A Multi-Domain Feature Fusion CNN for Myocardial Infarction Detection and Localization
Abstract
1. Introduction
- •
- The effective generation of frequency-domain spectrum and time-frequency images, combined with time-domain ECG signals, creates multi-domain information inputs. Leveraging S-transform for time-frequency representation and GAF for spectral analysis, this approach significantly enhances feature representation and generalization ability, thereby improving MI detection and localization accuracy.
- •
- Introduction of a novel MFF–CNN for automatically extracting deep features from input ECG signals, spectrum images derived from GAF, and time-frequency images generated by S-transform. This model integrates complementary characteristics from each domain, leveraging the strengths of both S-transform and GAF in capturing MI-related features, enabling effective multi-domain feature learning for MI detection and localization.
- •
- The MFF–CNN achieves superior results compared with existing methods, demonstrating its effectiveness in detecting and localizing MI. On the well-known PTB diagnostic ECG database, our proposed method sets a new benchmark in inter-patient evaluation with a detection accuracy of 99.98% and a location accuracy of 84.86%. Furthermore, in generalization tests using the PTB data for training and the PTB-XL data for validation, the MFF–CNN obtains a location accuracy of 78.21%, far surpassing that of single-domain models.
2. Materials and Methods
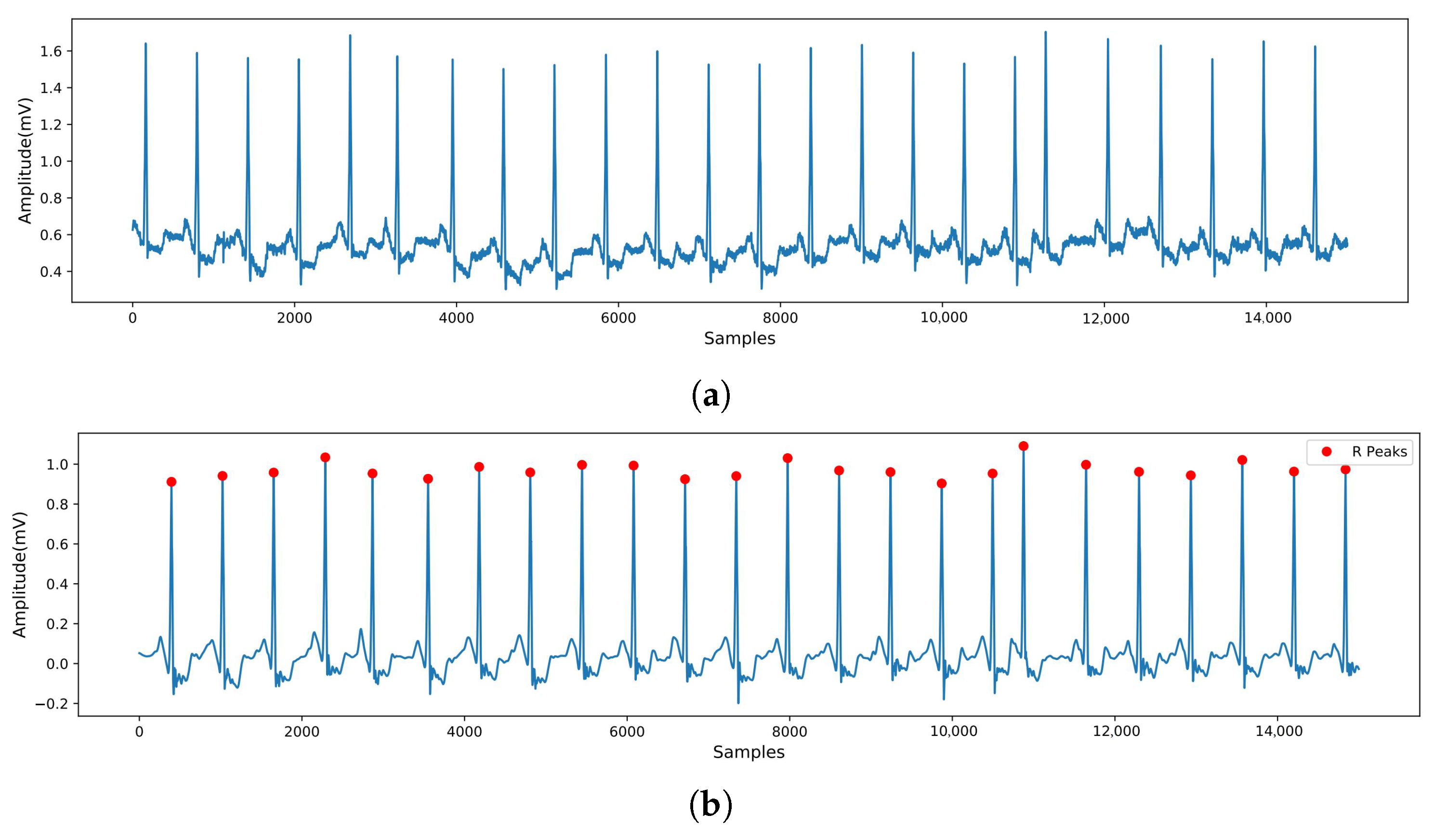
2.1. Data Preprocessing
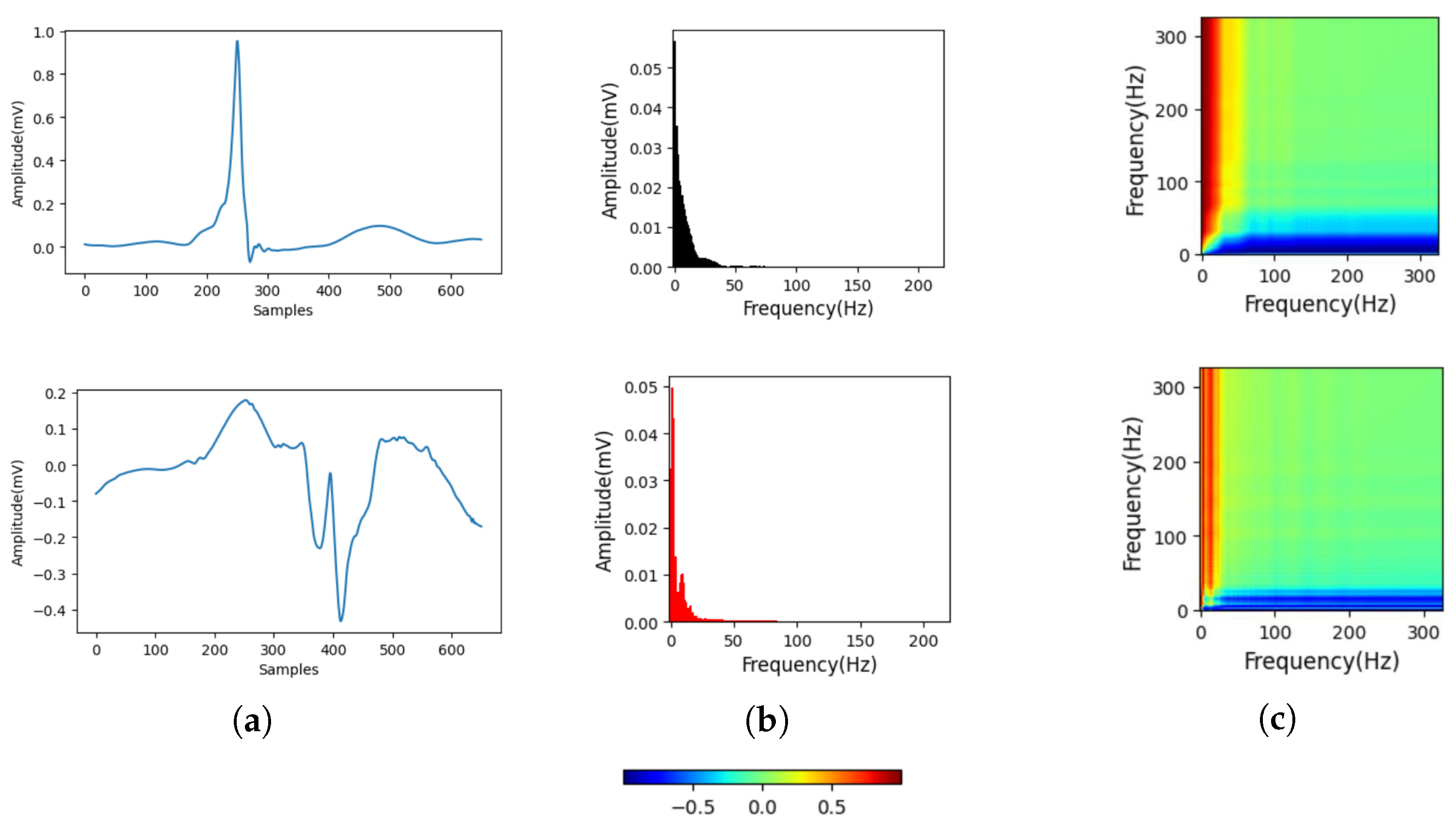
2.2. Gramian Angular Field for Spectrum Image Generation
2.3. S Transform
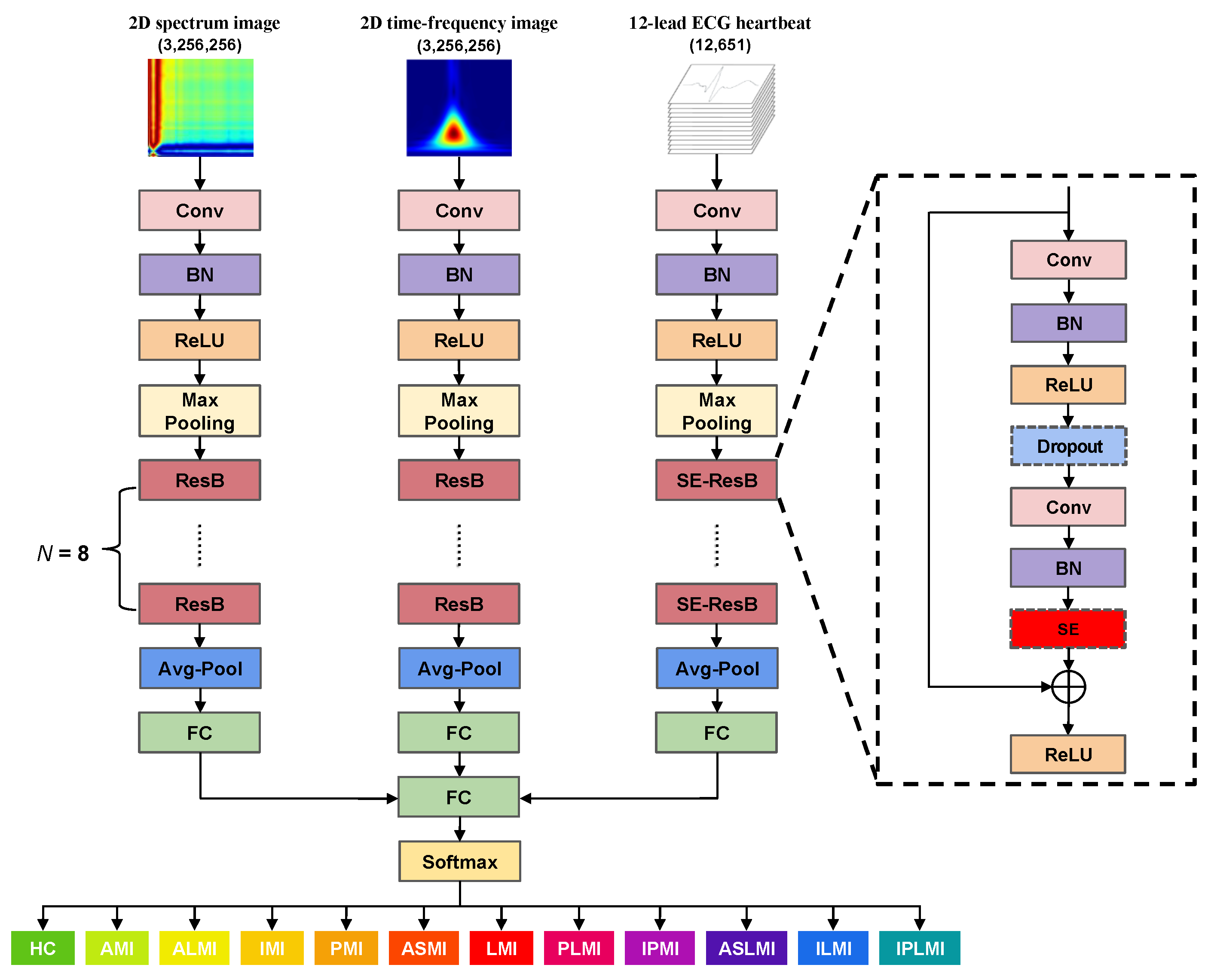
2.4. MFF–CNN Network
3. Experimental Results
3.1. Experimental Settings
3.2. Evaluation Indicators
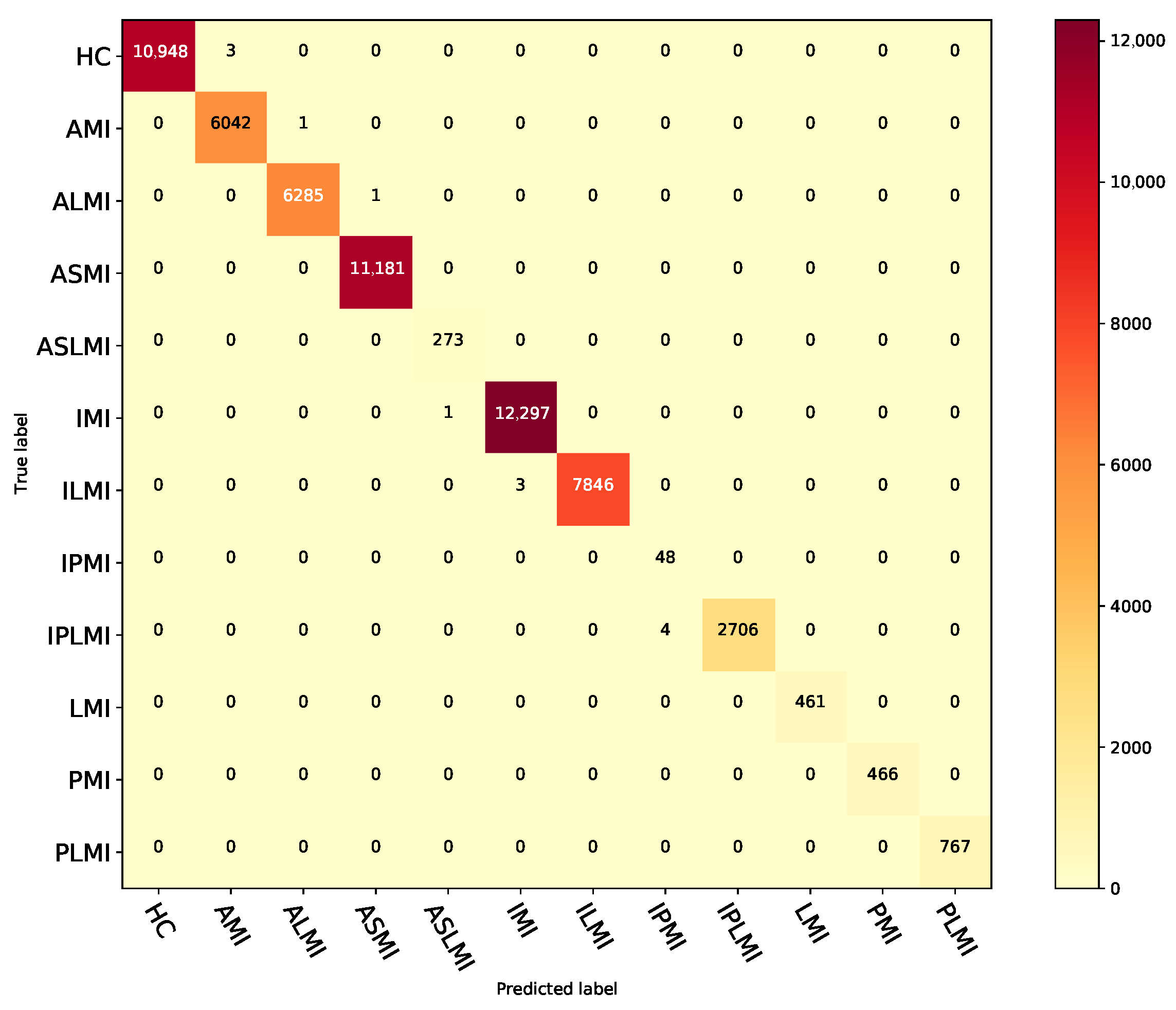
3.3. Intra-Patient Evaluation of the Performance of MI Detection and Localization on the PTB Database
3.4. Inter-Patient Evaluation of the Performance of MI Detection and Localization on the PTB Database
3.5. Ablation Experiments Under the Inter-Patient Paradigm
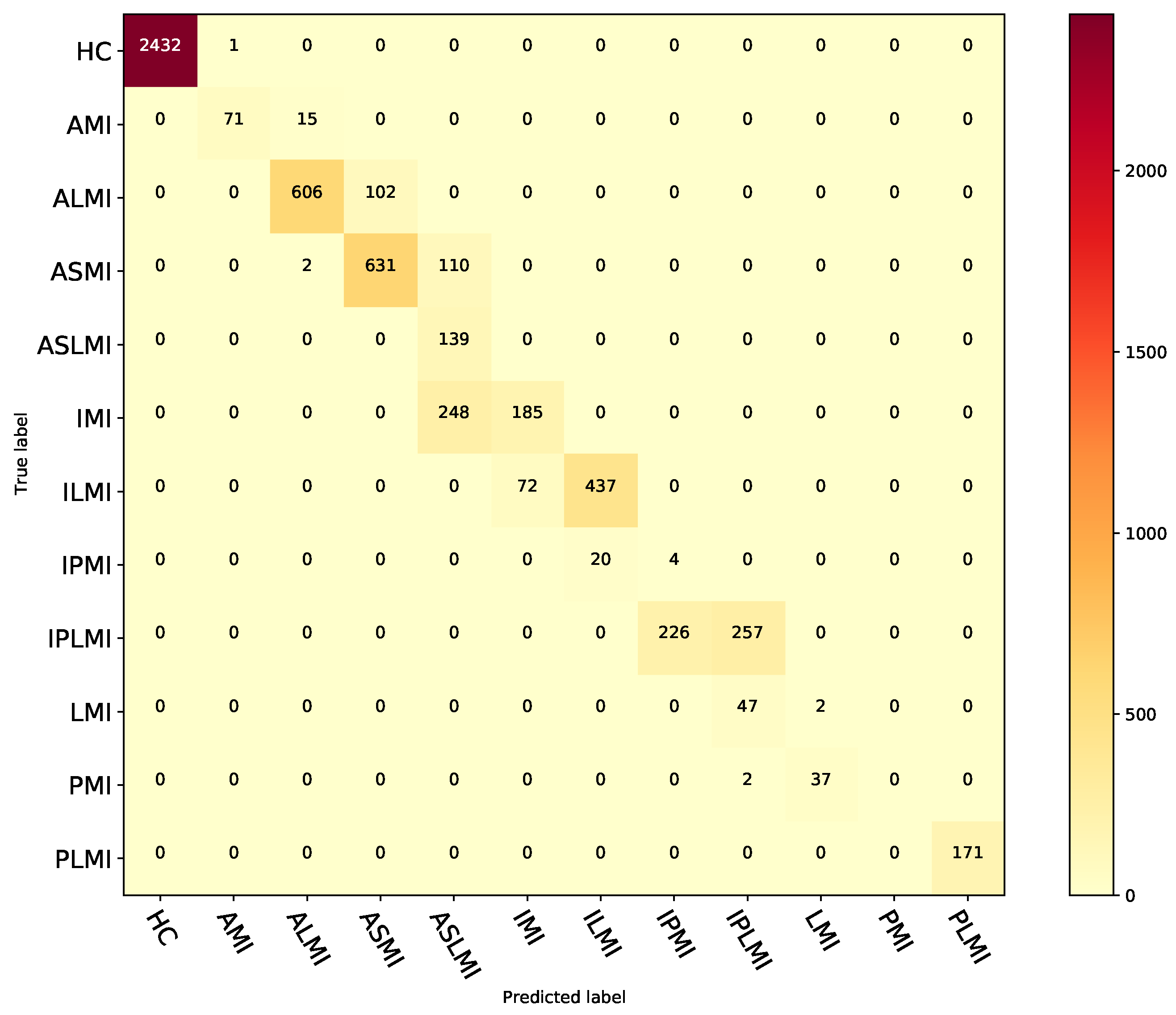
3.6. Generalizability Evaluation of the Proposed Method on the PTB-XL Dataset
3.7. Comparison Results with Existing Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wrold Health Organization. Global Health Estimates: Life Expectancy and Leading Causes of Death and Disability. Available online: https://www.who.int/data/gho/data/themes/mortality-and-global-health-estimates (accessed on 31 May 2023).
- Haddad, H.; Mielniczuk, L.; Davies, R.A. Recent advances in the management of chronic heart failure. Curr. Opin. Cardiol. 2012, 27, 161–168. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Han, Y. Current situation of ST-segment elevation myocardial infarction rescue in China and optimal management strategies we can use today. Zhonghua Xin Xue Guan Bing Za Zhi 2019, 47, 82–84. [Google Scholar] [PubMed]
- Benjamin, E.J.; Muntner, P.; Alonso, A.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Das, S.R.; et al. Heart disease and stroke statistics—2019 Update: A report from the American Heart Association. Circulation 2019, 139, e56–e528. [Google Scholar] [PubMed]
- Wan, X.; Liao, T.; Gong, W.; Liang, Y.; Wu, M.; Wang, B. A Precise Respiratory and Heart Rate Detection Method for Millimeter-Wave Radar. J. Mech. Med. Biol. 2024, 24, 2450004. [Google Scholar] [CrossRef]
- Sharma, L.; Tripathy, R.; Dandapat, S. Multiscale energy and eigenspace approach to detection and localization of myocardial infarction. IEEE Trans. Biomed. Eng. 2015, 62, 1827–1837. [Google Scholar] [CrossRef]
- Sridhar, C.; Lih, O.S.; Jahmunah, V.; Koh, J.E.; Ciaccio, E.J.; San, T.R.; Arunkumar, N.; Kadry, S.; Rajendra Acharya, U. Accurate detection of myocardial infarction using non linear features with ECG signals. J. Ambient Intell. Humaniz. Comput. 2021, 12, 3227–3244. [Google Scholar] [CrossRef]
- Fatimah, B.; Singh, P.; Singhal, A.; Pramanick, D.; Pranav, S.; Pachori, R.B. Efficient detection of myocardial infarction from single lead ECG signal. Biomed. Signal Process. Control 2021, 68, 102678. [Google Scholar] [CrossRef]
- Dohare, A.K.; Kumar, V.; Kumar, R. Detection of myocardial infarction in 12 lead ECG using support vector machine. Appl. Soft Comput. 2018, 64, 138–147. [Google Scholar] [CrossRef]
- Han, C.; Shi, L. Automated interpretable detection of myocardial infarction fusing energy entropy and morphological features. Comput. Methods Programs Biomed. 2019, 175, 9–23. [Google Scholar] [CrossRef]
- Wan, X.; Liu, Y.; Mei, X.; Ye, J.; Zeng, C.; Chen, Y. A novel atrial fibrillation automatic detection algorithm based on ensemble learning and multi-feature discrimination. Med. Biol. Eng. Comput. 2024, 62, 1809–1820. [Google Scholar] [CrossRef]
- Acharya, U.R.; Fujita, H.; Sudarshan, V.K.; Oh, S.L.; Adam, M.; Koh, J.E.; Tan, J.H.; Ghista, D.N.; Martis, R.J.; Chua, C.K.; et al. Automated detection and localization of myocardial infarction using electrocardiogram: A comparative study of different leads. Knowl. Based Syst. 2016, 99, 146–156. [Google Scholar] [CrossRef]
- Arif, M.; Malagore, I.A.; Afsar, F.A. Detection and localization of myocardial infarction using k-nearest neighbor classifier. J. Med. Syst. 2012, 36, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mei, X.; Chen, Y.; Wan, X. An effective premature ventricular contraction detection algorithm based on adaptive template matching and characteristic recognition. Signal Image Video Process. 2024, 18, 2811–2818. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, W.; Jia, D.; Hu, J.; Li, Z.; Yan, C.; You, T. Myocardial infarction detection based on multi-lead ensemble neural network. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 2614–2617. [Google Scholar]
- Feng, K.; Pi, X.; Liu, H.; Sun, K. Myocardial infarction classification based on convolutional neural network and recurrent neural network. Appl. Sci. 2019, 9, 1879. [Google Scholar] [CrossRef]
- Xiong, P.; Xue, Y.; Zhang, J.; Liu, M.; Du, H.; Zhang, H.; Hou, Z.; Wang, H.; Liu, X. Localization of myocardial infarction with multi-lead ECG based on DenseNet. Comput. Methods Programs Biomed. 2021, 203, 106024. [Google Scholar] [CrossRef]
- Strodthoff, N.; Strodthoff, C. Detecting and interpreting myocardial infarction using fully convolutional neural networks. Physiol. Meas. 2019, 40, 015001. [Google Scholar] [CrossRef]
- Qu, J.; Sun, Q.; Wu, W.; Zhang, F.; Liang, C.; Chen, Y.; Wang, C. An interpretable shapelets-based method for myocardial infarction detection using dynamic learning and deep learning. Physiol. Meas. 2024, 45, 035001. [Google Scholar] [CrossRef]
- Zhang, J.; Lin, F.; Xiong, P.; Du, H.; Zhang, H.; Liu, M.; Hou, Z.; Liu, X. Automated detection and localization of myocardial infarction with staked sparse autoencoder and treebagger. IEEE Access 2019, 7, 70634–70642. [Google Scholar] [CrossRef]
- Huang, J.; Chen, B.; Yao, B.; He, W. ECG arrhythmia classification using STFT-based spectrogram and convolutional neural network. IEEE Access 2019, 7, 92871–92880. [Google Scholar] [CrossRef]
- Al Rahhal, M.M.; Bazi, Y.; Al Zuair, M.; Othman, E.; BenJdira, B. Convolutional neural networks for electrocardiogram classification. J. Med. Biol. Eng. 2018, 38, 1014–1025. [Google Scholar] [CrossRef]
- Hao, P.; Gao, X.; Li, Z.; Zhang, J.; Wu, F.; Bai, C. Multi-branch fusion network for Myocardial infarction screening from 12-lead ECG images. Comput. Methods Programs Biomed. 2020, 184, 105286. [Google Scholar] [CrossRef] [PubMed]
- Swain, S.S.; Patra, D.; Singh, Y.O. Automated detection of myocardial infarction in ECG using modified Stockwell transform and phase distribution pattern from time-frequency analysis. Biocybern. Biomed. Eng. 2020, 40, 1174–1189. [Google Scholar] [CrossRef]
- Zhang, G.; Si, Y.; Wang, D.; Yang, W.; Sun, Y. Automated detection of myocardial infarction using a gramian angular field and principal component analysis network. IEEE Access 2019, 7, 171570–171583. [Google Scholar] [CrossRef]
- Yousuf, A.; Hafiz, R.; Riaz, S.; Farooq, M.; Riaz, K.; Rahman, M.M.U. Myocardial Infarction Detection from ECG: A Gramian Angular Field-based 2D-CNN Approach. arXiv 2023, arXiv:2302.13011. [Google Scholar]
- Bousseljot, R.; Kreiseler, D.; Schnabel, A. Nutzung der EKG-Signaldatenbank CARDIODAT der PTB über das Internet. Biomed. Tech./Biomed. Eng. 1995, 40, 317–318. [Google Scholar] [CrossRef]
- Wagner, P.; Strodthoff, N.; Bousseljot, R.; Samek, W.; Schaeffter, T. PTB-XL, a large publicly available electrocardiography dataset (version 1.0.3). PhysioNet 2022. [Google Scholar] [CrossRef]
- Reddy, G.U.; Muralidhar, M.; Varadarajan, S. ECG De-Noising using improved thresholding based on Wavelet transforms. Int. J. Comput. Sci. Netw. Secur. 2009, 9, 221–225. [Google Scholar]
- Donoho, D.L.; Johnstone, I.M. Ideal spatial adaptation by wavelet shrinkage. biometrika 1994, 81, 425–455. [Google Scholar] [CrossRef]
- Pan, J.; Tompkins, W.J. A real-time QRS detection algorithm. IEEE Trans. Biomed. Eng. 1985, 32, 230–236. [Google Scholar] [CrossRef]
- Wang, Z.; Oates, T. Imaging time-series to improve classification and imputation. arXiv 2015, arXiv:1506.00327. [Google Scholar]
- Keogh, E.J.; Pazzani, M.J. Scaling up dynamic time warping for datamining applications. In Proceedings of the Sixth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Boston, MA, USA, 20–23 August 2000; pp. 285–289. [Google Scholar]
- Stockwell, R.G.; Mansinha, L.; Lowe, R. Localization of the complex spectrum: The S transform. IEEE Trans. Signal Process. 1996, 44, 998–1001. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhang, Y.; Deng, Y.; Zhang, X. ECG authentication system design incorporating a convolutional neural network and generalized S-Transformation. Comput. Biol. Med. 2018, 102, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7132–7141. [Google Scholar]
- Fang, R.; Lu, C.C.; Chuang, C.T.; Chang, W.H. A visually interpretable detection method combines 3-D ECG with a multi-VGG neural network for myocardial infarction identification. Comput. Methods Programs Biomed. 2022, 219, 106762. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, F.; Huang, Q.; Chang, S.; Wang, H.; He, J. MFB-CBRNN: A hybrid network for MI detection using 12-lead ECGs. IEEE J. Biomed. Health Inform. 2019, 24, 503–514. [Google Scholar] [CrossRef]
- Han, C.; Shi, L. ML–ResNet: A novel network to detect and locate myocardial infarction using 12 leads ECG. Comput. Methods Programs Biomed. 2020, 185, 105138. [Google Scholar] [CrossRef]
- Fu, L.; Lu, B.; Nie, B.; Peng, Z.; Liu, H.; Pi, X. Hybrid network with attention mechanism for detection and location of myocardial infarction based on 12-lead electrocardiogram signals. Sensors 2020, 20, 1020. [Google Scholar] [CrossRef]
- Jian, J.Z.; Ger, T.R.; Lai, H.H.; Ku, C.M.; Chen, C.A.; Abu, P.A.R.; Chen, S.L. Detection of myocardial infarction using ECG and multi-scale feature concatenate. Sensors 2021, 21, 1906. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, W.; Zhang, S.; Xu, L.; Zhu, B.; Cui, H.; Geng, N.; Han, H.; Greenwald, S.E. Detection and localization of myocardial infarction based on multi-scale resnet and attention mechanism. Front. Physiol. 2022, 13, 783184. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, M.; Xiong, P.; Du, H.; Zhang, H.; Sun, G.; Hou, Z.; Liu, X. Automated Localization of Myocardial Infarction of Image-Based Multilead ECG Tensor With Tucker2 Decomposition. IEEE Trans. Instrum. Meas. 2022, 71, 2501215. [Google Scholar] [CrossRef]
- Han, C.; Sun, J.; Bian, Y.; Que, W.; Shi, L. Automated detection and localization of myocardial infarction with interpretability analysis based on deep learning. IEEE Trans. Instrum. Meas. 2023, 72, 2508512. [Google Scholar] [CrossRef]
- Guo, L.; Zhan, Q.; Yang, J.; An, Y.; Long, J.; Ma, N. Lead-grouped multi-stage learning for myocardial infarction localization. Methods 2025, 234, 315–323. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, C.; Ristaniemi, T.; Cong, F. Detection of Myocardial Infarction from Multi-lead ECG using Dual-Q Tunable Q-Factor Wavelet Transform. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 1496–1499. [Google Scholar]
- Campolo, M.; Labate, D.; La Foresta, F.; Morabito, F.; Lay-Ekuakille, A.; Vergallo, P. ECG-derived respiratory signal using Empirical Mode Decomposition. In Proceedings of the 2011 IEEE International Symposium on Medical Measurements and Applications, Bari, Italy, 30–31 May 2011; pp. 399–403. [Google Scholar]
- Liu, J.; Zhang, C.; Zhu, Y.; Ristaniemi, T.; Parviainen, T.; Cong, F. Automated detection and localization system of myocardial infarction in single-beat ECG using Dual-Q TQWT and wavelet packet tensor decomposition. Comput. Methods Programs Biomed. 2020, 184, 105120. [Google Scholar] [CrossRef]


| Types of MI | No. of Patients | No. of Records | No. of 12-Lead Heartbeats |
|---|---|---|---|
| Anterior MI (AMI) | 17 | 47 | 6043 |
| Anterior Lateral MI (ALMI) | 16 | 43 | 6286 |
| Anterior Septal MI (ASMI) | 27 | 77 | 11,181 |
| Anterior Septal Lateral MI (ASLMI) | 1 | 2 | 273 |
| Inferior MI (IMI) | 30 | 89 | 12,298 |
| Inferior Lateral MI (ILMI) | 23 | 56 | 7849 |
| Inferior Posterior MI (IPMI) | 1 | 1 | 48 |
| Inferior Posterior Lateral MI(IPLMI) | 8 | 19 | 2710 |
| Lateral MI (LMI) | 1 | 3 | 461 |
| Posterior MI (PMI) | 1 | 4 | 466 |
| Posterior Lateral MI (PLMI) | 2 | 5 | 767 |
| Healthy Control (HC) | 52 | 80 | 10,951 |
| Total | 179 | 426 | 59,333 |
| Types of MI | No. of Patients | No. of Records | No. of 12-Lead Heartbeats |
|---|---|---|---|
| Anterior MI (AMI) | 286 | 290 | 2434 |
| Anterior Lateral MI (ALMI) | 181 | 208 | 1585 |
| Anterior Septal MI (ASMI) | 1780 | 2017 | 16,590 |
| Inferior MI (IMI) | 2055 | 2331 | 17,815 |
| Inferior Lateral MI (ILMI) | 350 | 394 | 2898 |
| Inferior Posterior MI (IPMI) | 26 | 30 | 218 |
| Inferior Posterior Lateral MI(IPLMI) | 49 | 50 | 424 |
| Lateral MI (LMI) | 125 | 135 | 1050 |
| Posterior MI (PMI) | 14 | 14 | 137 |
| Healthy Control (HC) | 1967 | 2184 | 21,854 |
| Total | 6833 | 7653 | 65,005 |
| Layer Name | ResNet18 | SE-ResNet18 |
|---|---|---|
| Conv1 | 7 × 7, 64, stride 2 | (15,), 64, stride 2 |
| Conv2_x | 3 × 3, max pool, stride 2 | (3,), max pool, stride 2 |
| Conv3_x | ||
| Conv4_x | ||
| Conv5_x | ||
| Average pool, , softmax | Average pool, Dropout, , softmax |
| Model Input | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 99.98 | 99.98 | 99.98 | 99.94 | 99.98 |
| GAF image | 99.93 | 99.96 | 99.95 | 99.78 | 99.95 |
| ST image | 99.97 | 99.97 | 99.98 | 99.95 | 99.98 |
| ECG signal & GAF image & ST image (MFF–CNN) | 99.99 | 100 | 99.99 | 99.97 | 100 |
| Fold | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| 1 | 99.97 | 99.99 | 99.96 | 99.93 | 99.98 |
| 2 | 99.99 | 99.98 | 99.98 | 99.97 | 99.98 |
| 3 | 99.98 | 99.99 | 99.97 | 99.95 | 99.99 |
| 4 | 99.99 | 99.99 | 99.98 | 99.96 | 99.99 |
| 5 | 99.96 | 99.98 | 99.95 | 99.92 | 99.97 |
| 6 | 99.98 | 99.99 | 99.97 | 99.94 | 99.98 |
| 7 | 99.99 | 99.98 | 99.98 | 99.97 | 99.98 |
| 8 | 99.97 | 99.99 | 99.96 | 99.93 | 99.98 |
| 9 | 99.99 | 99.99 | 99.99 | 99.96 | 99.99 |
| 10 | 99.98 | 99.98 | 99.97 | 99.95 | 99.98 |
| Model Input | (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 99.91 | 99.75 | 99.92 | 99.99 | 99.83 |
| GAF image | 99.65 | 99.74 | 99.53 | 99.96 | 99.63 |
| ST image | 99.91 | 99.94 | 99.95 | 99.99 | 99.94 |
| ECG signal & GAF image & ST image (MFF–CNN) | 99.98 | 99.97 | 99.32 | 99.99 | 99.63 |
| Class | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| HC | 99.99 | 99.97 | 100 | 100 | 99.99 |
| AMI | 99.99 | 99.98 | 99.95 | 99.99 | 99.97 |
| ALMI | 100 | 99.98 | 99.98 | 100 | 99.98 |
| ASMI | 100 | 100 | 99.99 | 100 | 100 |
| ASLMI | 100 | 100 | 99.64 | 100 | 99.82 |
| IMI | 99.99 | 99.99 | 99.98 | 99.99 | 99.98 |
| ILMI | 99.99 | 99.96 | 100 | 100 | 99.98 |
| IPMI | 99.99 | 100 | 92.31 | 99.99 | 96 |
| IPLMI | 99.99 | 99.85 | 100 | 100 | 99.93 |
| LMI | 100 | 100 | 100 | 100 | 100 |
| PMI | 100 | 100 | 100 | 100 | 100 |
| PLMI | 100 | 100 | 100 | 100 | 100 |
| Average | 99.99 | 99.97 | 99.32 | 99.99 | 99.63 |
| Model Input | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 80.06 | 88.53 | 84.94 | 56.66 | 86.70 |
| GAF image | 73.18 | 91.59 | 76.51 | 22.31 | 83.37 |
| ST image | 79.19 | 90.00 | 83.07 | 49.35 | 86.40 |
| ECG signal & GAF image & ST image (MFF–CNN) | 99.98 | 100 | 99.97 | 99.95 | 99.98 |
| Fold | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| 1 | 99.97 | 100 | 99.96 | 99.92 | 99.98 |
| 2 | 99.99 | 99.99 | 99.98 | 99.97 | 99.99 |
| 3 | 99.98 | 100 | 99.97 | 99.95 | 99.99 |
| 4 | 99.99 | 99.99 | 99.98 | 99.96 | 99.98 |
| 5 | 99.96 | 99.98 | 99.94 | 99.92 | 99.97 |
| 6 | 99.98 | 100 | 99.97 | 99.94 | 99.98 |
| 7 | 99.99 | 99.99 | 99.98 | 99.97 | 99.99 |
| 8 | 99.98 | 99.99 | 99.96 | 99.93 | 99.97 |
| 9 | 99.99 | 99.98 | 99.99 | 99.97 | 99.99 |
| 10 | 99.98 | 99.99 | 99.97 | 99.96 | 99.98 |
| Model Input | (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 56.37 | 49.27 | 55.36 | 96.11 | 55.32 |
| GAF image | 58.65 | 52.53 | 54.11 | 95.45 | 50.79 |
| ST image | 61.66 | 55.37 | 54.64 | 96.20 | 63.89 |
| ECG signal & GAF image & ST image (MFF–CNN) | 84.86 | 62.96 | 64.03 | 98.68 | 60.59 |
| Class | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| HC | 99.98 | 99.96 | 100 | 100 | 99.98 |
| AMI | 99.72 | 82.56 | 99.98 | 98.61 | 89.87 |
| ALMI | 97.95 | 85.59 | 99.67 | 97.27 | 91.06 |
| ASMI | 96.32 | 84.93 | 97.99 | 86.08 | 85.5 |
| ASLMI | 93.85 | 100 | 93.69 | 27.97 | 43.71 |
| IMI | 94.5 | 42.73 | 98.66 | 71.98 | 53.62 |
| ILMI | 98.42 | 85.85 | 99.62 | 95.62 | 90.48 |
| IPMI | 95.77 | 16.67 | 96.1 | 1.74 | 3.15 |
| IPLMI | 95.27 | 53.21 | 99.08 | 83.99 | 65.15 |
| LMI | 98.56 | 4.08 | 99.36 | 5.13 | 4.55 |
| PMI | 99.33 | 0 | 100 | 0 | 0 |
| PLMI | 100 | 100 | 100 | 100 | 100 |
| Average | 97.47 | 62.96 | 98.68 | 64.03 | 60.59 |
| Model Input | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 56.37 | 49.27 | 55.36 | 96.11 | 55.32 |
| GAF image | 58.65 | 52.53 | 54.11 | 95.45 | 50.79 |
| ST image | 61.66 | 55.37 | 54.64 | 96.20 | 63.89 |
| ECG signal & GAF image | 68.42 | 56.21 | 58.37 | 97.12 | 59.84 |
| ECG signal & ST image | 72.15 | 59.83 | 61.24 | 97.85 | 63.52 |
| GAF image & ST image | 65.33 | 53.97 | 57.89 | 96.78 | 57.12 |
| ECG signal & GAF image & ST image (MFF–CNN) | 84.86 | 62.96 | 64.03 | 98.68 | 60.59 |
| Model Input | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 65.89 | 83.99 | 70.36 | 30.15 | 76.57 |
| GAF image | 71.95 | 86.66 | 74.98 | 42.92 | 80.40 |
| ST image | 66.51 | 85.30 | 70.46 | 29.41 | 77.17 |
| ECG signal & GAF image & ST image (MFF–CNN) | 91.57 | 100 | 88.73 | 74.93 | 94.83 |
| Model Input | (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| ECG signal | 28.73 | 12.90 | 12.95 | 91.14 | 14.99 |
| GAF image | 29.74 | 13.26 | 14.13 | 91.54 | 15.91 |
| ST image | 29.73 | 12.79 | 13.15 | 91.18 | 15.23 |
| ECG signal & GAF image & ST image (MFF–CNN) | 78.21 | 73.95 | 63.97 | 96.33 | 61.49 |
| Class | Acc (%) | Sen (%) | Pre (%) | Spe (%) | F1 (%) |
|---|---|---|---|---|---|
| HC | 91.57 | 74.94 | 100 | 100 | 85.67 |
| AMI | 90.30 | 65.74 | 22.62 | 91.25 | 33.66 |
| ALMI | 98.00 | 90.28 | 55.55 | 98.19 | 68.78 |
| ASMI | 98.68 | 95.76 | 99.03 | 99.68 | 97.37 |
| IMI | 78.61 | 24.17 | 91.58 | 99.16 | 38.25 |
| ILMI | 76.29 | 32.99 | 6.63 | 78.31 | 11.03 |
| IPMI | 96.89 | 79.36 | 8.05 | 96.95 | 14.62 |
| IPLMI | 99.79 | 86.79 | 82.14 | 99.88 | 84.40 |
| LMI | 99.81 | 93.81 | 94.26 | 99.91 | 94.03 |
| PMI | 99.94 | 95.62 | 79.88 | 99.95 | 87.04 |
| Average | 92.99 | 73.95 | 63.97 | 96.33 | 61.49 |
| Methods | Leads and Database | Detection or Location | Performance | |
|---|---|---|---|---|
| Intra-Patient | Inter-Patient | |||
| CNN and BiLSTM [38] | 12 leads PTB | Detection | Acc = 99.90% Se = 99.97% Sp = 99.54% | Acc = 93.08% Se = 94.42% Sp = 86.29% |
| CNN based on ResNet [39] | 12 leads PTB | Detection and location | Detection: Acc = 99.92% Se = 99.98% Location: Acc = 99.72% Se = 99.63% | Detection: Acc = 95.49% Se = 94.85% Location: Acc = 55.74% Se = 47.58% |
| CNN and BiGRU with attention [40] | 12 leads PTB | Detection and location | Detection: Acc = 99.93% Se = 99.99% Location: Acc = 99.11% Se = 99.02% | Detection: Acc = 96.50% Se = 97.10% Location Acc = 62.94% Se = 63.97% |
| Multi-scale feature [41] | 12 leads PTB | Detection and location | / | Detection: Acc = 95.76% Location: Acc = 61.82% |
| DenseNet [17] | 12 leads PTB | Location | Acc = 99.87% Se = 99.84% Sp = 99.98% | / |
| Multi-scale ResNet with attention [42] | 12 leads PTB | Detection and location | Detection: Acc = 99.98% Se = 99.94% Location: Acc = 99.79% Se = 99.88% | / |
| 3D ECG images [37] | 12 leads PTB | Detection | Acc = 100.00% Se = 100.00% Sp = 100.00% | Acc = 95.65% Se = 97.34% Sp = 90.80% |
| Tucker2 decomposition [43] | 12 leads PTB | Location | Acc = 99.67% Se = 99.98% Sp = 99.82% | Acc = 65.11% Se = 98.29% Sp = 71.91% |
| Multi-lead branch with ResNet with SE and LSTM [44] | 12 leads PTB | Detection and location | Detection: Acc = 99.94% Se = 99.99% Location: Acc = 99.69% Se = 99.58% | Detection: Acc = 96.55% Se = 96.17% Location: Acc = 67.89% Se = 63.16% |
| MFF–CNN (Our) | 12 leads PTB | Detection and location | Detection: Acc = 99.99% Se = 100.00% Location: Acc = 99.98% Se = 99.97% | Detection: Acc = 99.98% Se = 100.00% Location: Acc = 84.86% Se = 62.90% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Ye, J.; Li, Y.; Luo, Z.; Luo, J.; Wan, X. A Multi-Domain Feature Fusion CNN for Myocardial Infarction Detection and Localization. Biosensors 2025, 15, 392. https://doi.org/10.3390/bios15060392
Chen Y, Ye J, Li Y, Luo Z, Luo J, Wan X. A Multi-Domain Feature Fusion CNN for Myocardial Infarction Detection and Localization. Biosensors. 2025; 15(6):392. https://doi.org/10.3390/bios15060392
Chicago/Turabian StyleChen, Yunfan, Jinxing Ye, Yuting Li, Zhe Luo, Jieqiang Luo, and Xiangkui Wan. 2025. "A Multi-Domain Feature Fusion CNN for Myocardial Infarction Detection and Localization" Biosensors 15, no. 6: 392. https://doi.org/10.3390/bios15060392
APA StyleChen, Y., Ye, J., Li, Y., Luo, Z., Luo, J., & Wan, X. (2025). A Multi-Domain Feature Fusion CNN for Myocardial Infarction Detection and Localization. Biosensors, 15(6), 392. https://doi.org/10.3390/bios15060392





