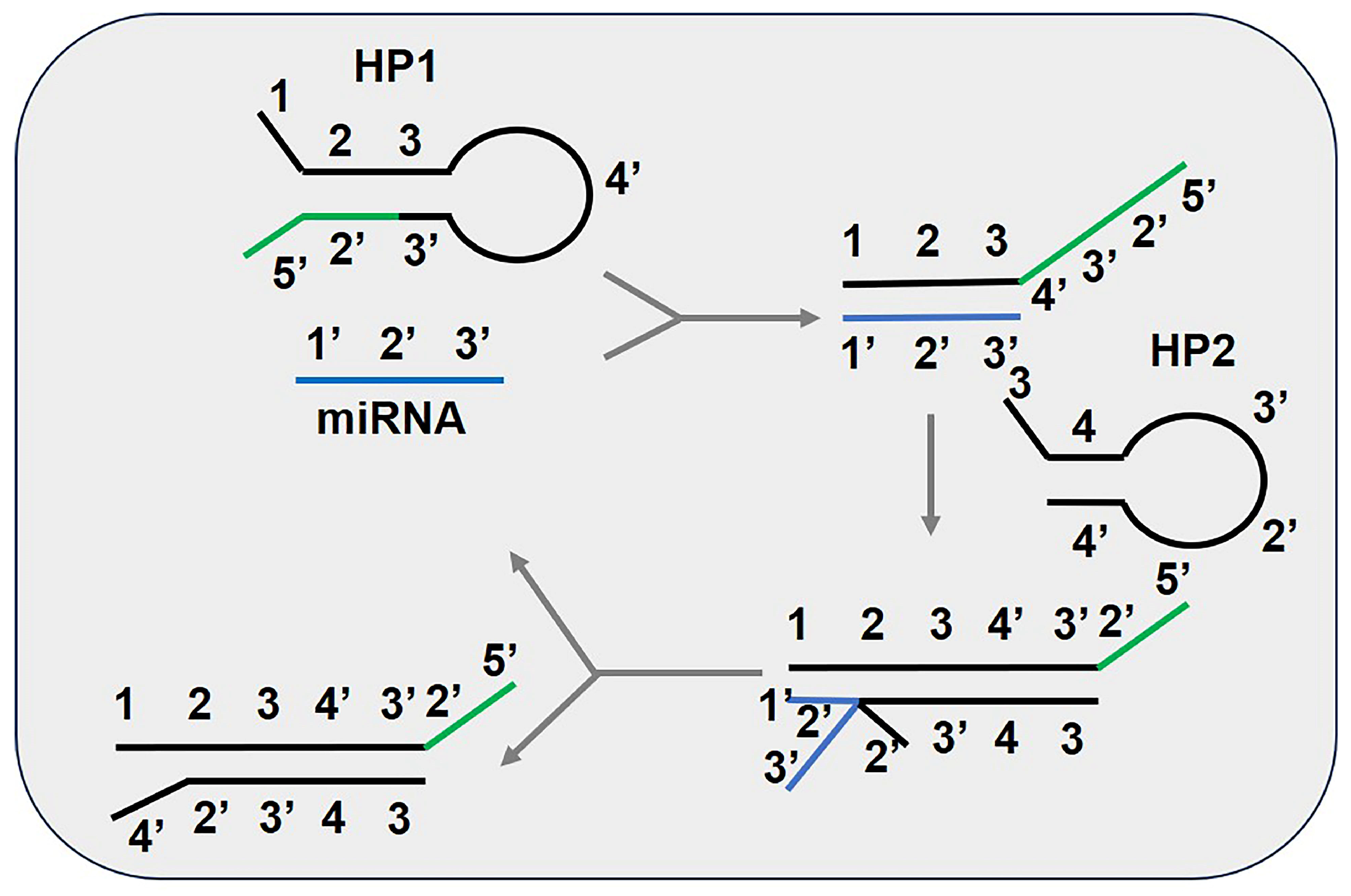
3.3. Catalytic Hairpin Assembly (CHA)
3.4. Strand-Displacement Amplification (SDA)
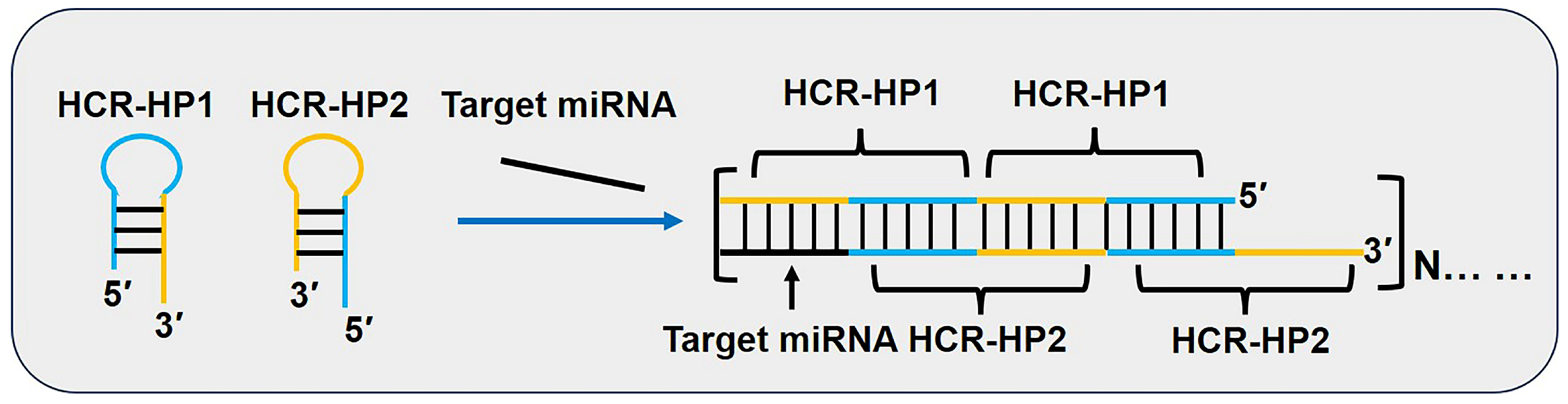
3.5. Hybridization Chain Reaction (HCR)
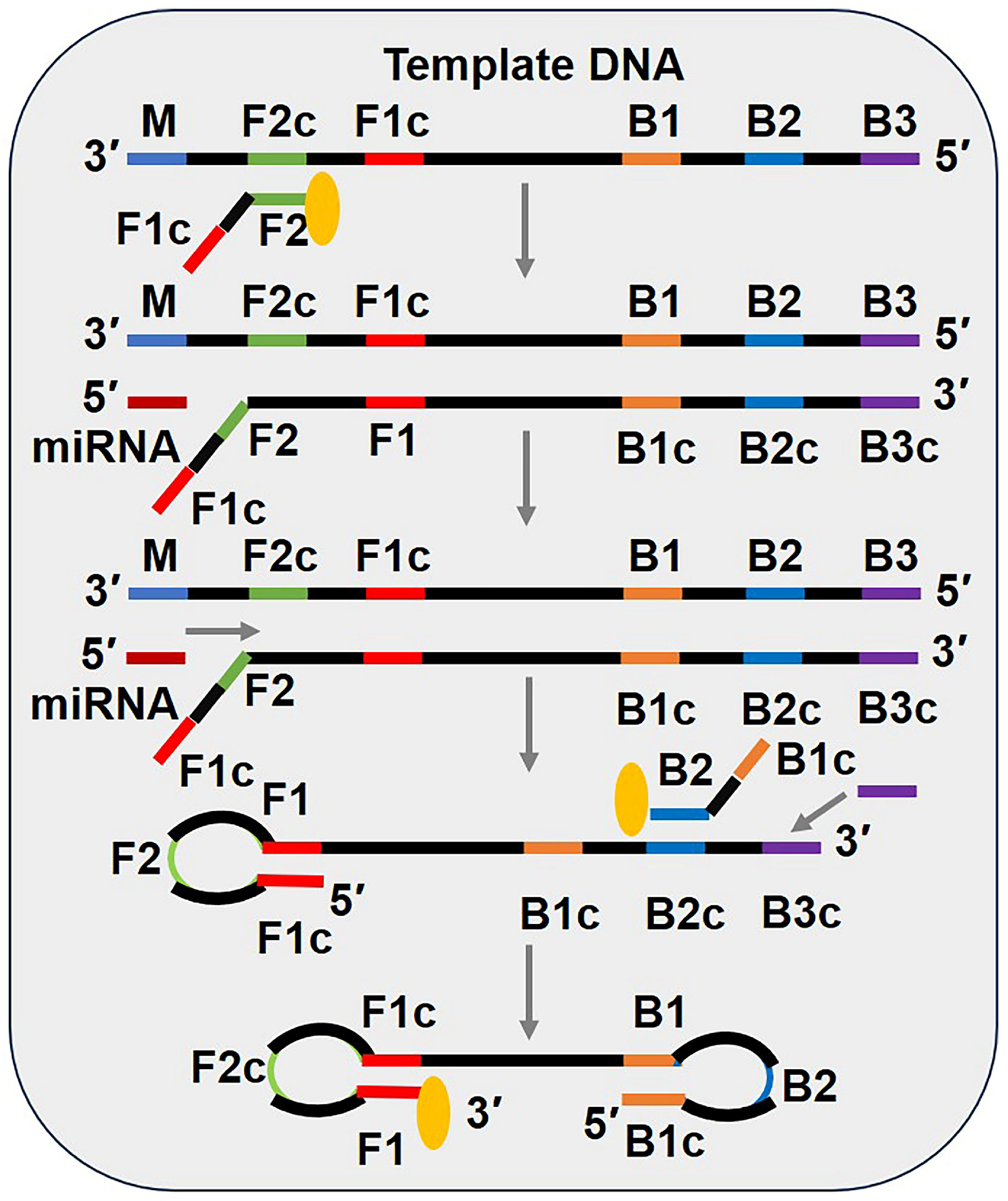
3.6. Loop-Mediated Isothermal Amplification (LAMP)
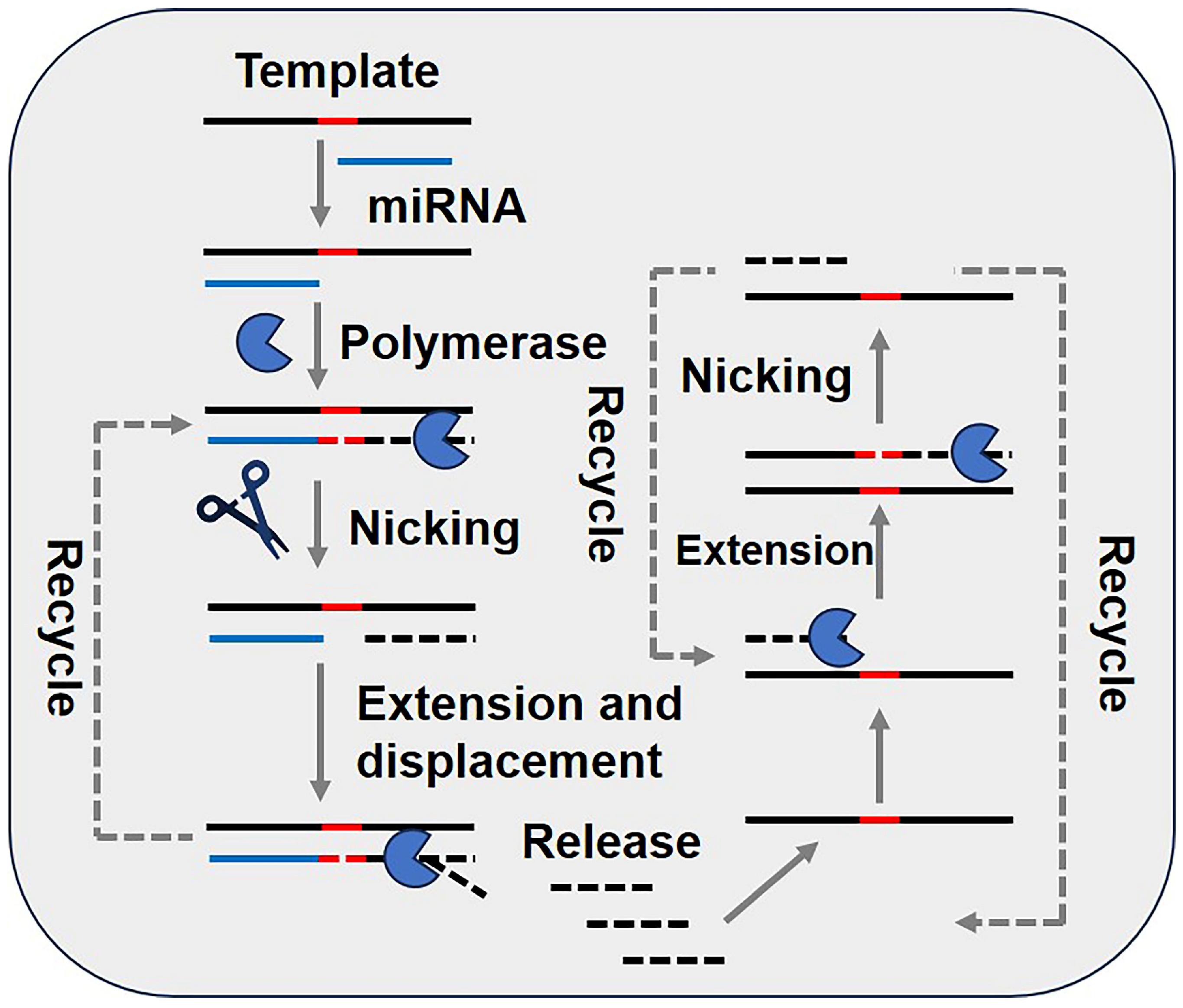
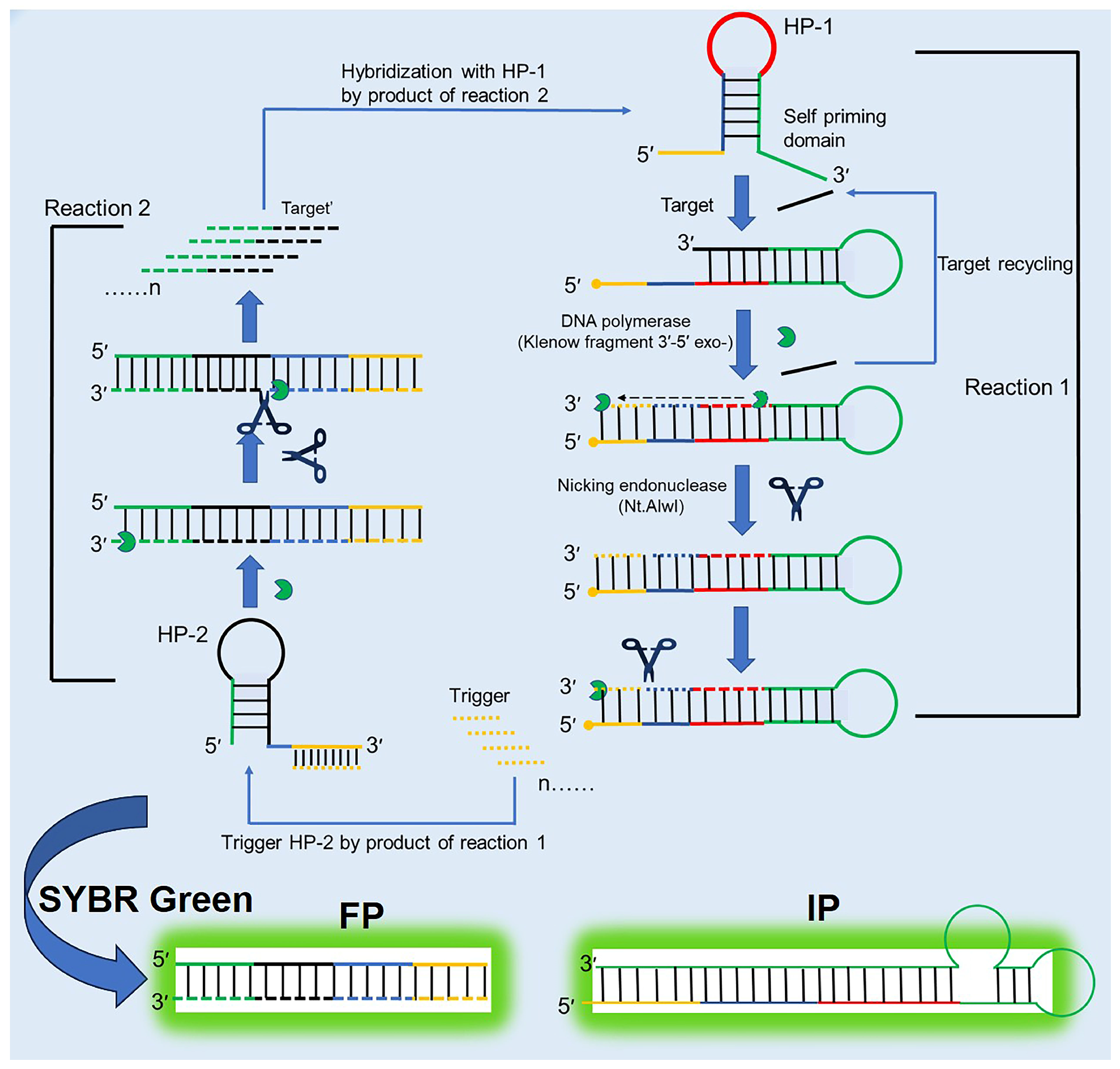
3.7. Exponential Amplification Reaction (EXPAR)
4. Clinical Sample Detection for Disease Diagnosis
4.1. The Application of miRNA Detection Methods in Disease Diagnosis
4.2. The Application of miRNA Detection Utilizing POCT for Disease Diagnosis
4.3. The Application of miRNA Detection Utilizing AI Assistance in Disease Diagnosis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MiRNA | MicroRNA |
| MS-ACPE | Magnetic-separation-assisted auto-cyclic primer extension |
| RT-qPCR | Reverse transcription quantitative PCR |
| ncRNA | Non-coding RNA |
| PTC | Papillary thyroid carcinoma |
| FNAB | Fine needle aspiration biopsy |
| POCT | Point-of-care testing |
| PICA | Product-induced catalysis amplification |
| RCA | Rolling circle amplification |
| DSNSA | Duplex-specific nuclease signal amplification |
| CHA | Catalytic hairpin assembly |
| SDA | Strand-displacement amplification |
| SPAAC | Strain-promoted azide–alkyne cycloaddition |
| HCR | Hybridization chain reaction |
| LAMP | Loop-mediated isothermal amplification |
| LOQ | Limit of quantitation |
| EXPAR | Exponential amplification reaction |
| ECL | Electrochemiluminescence |
| dNTPs | Deoxyribonucleoside triphosphates |
| AuNPs | Gold nanoparticles |
| AuNP@LH | Target-triggered locked hairpin DNA-functionalized Au nanoprobes |
| AIE | Aggregation-induced emission |
| FRET | Fluorescence resonance energy transfer |
| HPCL-FLD | High-performance liquid chromatography coupled with a fluorescence detector |
| ICP-MS | Inductively coupled plasma–mass spectrometry |
| HP | Hairpin primer |
| dsDNA | Double-stranded DNA |
| FIP | Forward inner primer |
| BIP | Backward inner primer |
References
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.M.; Yu, X.M.; Hu, S.N.; Yu, J. A brief review on the mechanisms of miRNA regulation. Genom. Proteom. Bioinform. 2009, 7, 147–154. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.D.; Li, Q.; Zhang, R.S.; Dai, X.L.; Chen, W.J.; Xing, D.M. Circulating microRNAs: Biomarkers of disease. Clin. Chim. Acta 2021, 516, 46–54. [Google Scholar] [CrossRef]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Kilic, T.; Erdem, A.; Ozsoz, M.; Carrara, S. MicroRNA biosensors: Opportunities and challenges among conventional and commercially available techniques. Biosens. Bioelectron. 2018, 99, 525–546. [Google Scholar] [CrossRef]
- Kondrotiene, A.; Dauksa, A.; Pamedytyte, D.; Kazokaite, M.; Zvirbliene, A.; Dauksiene, D.; Simanaviciene, V.; Klimaite, R.; Golubickaite, I.; Stakaitis, R.; et al. Plasma-derived miRNA-222 as a candidate marker for papillary thyroid cancer. Int. J. Mol. Sci. 2020, 21, 6445. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Lim, Y.S.; Lee, J.C.; Wang, S.G.; Park, H.Y.; Kim, S.Y.; Lee, B.J. Differential expression levels of plasma-derived miR-146b and miR-155 in papillary thyroid cancer. Oral. Oncol. 2015, 51, 77–83. [Google Scholar] [CrossRef]
- Santos, D.A.R.; Gaiteiro, C.; Santos, M.; Santos, L.; Dinis-Ribeiro, M.; Lima, L. MicroRNA biomarkers as promising tools for early colorectal cancer screening-A comprehensive review. Int. J. Mol. Sci. 2023, 24, 11023. [Google Scholar] [CrossRef]
- Petkevich, A.A.; Abramov, A.A.; Pospelov, V.I.; Malinina, N.A.; Kuhareva, E.I.; Mazurchik, N.V.; Tarasova, O.I. Exosomal and non-exosomal miRNA expression levels in patients with HCV-related cirrhosis and liver cancer. Oncotarget 2021, 12, 1697–1706. [Google Scholar] [CrossRef]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, L.; Medyany, V.; Ezic, J.; Lotfi, R.; Niesler, B.; Roeth, R.; Engelhardt, D.; Laban, S.; Schuler, P.J.; Hoffmann, T.K.; et al. Cargo and functional profile of saliva-derived exosomes reveal biomarkers specific for head and neck cancer. Front. Med. 2022, 9, 904295. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Trachtenberg, A.J.; Kuo, W.P.; Cheng, Y.S. Genomewide study of salivary microRNAs for detection of oral cancer. J. Dent. Res. 2014, 93, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Rapado-Gonzalez, O.; Majem, B.; Alvarez-Castro, A.; Diaz-Peña, R.; Abalo, A.; Suarez-Cabrera, L.; Gil-Moreno, A.; Santamaria, A.; Lopez-Lopez, R.; Muinelo-Romay, L.; et al. A novel saliva-based miRNA signature for colorectal cancer diagnosis. J. Clin. Med. 2020, 8, 2029. [Google Scholar] [CrossRef]
- Chen, D.; Chen, N.; Liu, F.N.; Wang, Y.M.; Liang, H.G.; Yang, Y.B.; Yuan, Q. Flexible point-of-care electrodes for ultrasensitive detection of bladder tumor-relevant miRNA in urine. Anal. Chem. 2023, 95, 1847–1855. [Google Scholar] [CrossRef] [PubMed]
- Lekchnov, E.A.; Amelina, E.V.; Bryzgunova, O.E.; Zaporozhchenko, I.A.; Konoshenko, M.Y.; Yarmoschuk, S.V.; Murashov, I.S.; Pashkovskaya, O.A.; Gorizkii, A.M.; Zheravin, A.A.; et al. Searching for the novel specific predictors of prostate cancer in urine: The analysis of 84 miRNA expression. Int. J. Mol. Sci. 2018, 19, 4088. [Google Scholar] [CrossRef]
- Bustos, M.A.; Gottlieb, J.; Choe, J.; Suyeon, R.; Lin, S.Y.; Allen, W.M.; Krasne, D.L.; Wilson, T.G.; Hoon, D.S.B.; Linehan, J.A. Diagnostic miRNA signatures in paired tumor, plasma, and urine specimens from renal cell carcinoma patients. Clin. Chem. 2024, 70, 261–272. [Google Scholar] [CrossRef]
- Fakhr, Z.A.; Zhu, X.L.; Wang, H.C.; Ma, R.Y.; Lin, Z.W.; Shen, X.D.; Liu, J.T.; Zeng, S.; Cai, S. Embarking on a journey through Micro-RNA and Circular-RNA detection methods. TrAC-Trends Anal. Chem. 2024, 181, 118035. [Google Scholar] [CrossRef]
- Deng, R.J.; Zhang, K.X.; Li, J.H. Isothermal amplification for microRNA detection: From the test tube to the cell. Accounts Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef]
- Byron, S.A.; Van Keuren-Jensen, K.R.; Engelthaler, D.M.; Carpten, J.D.; Craig, D.W. Translating RNA sequencing into clinical diagnostics: Opportunities and challenges. Nat. Rev. Genet. 2016, 17, 257–271. [Google Scholar] [CrossRef]
- De Felice, M.; De Falco, M.; Antonacci, A.; Colella, S.; Vedi, V.; Isticato, R.; Romano, A.M.; Nocerino, V.; Miranda, B.; De Stefano, L.; et al. Latest trends in biosensors powered by nucleic acid isothermal amplification for the diagnosis of joint infections: From sampling to identification towards the point-of-care. TrAC-Trends Anal. Chem. 2024, 181, 118036. [Google Scholar] [CrossRef]
- Yin, W.H.; Zhuang, J.J.; Li, J.L.; Xia, L.P.; Hu, K.; Yin, J.X.; Mu, Y. Digital recombinase polymerase amplification, digital loop-mediated isothermal amplification, and digital CRISPR-Cas assisted assay: Current status, challenges, and perspectives. Small 2023, 19, 2303398. [Google Scholar] [CrossRef]
- Zhao, Y.X.; Chen, F.; Li, Q.; Wang, L.H.; Fan, C.H. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Bialy, R.M.; Mainguy, A.; Li, Y.F.; Brennan, J.D. Functional nucleic acid biosensors utilizing rolling circle amplification. Chem. Soc. Rev. 2022, 51, 9009–9067. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.B.; Yang, L.T. Recent progress in molecular diagnostics: The synergy of rolling circle amplification and CRISPR/Cas systems (2018-2024)—A concise review. TrAC-Trends Anal. Chem. 2024, 180, 117902. [Google Scholar] [CrossRef]
- Kuhn, H.; Frank-Kamenetskii, M.D. Template-independent ligation of single-stranded DNA by T4 DNA ligase. FEBS J. 2005, 272, 5991–6000. [Google Scholar] [CrossRef]
- Jiang, W.Y.; Chen, Z.P.; Lu, J.; Ren, X.; Ma, Y. Ultrasensitive visual detection of miRNA-143 using a CRISPR/Cas12a-based platform coupled with hyperbranched rolling circle amplification. Talanta 2022, 251, 123784. [Google Scholar] [CrossRef]
- Yin, B.C.; Liu, Y.Q.; Ye, B.C. One-step, multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc. 2012, 134, 5064–5067. [Google Scholar] [CrossRef]
- Wu, Y.D.; Cui, S.; Li, Q.; Zhang, R.S.; Song, Z.M.; Gao, Y.Z.; Chen, W.J.; Xing, D.M. Recent advances in duplex-specific nuclease-based signal amplification strategies for microRNA detection. Biosens. Bioelectron. 2020, 165, 112449. [Google Scholar] [CrossRef]
- Shen, W.; Yeo, K.H.; Gao, Z.Q. A simple and highly sensitive fluorescence assay for microRNAs. Analyst 2015, 140, 1932–1938. [Google Scholar] [CrossRef]
- Qi, T.; Song, C.; He, J.; Shen, W.; Kong, D.Z.; Shi, H.W.; Tan, L.; Pan, R.R.; Tang, S.; Lee, H.K. Highly sensitive detection of multiple microRNAs by high-performance liquid chromatography coupled with long and short probe-based recycling amplification. Anal. Chem. 2020, 92, 5033–5040. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.Q.; Cao, J.X.; Xu, F.F.; Chen, Y. Duplex-specific nuclease-mediated amplification strategy for mass spectrometry quantification of miRNA-200c in breast cancer stem cells. Anal. Chem. 2019, 91, 8820–8826. [Google Scholar] [CrossRef]
- Zhang, S.X.; Liu, R.; Xing, Z.; Zhang, S.C.; Zhang, X.R. Multiplex miRNA assay using lanthanide-tagged probes and the duplex-specific nuclease amplification strategy. Chem. Commun. 2016, 52, 14310–14313. [Google Scholar] [CrossRef] [PubMed]
- Li, B.L.; Ellington, A.D.; Chen, X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011, 39, e110. [Google Scholar] [CrossRef]
- Yin, P.; Choi, H.M.T.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.L.; Yin, Y.; Du, S.M.; Kong, L.Q.; Yang, Z.H.; Chang, Y.Y.; Chai, Y.Q.; Yuan, R. Programmable high-speed and hyper-efficiency DNA signal magnifier. Adv. Sci. 2022, 9, 2104084. [Google Scholar] [CrossRef]
- Wang, H.C.; Shen, M.Z.; Shen, X.D.; Liu, J.T.; Huang, W.W.; Jiang, X.F.; Liu, H.; Zeng, S.; Nan, K.W.; Cai, S. An enzyme-free sensing platform for miRNA detection and in situ imaging in clinical samples based on DNAzyme cleavage-triggered catalytic hairpin assembly. Biosens. Bioelectron. 2024, 256, 116279. [Google Scholar] [CrossRef]
- Li, J.; Luo, H.X. Nicking site enzyme assisted catalytic hairpin assembly based scaffold for sensitive monitoring of miRNA-21. Microchem. J. 2022, 174, 107059. [Google Scholar] [CrossRef]
- Luo, Z.W.; Li, Y.X.; Zhang, P.; He, L.; Feng, Y.T.; Feng, Y.Q.; Qian, C.; Tian, Y.H.; Duan, Y.X. Catalytic hairpin assembly as cascade nucleic acid circuits for fluorescent biosensor: Design, evolution and application. TrAC-Trends Anal. Chem. 2022, 151, 116582. [Google Scholar] [CrossRef]
- Xu, M.D.; Lin, L.; Li, N.; Jiang, X.Y.; Li, J.L.; Gong, L.Z.; Zhuang, J.Y. Nanoscale assembly line composed of dual DNA-machines enabling sensitive microRNA detection using upconversion nanoparticles probes. J. Pharm. Biomed. Anal. 2021, 195, 113842. [Google Scholar] [CrossRef]
- Li, G.; Niu, P.; Ge, S.J.; Cao, D.W.; Sun, A.D. SERS based lateral flow assay for rapid and ultrasensitive quantification of dual laryngeal squamous cell carcinoma-related miRNA biomarkers in human serum using Pd-Au core-shell nanorods and catalytic hairpin assembly. Front. Mol. Biosci. 2022, 8, 813007. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.R.; Weng, B.R.; Liu, S.J.; Kang, N.N.; Ran, J.B.; Deng, Z.S.; Wang, H.M.; Yang, C.Y.; Wang, F. Acid-improved DNAzyme-based chemiluminescence miRNA assay coupled with enzyme-free concatenated DNA circuit. Biosens. Bioelectron. 2022, 204, 114060. [Google Scholar] [CrossRef]
- Zhang, X.L.; Yin, Y.; Du, S.M.; Kong, L.Q.; Chai, Y.Q.; Li, Z.H.; Yuan, R. Dual 3D DNA nanomachine-mediated catalytic hairpin assembly for ultrasensitive detection of microRNA. Anal. Chem. 2021, 93, 13952–13959. [Google Scholar] [CrossRef]
- Han, Y.; Hu, H.H.; Yu, L.S.; Zeng, S.; Min, J.Z.; Cai, S. A duplex-specific nuclease (DSN) and catalytic hairpin assembly (CHA)-mediated dual amplification method for miR-146b detection. Analyst 2023, 148, 556–561. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Han, S.; Ren, T.; Han, L.; Ma, X.Y.; Huang, L.J.; Sun, X. A dual-cycle isothermal amplification method for microrna detection: Combination of a duplex-specific nuclease enzyme-driven dna walker with improved catalytic hairpin assembly. Int. J. Mol. Sci. 2025, 26, 689. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Shen, X.D.; Hu, H.H.; Zeng, S.; Min, J.Z.; Li, J.B.; Cai, S. A dual-cycle DNA walker sensor for sensitive clinical detection of microRNAs. Anal. Chim. Acta 2025, 1352, 343935. [Google Scholar] [CrossRef]
- Walker, G.T.; Little, M.C.; Nadeau, J.G.; Shank, D.D. Isothermal invitro amplification of DNA by a restriction enzyme DNA-polymerase system. Proc. Natl. Acad. Sci. USA 1992, 89, 392–396. [Google Scholar] [CrossRef]
- Li, H.K.; Cai, Q.Q.; Wu, D.; Jie, G.F.; Zhou, H. Fluorescence energy transfer biosensing platform based on hyperbranched rolling circle amplification and multi-site strand displacement for ultrasensitive detection of miRNA. Anal. Chim. Acta 2022, 1222, 340190. [Google Scholar] [CrossRef]
- Choi, H.M.T.; Chang, J.Y.; Trinh, L.A.; Padilla, J.E.; Fraser, S.E.; Pierce, N.A. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 2010, 28, 1208–1212. [Google Scholar] [CrossRef]
- Zhu, L.J.; Shao, X.L.; Luo, Y.B.; Huang, K.L.; Xu, W.T. Two-way gold nanoparticle label-free sensing of specific sequence and small molecule targets using switchable concatemers. ACS Chem. Biol. 2017, 12, 1373–1380. [Google Scholar] [CrossRef]
- Yang, L.; Liu, C.H.; Ren, W.; Li, Z.P. Graphene surface-anchored fluorescence sensor for sensitive detection of microRNA coupled with enzyme-free signal amplification of hybridization chain reaction. ACS Appl. Mater. Interfaces 2012, 4, 6450–6453. [Google Scholar] [CrossRef]
- Cai, S.; Cao, Z.J.; Lau, C.W.; Lu, J.Z. Label-free technology for the amplified detection of microRNA based on the allosteric hairpin DNA switch and hybridization chain reaction. Analyst 2014, 139, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Li, J.L.; Li, M.; An, R.; Zhang, X.; Cai, S. A chemiluminescence signal amplification method for microRNA detection: The combination of molecular aptamer beacons with enzyme-free hybridization chain reaction. Molecules 2024, 29, 5782. [Google Scholar] [CrossRef]
- Xue, Q.W.; Liu, C.X.; Li, X.; Dai, L.; Wang, H.S. Label-free fluorescent DNA dendrimers for microRNA detection based on nonlinear hybridization chain reaction-mediated multiple G-quadruplex with low background signal. Bioconjug. Chem. 2018, 29, 1399–1405. [Google Scholar] [CrossRef] [PubMed]
- Li, C.P.; Li, Z.P.; Jia, H.X.; Yan, J.L. One-step ultrasensitive detection of microRNAs with loop-mediated isothermal amplification (LAMP). Chem. Commun. 2011, 47, 2595–2597. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.X.; Li, Z.P.; Liu, C.H.; Cheng, Y.Q. Ultrasensitive detection of microRNAs by exponential isothermal amplification. Angew. Chem.-Int. Edit. 2010, 49, 5498–5501. [Google Scholar] [CrossRef]
- Shi, C.; Liu, Q.; Ma, C.P.; Zhong, W.W. Exponential strand-displacement amplification for detection of microRNAs. Anal. Chem. 2014, 86, 336–339. [Google Scholar] [CrossRef]
- Song, J.Y.; Jung, Y.J.; Lee, S.; Park, H.G. Self-priming hairpin-utilized isothermal amplification enabling ultrasensitive nucleic acid detection. Anal. Chem. 2020, 92, 10350–10356. [Google Scholar] [CrossRef]
- Wang, R.L.; Lan, L.; Liu, L.; Cheng, L. Asymmetric polymerase chain reaction and loop-mediated isothermal amplification (AP-LAMP) for ultrasensitive detection of microRNAs. Chin. Chem. Lett. 2020, 31, 159–162. [Google Scholar] [CrossRef]
- Tian, W.M.; Li, P.J.; He, W.L.; Liu, C.H.; Li, Z.P. Rolling circle extension-actuated loop-mediated isothermal amplification (RCA-LAMP) for ultrasensitive detection of microRNAs. Biosens. Bioelectron. 2019, 128, 17–22. [Google Scholar] [CrossRef]
- Chen, Y.X.; Wu, X.; Huang, K.J. A sandwich-type electrochemical biosensing platform for microRNA-21 detection using carbon sphere-MoS2 and catalyzed hairpin assembly for signal amplification. Sens. Actuator B Chem. 2018, 270, 179–186. [Google Scholar] [CrossRef]
- Hosseinzadeh, E.; Ravan, H.; Mohammadi, A.; Mohammad-rezaei, R.; Norouzi, A.; Hosseinzadeh, H. Target-triggered three-way junction in conjugation with catalytic concatemers-functionalized nanocomposites provides a highly sensitive colorimetric method for miR-21 detection. Biosens. Bioelectron. 2018, 117, 567–574. [Google Scholar] [CrossRef]
- Zhang, S.S.; Xu, S.L.; Li, X.; Ma, R.N.; Cheng, G.G.; Xue, Q.W.; Wang, H.S. Double-signal mode based on metal-organic framework coupled cascaded nucleic acid circuits for accurate and sensitive detection of serum circulating miRNAs. Chem. Commun. 2020, 56, 4288–4291. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Li, J.Q.; Zuo, C.; Tao, Y.Y.; Bai, S.L.; Li, J.L.; Zhang, Z.; Xie, G.M. Specific discrimination and universal signal amplification for RNA detection by coupling toehold exchange with RCA through nucleolytic conversion of a structure-switched hairpin probe. Anal. Chim. Acta 2019, 1068, 96–103. [Google Scholar] [CrossRef]
- Qin, H.J.; Chen, Z.Y.; Zuo, F.J.; Cao, R.F.; Wang, F.Y.; Wu, H.P.; Wang, S.J.; Xie, Y.J.; Ding, S.J.; Min, X. “DSN-mismatched CRISPR” sensor for highly selective and sensitive detection of under-expressed miR-let-7a. Anal. Chim. Acta 2024, 1295, 342273. [Google Scholar] [CrossRef]
- Liao, T.B.; Luo, K.X.; Tu, J.Y.; Zhang, Y.L.; Zhang, G.J.; Sun, Z.Y. DSN signal amplification strategy based nanochannels biosensor for the detection of miRNAs. Bioelectrochemistry 2024, 160, 108771. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, B.B.; He, M.; Yuan, G.L.; Hu, B. Dual-amplification single-particle ICP-MS strategy based on strand displacement amplification-CRISPR/Cas12a amplification for homogeneous detection of miRNA. Anal. Chem. 2024, 97, 811–817. [Google Scholar] [CrossRef]
- Ma, Y.Z.; Wang, T.T.; Qu, X.L.; Yan, R.H.; Miao, P. Electrochemical quantification of miRNA based on strain-promoted azide-alkyne cycloaddition ligated tetrahedral DNA nanotags. Anal. Chem. 2024, 96, 20348–20353. [Google Scholar] [CrossRef]
- Yu, K.H.; Wu, Z.Y.; Yang, L.Z. Product-induced catalytic amplification strategy based on DNA tetrahedron for detection of miRNA-21 in colorectal cancer. Talanta 2025, 285, 127354. [Google Scholar] [CrossRef]
- Cui, F.W.; Chen, W.W.; Wang, P.L.; Fan, J.W.; Si, D.Y.; Ma, Q.; Shi, J.W.; He, Y.Q. Gold metallene-based ECL biosensor to detect miRNA-126 for coronary artery calcification diagnosis. Biosens. Bioelectron. 2025, 271, 116993. [Google Scholar] [CrossRef]
- Li, J.; Zhang, W.; Chang, R.R.; Lan, Y.T.; Qiu, D.X.; Wang, K.M.; Huang, J.; Xu, Q. A magnetic separation-assisted auto-cyclic primer extension for OSCC-associated salivary miRNA detection. Biosens. Bioelectron. 2024, 269, 116936. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.N.; Liu, F.M.; Du, X.J.; Zhao, X.L. Dual-mode, signal-amplified DNA biosensor for label-free, reliable assay of gestational diabetes mellitus-related miRNA (miR-135a). J. Pharm. Biomed. Anal. 2024, 253, 116565. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.J.; Xie, T.F.; Li, J.Y.; Wang, Z.T.; Zhang, P.F.; Sui, X.L.; Chen, J.H. Split G-quadruplex programmed recyclable AIE-biosensor for label-free detection of miRNA in acute kidney injury. Anal. Chem. 2024, 96, 17814–17823. [Google Scholar] [CrossRef]
- Chen, X.F.; Xiang, Q.Y.; Yan, S.H.; Wang, Y.Y.; Su, N.; Yang, X.; Gao, M.X.; Zhang, X.M. Simultaneous multi-miRNA detection in urinary small extracellular vesicles using target-triggered locked hairpin DNA-functionalized Au nanoprobes for systemic lupus erythematosus diagnosis. Anal. Chem. 2024, 96, 16370–16378. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Yuan, P.P.; Gan, Y.Q.; Long, X.; Deng, Z.W.; Tang, Y.L.; Yang, Y.J.; Zhong, S. A one-pot isothermal Fluorogenic Mango II arrays-based assay for label-free detection of miRNA. Talanta 2024, 281, 126920. [Google Scholar] [CrossRef]
- Wang, P.L.; Liang, Z.H.; Li, Z.R.; Wang, D.Y.; Ma, Q. Plasmonic nanocavity-modulated electrochemiluminescence sensor for gastric cancer exosomal miRNA detection. Biosens. Bioelectron. 2023, 245, 115847. [Google Scholar] [CrossRef]
- Wang, N.; Zhang, J.; Xiao, B.; Chen, A.L. Microfluidic-assisted integrated nucleic acid test strips for POCT. Talanta 2024, 267, 125150. [Google Scholar] [CrossRef]
- Dorta-Gorrin, A.; Navas-Mendez, J.; Gozalo-Marguello, M.; Miralles, L.; Garcia-Hevia, L. Detection of SARS-CoV-2 based on nucleic acid amplification tests (NAATs) and its integration into nanomedicine and microfluidic devices as point-of-care testing (POCT). Int. J. Mol. Sci. 2023, 24, 10233. [Google Scholar] [CrossRef]
- Yin, B.F.; Wan, X.H.; Sohan, A.S.M.M.F.; Lin, X.D. Microfluidics-based POCT for SARS-CoV-2 diagnostics. Micromachines 2022, 13, 1238. [Google Scholar] [CrossRef]
- Zeng, X.M.; Wang, L.N.; Liu, C.; Zhang, J.H.; Shi, H.W.; Shen, W.; Kong, D.Z.; Huang, C.; Lee, H.K.; Tang, S. An integrated liposome-based microfluidic strategy for rapid colorimetric analysis: A case study of microRNA-21 detection. Talanta 2024, 272, 125838. [Google Scholar] [CrossRef]
- Yao, C.Y.; Liu, X.L.; Lu, X.H.; Wang, L.; Jia, J.; Li, Z. Smartphone-based fluorescent profiling of quaternary microRNAs in urine for rapid diagnosis of urological cancers using a multiplexed isothermal exponential amplification reaction. Anal. Chem. 2024, 96, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.J.; Ya, Y.; Cen, X.T.; Tang, D.Y.; Shi, J.Y.; Wu, Y.Y.; Luo, H.; Huang, K.J.; Tan, X.C.; Yan, F.Y. Multiple signal amplification strategy induced by biomarkers of lung cancer: A self-powered biosensing platform adapted for smartphones. Int. J. Biol. Macromol. 2024, 264, 130661. [Google Scholar] [CrossRef]
- Yan, H.; Wen, Y.J.; Tian, Z.M.; Hart, N.; Han, S.; Hughes, S.J.; Zeng, Y.A. one-pot isothermal Cas12-based assay for the sensitive detection of microRNAs. Nat. Biomed. Eng. 2023, 7, 1583–1601. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.Y.; Wang, D.W.; Zhou, P.; Pan, Y.B.; Wan, X.Y.; Pan, W.; Li, N.; Tang, B. A lateral flow assay strip for simultaneous detection of miRNA and exosomes in liver cancer. Chem. Commun. 2024, 60, 7491–7494. [Google Scholar] [CrossRef]
- Kim, J.; Campbell, A.S.; de Avila, B.E.F.; Wang, J. Wearable biosensors for healthcare monitoring. Nat. Biotechnol. 2019, 37, 389–406. [Google Scholar] [CrossRef]
- Gao, J.W.; Gao, Y.K.; Han, Y.K.; Pang, J.B.; Wang, C.; Wang, Y.H.; Liu, H.; Zhang, Y.; Han, L. Ultrasensitive label-free miRNA sensing based on a flexible graphene field-effect transistor without functionalization. ACS Appl. Electron. Mater. 2020, 2, 1090–1098. [Google Scholar] [CrossRef]
- Sun, M.Y.; Wang, S.; Zhang, Y.H.; Zhang, Z.; Wang, S.; Wang, Z.H.; Chen, X.S.; Liu, H.; Zhang, Y.; Han, L. An ultrasensitive flexible biosensor enabled by high-performance graphene field-effect transistors with defect-free van der Waals contacts for breast cancer miRNA fast detection. Talanta 2025, 287, 127637. [Google Scholar] [CrossRef]
- Harvey, J.D.; Jena, P.V.; Baker, H.A.; Zerze, G.H.; Williams, R.M.; Galassi, T.V.; Roxbury, D.; Mittal, J.; Heller, D.A. A carbon nanotube reporter of microRNA hybridization events in vivo. Nat. Biomed. Eng. 2017, 1, 0041. [Google Scholar] [CrossRef]
- Lee, J.; Park, J.; Lee, J.Y.; Yeo, J.S. Contact transfer printing of side edge prefunctionalized nanoplasmonic arrays for flexible microRNA biosensor. Adv. Sci. 2016, 2, 1500121. [Google Scholar] [CrossRef]
- Jin, Y.X.; Wu, Z.; Li, L.; Yan, R.Q.; Zhu, J.L.; Wen, W.; Zhang, X.H.; Wang, S.F. Zinc-air battery-based self-powered sensor with high output power for ultrasensitive microRNA let-7a detection in cancer cells. Anal. Chem. 2022, 94, 14368–14376. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, H.; Wang, H. Machine learning and AI in cancer prognosis, prediction, and treatment selection: A critical approach. J. Multidiscip. Healthc. 2023, 16, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Sarker, I.H. Machine learning: Algorithms, real-world applications and research directions. SN Comput. Sci. 2021, 2, 160. [Google Scholar] [CrossRef]
- Zhao, H.Y.; Yan, Y.M.; Zhang, L.H.; Li, X.; Jia, L.; Ma, L.; Su, X. DNA molecular computing with weighted signal amplification for cancer miRNA biomarker diagnostics. Adv. Sci. 2025, 12, 2416490. [Google Scholar] [CrossRef]
- Zhang, Z.W.; Liu, X.Y.; Peng, C.Y.; Du, R.; Hong, X.Q.; Xu, J.; Chen, J.M.; Li, X.M.; Tang, Y.J.; Li, Y.W.; et al. Machine learning-aided identification of fecal extracellular vesicle microRNA signatures for noninvasive detection of colorectal cancer. ACS Nano 2025, 19, 10013–10025. [Google Scholar] [CrossRef]
- Wang, T.T.; Zhang, Y.; Su, H.N.; Yu, X.N.; Li, Q.; Liu, Y.; Cui, C.Y.; Huang, X.F.; Qing, L.S.; Luo, P. Target-triggered catalytic hairpin assembly of miR-21/155/1 coupled with dsDNA-reporter amplified detection for prediction of clinically significant coronary artery disease. Sens. Actuator B Chem. 2025, 426, 137053. [Google Scholar] [CrossRef]
- Pei, J.W.; Li, L.; Li, C.; Li, Z.Y.; Wu, Y.; Kuang, H.Y.; Ma, P.; Huang, L.; Liu, J.B.; Tian, G. Dumbbell probe-bridged CRISPR/Cas13a and nicking-mediated DNA cascade reaction for highly sensitive detection of colorectal cancer-related microRNAs. Biosens. Bioelectron. 2025, 273, 117190. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Y.; Wang, Y.F.; Wei, Y.; Lu, Z.D.; Ju, H.X.; Yan, F.; Liu, Y. Size-coded hydrogel microbeads for extraction-free serum multi-miRNAs quantifications with machine-learning-aided lung cancer subtypes classification. Nano Lett. 2024, 25, 453–460. [Google Scholar] [CrossRef]
- Zhang, X.W.; Qi, G.X.; Liu, M.X.; Yang, Y.F.; Wang, J.H.; Yu, Y.L.; Chen, S. Deep learning promotes profiling of multiple miRNAs in single extracellular vesicles for cancer diagnosis. ACS Sens. 2024, 9, 1555–1564. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lu, H.; Fang, D.; Zhang, Y.Y.; Tang, Y.T.; Zhao, S.S.; Yan, J.; Qin, X.J.; Shen, J.L.; Yang, F. DNA-encoded plasmonic bubbles aggregating dual-microRNA SERS signals for cancer diagnosis. Aggregate 2024, 5, e636. [Google Scholar] [CrossRef]
- Wen, Y.; Zhang, X.W.; Li, Y.Y.; Chen, S.; Yu, Y.L.; Wang, J.H. Ultramultiplex NaLnF4 nanosatellites combined with ICP-MS for exosomal multi-miRNA analysis and cancer classification. Anal. Chem. 2022, 94, 16196–16203. [Google Scholar] [CrossRef]
- Zhao, X.; Wang, Z.X.; Ji, X.L.; Bu, S.Y.; Fang, P.L.; Wang, Y.; Wang, M.X.; Yang, Y.; Zhang, W.J.; Leung, A.Y.H.; et al. Discrete single-cell microRNA analysis for phenotyping the heterogeneity of acute myeloid leukemia. Biomaterials 2022, 291, 121869. [Google Scholar] [CrossRef] [PubMed]

| Types of Biological Fluids | Diagnostic Potential for Cancer | Main Source | Advantages and Limitations | Reference |
|---|---|---|---|---|
| Serum | Various cancers | Encapsulated in exosomes or bound to proteins | This method avoids degradation of endogenous RNase, and miRNA can be specifically expressed | [9] |
| Passive leakage | ||||
| Active secretion | This method requires complex sample pretreatment and biological matrices | |||
| Saliva | Oral squamous cell carcinoma, colorectal cancer, liver cancer, head-and-neck cancer | Salivary glands, gingival crevice fluid, and desquamated oral epithelial cells | Saliva collection is noninvasive and painless | [10,11,12,13,14] |
| There is a risk of viral/bacterial contamination, and there are no dedicated miRNA isolation kits | ||||
| Urine | Renal tumors, bladder tumors, prostate cancer | Active secretion by various tissues and cells | Such miRNAs yield results directly related to urinary system disease; miRNAs from urine are stable | [15,16,17] |
| Secretion of urinary tract cells | There is a low limit of detection and a lack of internal references |
| Analytical Methods | Target | Sample | Required Enzymes | Template/Primers | Temperature (°C) | Time (min) | LOD | Ref |
|---|---|---|---|---|---|---|---|---|
| DSNSA (HPLC-FLD) | miR-21, miR-122, miR-155 | Human serum | 1: DSN enzyme | 3 | 40 | 200 | 0.26–0.39 fM | [31] |
| CHA-HCR-DNAzyme (chemiluminescence) | miR-21 | 10% human serum | 1: DNAzyme | 6 | 120 | 20.5 pM | [42] | |
| AP-LAMP (fluorescence) | miR-34a | MCF7 cell | 2: DNA polymerase and Bst DNA polymerase | 6 | 60–65 | 90 | 10 aM | [59] |
| RCA-LAMP (fluorescence) | miR-let7a | HCT-116 cell | 2: T4 RNA ligase and Bst DNA polymerase | 4 | 39 and 65 | 110 | 10 aM | [60] |
| CS-MoS2-CHA (electrochemistry) | miR-21 | Human serum | None | 4 | 37 | 210 | 16 aM | [61] |
| DNAzyme-CHA (electrochemistry) | miR-21 | Human serum | 1: DNAzyme | 6 | 37 | 285 | 1 aM | [62] |
| Gox and MOF nanozyme (ratiometric fluorescence) | miR-21 | Human serum | None | 4 | 37 | 280 | 0.8 aM | [63] |
| RCA-CDT (fluorescence) | miR-let7d | A549 cells | 1: pi29 DNA polymerase | 2 | 37 | 90 | 0.46 fM | [64] |
| DSN-mismatched CRISPR/Cas12a (fluorescence) | miR-let7a | Human serum | 2. Duplex-specific nuclease and Cas12a | 4 | 37 and 50 | 110 | 64.17 fM | [65] |
| DSN-based DNA-modified nanochannels (nanochannel) | miR-21 | MCF-7 and HeLa cells | 1. Duplex-specific nuclease | 1 | 50 | 120 | 1 fM | [66] |
| Analytical Methods | Diseases and Target miRNAs | Sample | Sample Preparation | Determination Time (min) | LOQ | Ref |
|---|---|---|---|---|---|---|
| SDA and CRISPR/Cas12a (spICP-MS) | Breast cancer, miR-21 | Human serum and blood | Total RNA extraction | 145 | 0.5 fM | [67] |
| SDA and SPAAC ligation (electrochemistry) | Non-small-cell lung cancer, miR-21 | Human tissue | Total RNA extraction | 110 | 100 aM | [68] |
| PICA (fluorescence) | Colorectal cancer, miR-21 | 10% Human serum | Total RNA extraction | 30 | 20 pM | [69] |
| Au metallene/luminescence nanovesicle (electrochemiluminescence) | Coronary artery calcification, miR-126-3p | Human blood | Total RNA extraction | 137 | 1 fM | [70] |
| MS-ACPE (fluorescence) | Oral squamous cell carcinoma, miR-31 | Human saliva | High-speed centrifuging | 440 | 10 pM | [71] |
| Dual-mode signal amplification (fluorescence and colorimetric) | Gestational diabetes mellitus, miR-135a | Human plasma | Total RNA extraction | 37 | 0.56 and 8.3 nM | [72] |
| AIE-split G-quadruplex (fluorescence) | Acute kidney injury, miR-21 | 10% Human urine | Centrifugation | 30 | 10.36 fM | [73] |
| AuNP@LH (fluorescence) | Systemic lupus erythematosus, miR-146a, miR-29c, and miR-150 | Human urine | Centrifugal Filter (100 kDa) | 110 | 50 pM | [74] |
| Mango II arrays (fluorescence) | Lung and liver cancer, miR-21 | 50% Human serum | Centrifugation and incubated at 95 °C | 180 | 10 fM | [75] |
| Plasmonic nanocavity-modulated ECL (electrochemiluminescence) | Gastric cancer, exosomal miR-223-3p | Human ascites | Centrifugation and exosome Kit | Not mentioned | 1 fM | [76] |
| Analytical Methods | Diseases and Target miRNAs | Sample | Sample Preparation Method | Determination Time (min) | LOQ | Ref |
|---|---|---|---|---|---|---|
| Microfluidics and smartphone (DSNSA) | Type 2 diabetes, miR-21 | Human serum | Total RNA extraction | 150 | 1 pM | [80] |
| Smartphone (EXPAR) | Bladder cancer, prostate cancer, miR-223, and miR-155 | Human urine | Total RNA extraction | 22.5 | 1 pM | [81] |
| Smartphone (SDA and HCR) | Lung cancer and miR-21 | 1% Human serum | Not mentioned | 60 | 0.1 fM | [82] |
| Lateral flow assay and smartphone (RCA with CRISPR–Cas12a) | Pancreatic cancer, miR-21, miR-451a, and miR-1246 | Human plasma | Total RNA extraction | 20 to 180 | 20, 10, and 50 fM | [83] |
| Lateral flow assay (CHA and AuNPs) | Liver cancer and miR-223 | Human serum | Not mentioned | 40 | 1 pM | [84] |
| Analytical Methods | Diseases and Target miRNAs | Sample | Diagnosis Accuracy | Diagnosis Time (min) | AI Types | Ref |
|---|---|---|---|---|---|---|
| Localized CHA (LCHA) and PMSD | Non-small-cell lung cancer, miR-182, miR-21, miR-148, let-7b, miR-143, and miR-30a | Human tissues | 92.86% | 150 (sample to diagnosis) | Machine learning | [93] |
| CRISPR/Cas13a | Colorectal cancer, miR-16–2, miR-375, miR-378a, and miR-7 | Human fecal | 97.4% | 100 | Machine learning | [94] |
| Target-triggered catalytic hairpin assembly | Coronary artery disease, miR-21, miR-155, miR-1 | 50% Human serum | 87.5% | 90 | Machine learning | [95] |
| Dumbbell probe-mediated CRISPR/Cas13a with nicking-induced DNA cascade reaction (DP-bridged Cas13a/NDCR) | Colorectal cancer, miR-17, miR-21, miR-182, and miR-223 | Human blood plasma | 100% | 120 | Machine learning | [96] |
| Size-coded hydrogel microbeads | Lung cancer, miR-21, miR-205, and miR-375 | Human serum | 80% (lung cancer sub-type prediction) | 120 | Machine learning | [97] |
| Nanoflare probe, CHA amplification, and total internal reflection fluorescence (TIRF) | Lung cancer, breast cancers, colon cancers, and cervical cancers, miR-21, miR-122, mir-375 | Human plasma | 100% | 120 | Deep learning | [98] |
| DNA-encoded plasmonic-bubble-driven SERS | Liver cancer, miR-21, and miR-155 | Human blood | 83.3% | Not mentioned | Decision-tree-based classifier algorithms | [99] |
| Nanosatellites (magnetic beads (MBs) @ NaLnF4) and CHA amplification in combination with ICP-MS | Breast cancer, lung cancer, stomach cancer, colon cancer, and cervical cancer, miR-200b, miR-21, miR-151, miR-155, miR-139, let-7b, miR-191, miR-214, let-7f, and miR-30e | Human plasma | 100% | 90 | Linear discriminant analysis (LDA) | [100] |
| Nanoneedle-based discrete analysis | Acute myeloid leukemia (AML), miR-155, miR-21, miR-125b, miR-99a, miR-223, miR-29a, miR-126, miR-181a, and miR-196b | Living AML cells | 90% | 120 | Machine learning | [101] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Sun, X.; Cai, S. Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis. Biosensors 2025, 15, 395. https://doi.org/10.3390/bios15060395
Han Y, Sun X, Cai S. Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis. Biosensors. 2025; 15(6):395. https://doi.org/10.3390/bios15060395
Chicago/Turabian StyleHan, Yu, Xin Sun, and Sheng Cai. 2025. "Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis" Biosensors 15, no. 6: 395. https://doi.org/10.3390/bios15060395
APA StyleHan, Y., Sun, X., & Cai, S. (2025). Advances in Research on Isothermal Signal Amplification Mediated MicroRNA Detection of Clinical Samples: Application to Disease Diagnosis. Biosensors, 15(6), 395. https://doi.org/10.3390/bios15060395








