Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analysis of CRC Patients
2.2. Design of KRAS Templates, Primers, and Probes
2.3. qPCR Experimental Protocol
2.4. Standard Curve Establishment
2.5. Tissue DNA Extraction and KRAS Mutation Detection Using LBDA Technology
3. Results
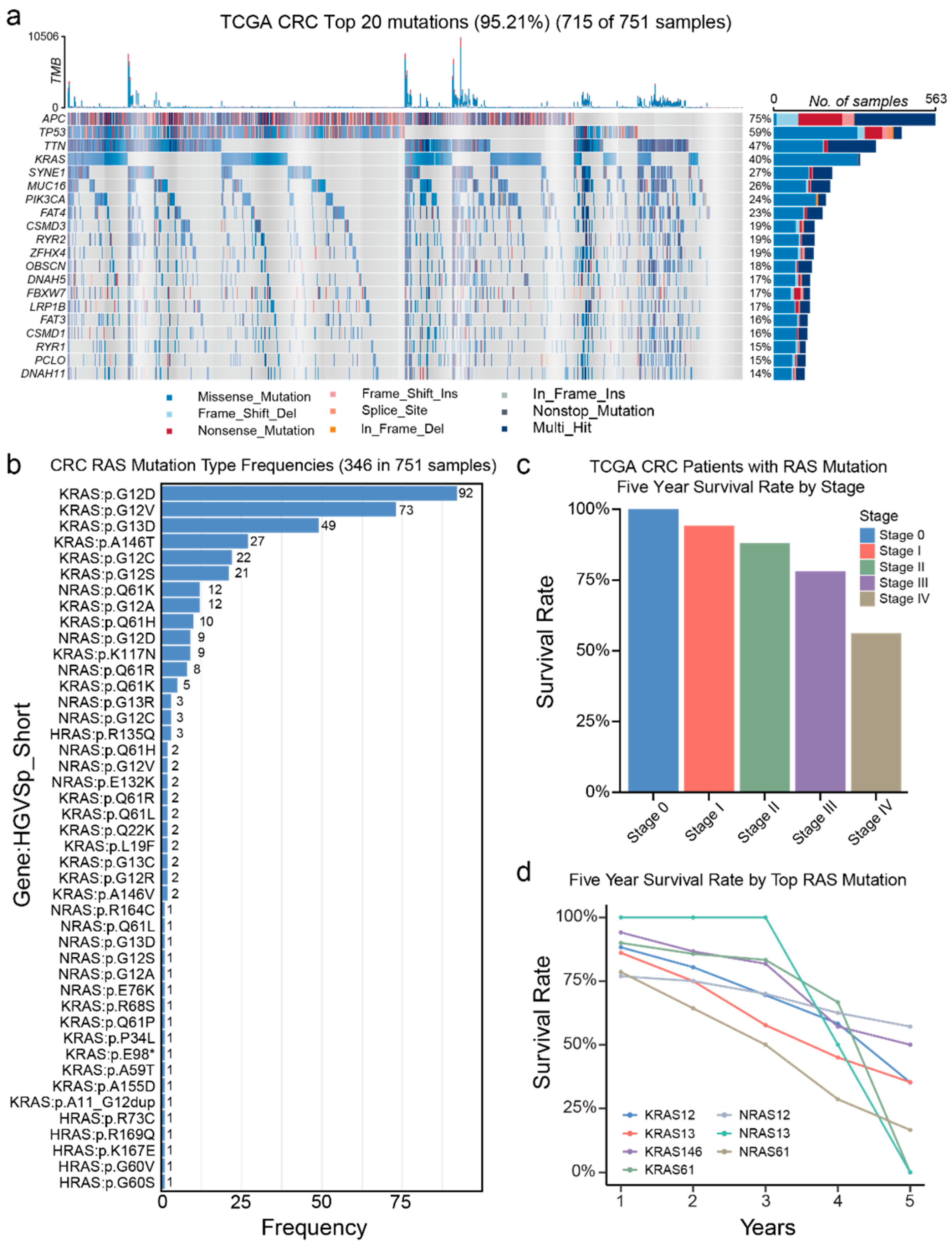
3.1. Genetic Alterations in TCGA
3.2. Genetic Alterations in a Chinese CRC Patient Group
3.3. Comparison of the Mutation Gene Signatures Between TCGA and Chinese Patient Population
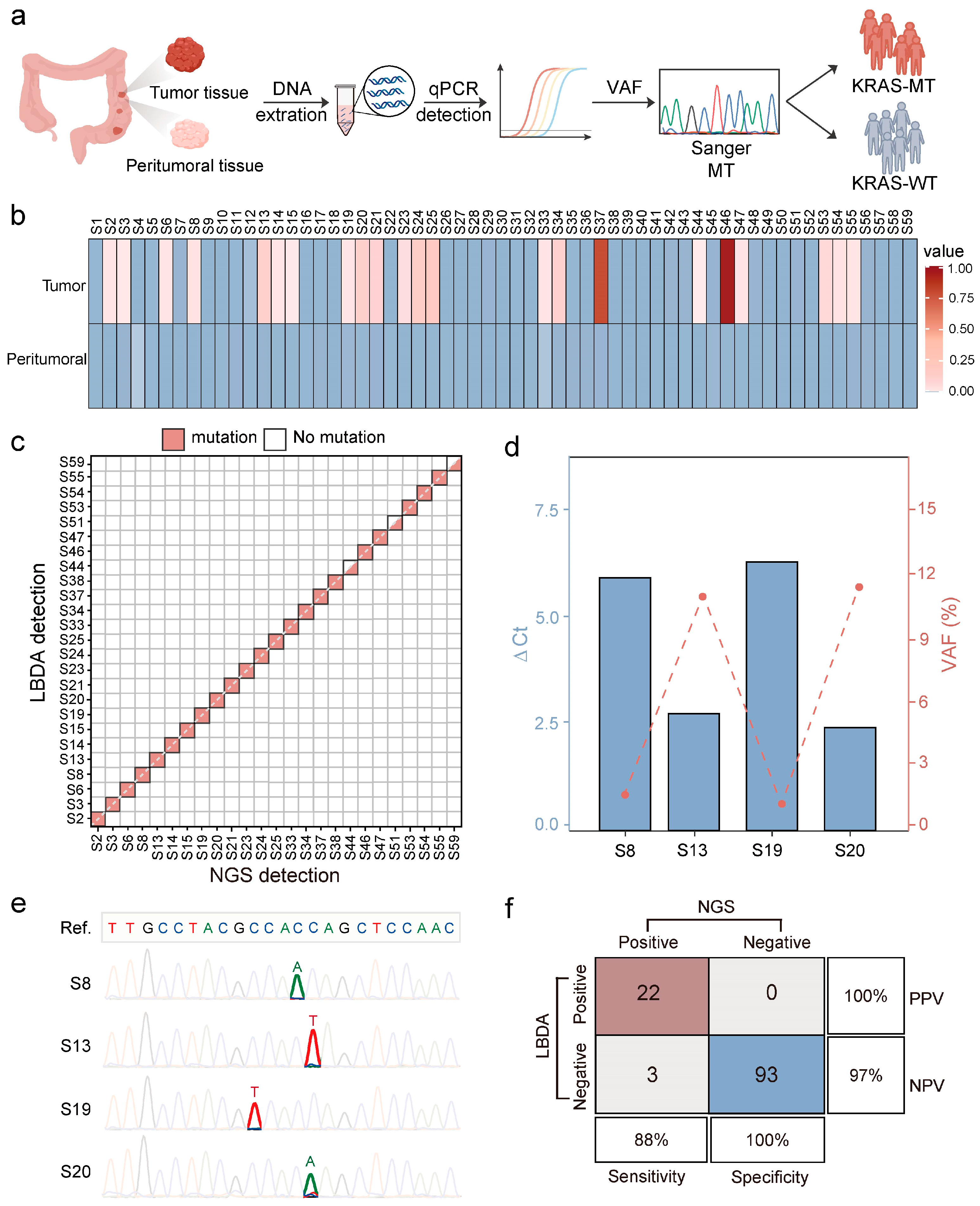
3.4. Detection of KRAS Mutations Using LBDA Technology
3.5. Clinical Application of the LBDA Method for Detecting KRAS Mutations in CRC Tissues
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mármol, I.; Sánchez-de-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M.J. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- Beniwal, S.S.; Lamo, P.; Kaushik, A.; Lorenzo-Villegas, D.L.; Liu, Y.; MohanaSundaram, A. Current Status and Emerging Trends in Colorectal Cancer Screening and Diagnostics. Biosensors 2023, 13, 926. [Google Scholar] [CrossRef]
- Han, B.; Zheng, R.; Zeng, H.; Wang, S.; Sun, K.; Chen, R.; Li, L.; Wei, W.; He, J. Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 2024, 4, 47–53. [Google Scholar] [CrossRef]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Maomao, C.; He, L.; Dianqin, S.; Siyi, H.; Xinxin, Y.; Fan, Y.; Shaoli, Z.; Changfa, X.; Lin, L.; Ji, P.; et al. Current cancer burden in China: Epidemiology, etiology, and prevention. Cancer Biol. Med. 2022, 19, 1121–1138. [Google Scholar] [CrossRef]
- Morris, V.K.; Kennedy, E.B.; Baxter, N.N.; Benson, A.B., 3rd; Cercek, A.; Cho, M.; Ciombor, K.K.; Cremolini, C.; Davis, A.; Deming, D.A.; et al. Treatment of Metastatic Colorectal Cancer: ASCO Guideline. J. Clin. Oncol. 2023, 41, 678–700. [Google Scholar] [CrossRef]
- Nunes, L.; Li, F.; Wu, M.; Luo, T.; Hammarström, K.; Torell, E.; Ljuslinder, I.; Mezheyeuski, A.; Edqvist, P.-H.; Löfgren-Burström, A.; et al. Prognostic genome and transcriptome signatures in colorectal cancers. Nature 2024, 633, 137–146. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Y.; Yang, W.; Wang, C.; Guo, T.; Yang, J.; Shao, Z.; Cai, G.; Cai, S.; Zhang, L.; et al. Molecular Profiling Provides Clinical Insights Into Targeted and Immunotherapies as Well as Colorectal Cancer Prognosis. Gastroenterology 2023, 165, 414–428.e417. [Google Scholar] [CrossRef]
- Zhu, G.; Pei, L.; Xia, H.; Tang, Q.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef]
- Modest, D.P.; Ricard, I.; Heinemann, V.; Hegewisch-Becker, S.; Schmiegel, W.; Porschen, R.; Stintzing, S.; Graeven, U.; Arnold, D.; von Weikersthal, L.F.; et al. Outcome according to KRAS-, NRAS- and BRAF-mutation as well as KRAS mutation variants: Pooled analysis of five randomized trials in metastatic colorectal cancer by the AIO colorectal cancer study group. Ann. Oncol. 2016, 27, 1746–1753. [Google Scholar] [CrossRef]
- Heinemann, V.; von Weikersthal, L.F.; Decker, T.; Kiani, A.; Vehling-Kaiser, U.; Al-Batran, S.E.; Heintges, T.; Lerchenmüller, C.; Kahl, C.; Seipelt, G.; et al. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): A randomised, open-label, phase 3 trial. Lancet Oncol. 2014, 15, 1065–1075. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Lenz, H.J.; Köhne, C.H.; Heinemann, V.; Tejpar, S.; Melezínek, I.; Beier, F.; Stroh, C.; Rougier, P.; van Krieken, J.H.; et al. Fluorouracil, leucovorin, and irinotecan plus cetuximab treatment and RAS mutations in colorectal cancer. J. Clin. Oncol. 2015, 33, 692–700. [Google Scholar] [CrossRef]
- Huang, D.; Sun, W.; Zhou, Y.; Li, P.; Chen, F.; Chen, H.; Xia, D.; Xu, E.; Lai, M.; Wu, Y.; et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. 2018, 37, 173–187. [Google Scholar] [CrossRef]
- Cornish, A.J.; Gruber, A.J.; Kinnersley, B.; Chubb, D.; Frangou, A.; Caravagna, G.; Noyvert, B.; Lakatos, E.; Wood, H.M.; Thorn, S.; et al. The genomic landscape of 2,023 colorectal cancers. Nature 2024, 633, 127–136. [Google Scholar] [CrossRef]
- Raskov, H.; Søby, J.H.; Troelsen, J.; Bojesen, R.D.; Gögenur, I. Driver Gene Mutations and Epigenetics in Colorectal Cancer. Ann. Surg. 2020, 271, 75–85. [Google Scholar] [CrossRef]
- Cai, Z.X.; Tang, X.D.; Gao, H.L.; Tang, C.; Nandakumar, V.; Jones, L.; Ye, H.; Lou, F.; Zhang, D.; Sun, H.; et al. APC, FBXW7, KRAS, PIK3CA, and TP53 Gene Mutations in Human Colorectal Cancer Tumors Frequently Detected by Next-Generation DNA Sequencing. J. Mol. Genet. Genom. Med. 2014, 8, 862–1747. [Google Scholar]
- Wu, Y.; Guo, J.; Li, W.; Xiu, X.; Thirunavukarasu, D.; Wang, Y.; Wang, K.; Chen, W.; Yu Zhang, D.; Yang, X.; et al. Enhanced Detection of Novel Low-Frequency Gene Fusions via High-Yield Ligation and Multiplexed Enrichment Sequencing. Angew. Chem. Int. Ed. 2024, 63, e202316484. [Google Scholar] [CrossRef] [PubMed]
- Weng, Z.; Li, J.; Wu, Y.; Xiu, X.; Wang, F.; Zuo, X.; Song, P.; Fan, C. Massively parallel homogeneous amplification of chip-scale DNA for DNA information storage (MPHAC-DIS). Nat. Commun. 2025, 16, 667. [Google Scholar] [CrossRef]
- Shen, H.; Weng, Z.; Zhao, H.; Song, H.; Wang, F.; Fan, C.; Song, P. Random Sanitization in DNA Information Storage Using CRISPR-Cas12a. J. Am. Chem. Soc. 2024, 146, 35155–35164. [Google Scholar] [CrossRef]
- Su, X.; Zheng, Q.; Xiu, X.; Zhao, Q.; Wang, Y.; Han, D.; Song, P. Challenges and prospects in utilizing technologies for gene fusion analysis in cancer diagnostics. Med-X 2024, 2, 14. [Google Scholar] [CrossRef]
- Song, P.; Chen, S.X.; Yan, Y.H.; Pinto, A.; Cheng, L.Y.; Dai, P.; Patel, A.A.; Zhang, D.Y. Selective multiplexed enrichment for the detection and quantitation of low-fraction DNA variants via low-depth sequencing. Nat. Biomed. Eng. 2021, 5, 690–701. [Google Scholar] [CrossRef]
- Si, Y.; Wang, X.; Su, X.; Weng, Z.; Hu, Q.; Li, Q.; Fan, C.; Zhang, D.Y.; Wang, Y.; Luo, S.; et al. Extended Enrichment for Ultrasensitive Detection of Low-Frequency Mutations by Long Blocker Displacement Amplification. Angew. Chem. Int. Ed. 2024, 63, e202400551. [Google Scholar] [CrossRef]
- Fodde, R. The APC gene in colorectal cancer. Eur. J. Cancer 2002, 38, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Iacopetta, B. TP53 mutation in colorectal cancer. Hum. Mutat. 2003, 21, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Naccarati, A.; Polakova, V.; Pardini, B.; Vodickova, L.; Hemminki, K.; Kumar, R.; Vodicka, P. Mutations and polymorphisms in TP53 gene--an overview on the role in colorectal cancer. Mutagenesis 2012, 27, 211–218. [Google Scholar] [CrossRef]
- Oh, J.H.; Jang, S.J.; Kim, J.; Sohn, I.; Lee, J.Y.; Cho, E.J.; Chun, S.M.; Sung, C.O. Spontaneous mutations in the single TTN gene represent high tumor mutation burden. NPJ Genom. Med. 2020, 5, 33. [Google Scholar] [CrossRef]
- Cheng, X.; Yin, H.; Fu, J.; Chen, C.; An, J.; Guan, J.; Duan, R.; Li, H.; Shen, H. Aggregate analysis based on TCGA: TTN missense mutation correlates with favorable prognosis in lung squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2019, 145, 1027–1035. [Google Scholar] [CrossRef]
- Jancík, S.; Drábek, J.; Radzioch, D.; Hajdúch, M. Clinical relevance of KRAS in human cancers. J. Biomed. Biotechnol. 2010, 2010, 150960. [Google Scholar] [CrossRef]
- Edkins, S.; O’Meara, S.; Parker, A.; Stevens, C.; Reis, M.; Jones, S.; Greenman, C.; Davies, H.; Dalgliesh, G.; Forbes, S.; et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol. Ther. 2006, 5, 928–932. [Google Scholar] [CrossRef]
- Petrosino, M.; Novak, L.; Pasquo, A.; Chiaraluce, R.; Turina, P.; Capriotti, E.; Consalvi, V. Analysis and Interpretation of the Impact of Missense Variants in Cancer. Int. J. Mol. Sci. 2021, 22, 5416. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Miteva, M.A.; Wang, L.; Alexov, E. Analyzing effects of naturally occurring missense mutations. Comput. Math. Methods Med. 2012, 2012, 805827. [Google Scholar] [CrossRef]
- Gerasimavicius, L.; Livesey, B.J.; Marsh, J.A. Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure. Nat. Commun. 2022, 13, 3895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, J.; Yang, Y.; Lu, J.; Gao, J.; Lu, T.; Sun, J.; Jiang, H.; Zhu, Y.; Zheng, Y.; et al. Molecular spectrum of KRAS, NRAS, BRAF and PIK3CA mutations in Chinese colorectal cancer patients: Analysis of 1110 cases. Sci. Rep. 2015, 5, 18678. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, C.; Li, X.; Luo, W.; Roy, B.; Xiong, T.; Zhang, X.; Yang, H.; Wang, J.; Ye, Z.; et al. The landscape of somatic mutation in sporadic Chinese colorectal cancer. Oncotarget 2018, 9, 27412–27422. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.; Rocha, P.; Veiga, I.; Guedes, J.; Pinheiro, M.; Peixoto, A.; Pinto, C.; Fragoso, M.; Sanches, E.; Araújo, A.; et al. Comparison of methodologies for KRAS mutation detection in metastatic colorectal cancer. Cancer Genet. 2011, 204, 439–446. [Google Scholar] [CrossRef]
- Carotenuto, P.; Roma, C.; Rachiglio, A.M.; Tatangelo, F.; Pinto, C.; Ciardiello, F.; Nappi, O.; Iaffaioli, R.V.; Botti, G.; Normanno, N. Detection of KRAS mutations in colorectal carcinoma patients with an integrated PCR/sequencing and real-time PCR approach. Pharmacogenomics 2010, 11, 1169–1179. [Google Scholar] [CrossRef]
- Dono, M.; Massucco, C.; Chiara, S.; Sonaglio, C.; Mora, M.; Truini, A.; Cerruti, G.; Zoppoli, G.; Ballestrero, A.; Truini, M.; et al. Low percentage of KRAS mutations revealed by locked nucleic acid polymerase chain reaction: Implications for treatment of metastatic colorectal cancer. Mol. Med. 2013, 18, 1519–1526. [Google Scholar] [CrossRef]
- Chubarov, A.S.; Oscorbin, I.P.; Filipenko, M.L.; Lomzov, A.A.; Pyshnyi, D.V. Allele-Specific PCR for KRAS Mutation Detection Using Phosphoryl Guanidine Modified Primers. Diagnostics 2020, 10, 872. [Google Scholar] [CrossRef]
- Lyu, N.; Rajendran, V.K.; Li, J.; Engel, A.; Molloy, M.P.; Wang, Y. Highly specific detection of KRAS single nucleotide polymorphism by asymmetric PCR/SERS assay. Analyst 2021, 146, 5714–5721. [Google Scholar] [CrossRef]
- Ren, J.; Xu, G.; Liu, H.; He, N.; Zhao, Z.; Wang, M.; Gu, P.; Chen, Z.; Deng, Y.; Wu, D.; et al. A Chamber-Based Digital PCR Based on a Microfluidic Chip for the Absolute Quantification and Analysis of KRAS Mutation. Biosensors 2023, 13, 778. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wu, L.R.; Yan, Y.H.; Zhang, J.X.; Chu, T.; Kwong, L.N.; Patel, A.A.; Zhang, D.Y. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 2022, 6, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Weng, Z.; Huang, Z.; Li, Q.; Fan, C.; Luo, S.; Song, P. Selective Enrichment of Low-Abundance DNA Variants Based on Programmable Peptide Nucleic Acid Probes. Chin. J. Chem. 2024, 42, 2572–2580. [Google Scholar] [CrossRef]
- Zhao, H.; Xiu, X.; Li, M.; Dai, S.; Gou, M.; Tao, L.; Zuo, X.; Fan, C.; Tian, Z.; Song, P. Programming Super DNA-Enzyme Molecules for On-Demand Enzyme Activity Modulation. Angew. Chem. Int. Ed. 2023, 62, e202214450. [Google Scholar] [CrossRef]



| Sequence Name | Sequence |
|---|---|
| KRAS-WT | TGACTGAATATAAACTTGTGGTAGTGGAGCTGGTGGCGTAAGCAAGAGTGCCTTGACGATACAGCTAATTCAGAATCATTTTGTGGACGAATATGATCCAACAATAGAGGTAAATCTTGTTTTATATGCATATTACTGGTGCAGGACCATTCTTTGATACAGA |
| KRAS-MT G13C (G > T) | TGACTGAATATAAACTTGTGGTAGTGGAGCTGGTTGCGTAAGCAAGAGTGCCTTGACGATACAGCTAATTCAGAATCATTTTGTGGACGAATATGATCCAACAATAGAGGTAAATCTTGTTTTATATGCATATTACTGGTGCAGGACCATTCTTTGATACAGA |
| Sequence Name | Sequence |
|---|---|
| KRAS-FP | 5′-GCTCTTCCTCTCACATCTTTATTTAACC-3′ |
| KRAS-RP | 5′-TCCACACTGCAGTGTGAACAG-3′ |
| KRAS-Blocker | GTAGTTGGAGCTGGTGGCGTAGGCAAGAGT/iSpC3//iSpC3/CA |
| Method | LoD (VAF %) | Throughput (Per Test) | Mutation Coverage | Time (h) | Cost ($) | Reference |
|---|---|---|---|---|---|---|
| LBDA (this work) | 0.08 | >81 | Known + novel | 3 | ~2 | [23] |
| AS-PCR | 0.1~1 | 1 | Known only | ~5 | ~5 | [39] |
| Asy-PCR/SERS | 0.1 | 1 | Known only | ~5 | ~6 | [40] |
| Chamber-Based Digital PCR | 0.2 | 3 | Known only | ~5 | ~28 | [41] |
| NGS | 0.2 | high | Known + novel | ~168 | ~1112 | [42] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, P.; Su, X.; Leong, S.; Xiu, X.; Song, P.; Peng, J.; Si, Y. Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection. Biosensors 2025, 15, 308. https://doi.org/10.3390/bios15050308
Lu P, Su X, Leong S, Xiu X, Song P, Peng J, Si Y. Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection. Biosensors. 2025; 15(5):308. https://doi.org/10.3390/bios15050308
Chicago/Turabian StyleLu, Ping, Xinglei Su, Sirui Leong, Xuehao Xiu, Ping Song, Junjie Peng, and Yunpei Si. 2025. "Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection" Biosensors 15, no. 5: 308. https://doi.org/10.3390/bios15050308
APA StyleLu, P., Su, X., Leong, S., Xiu, X., Song, P., Peng, J., & Si, Y. (2025). Analysis of Colorectal Cancer Gene Mutations and Application of Long Blocker Displacement Amplification Technology for High-Throughput Mutation Detection. Biosensors, 15(5), 308. https://doi.org/10.3390/bios15050308






