Solid-State Nanopore Readout of Programmable DNA and Peptide Nanostructures for Scalable Digital Data Storage
Abstract
1. Introduction
2. Results
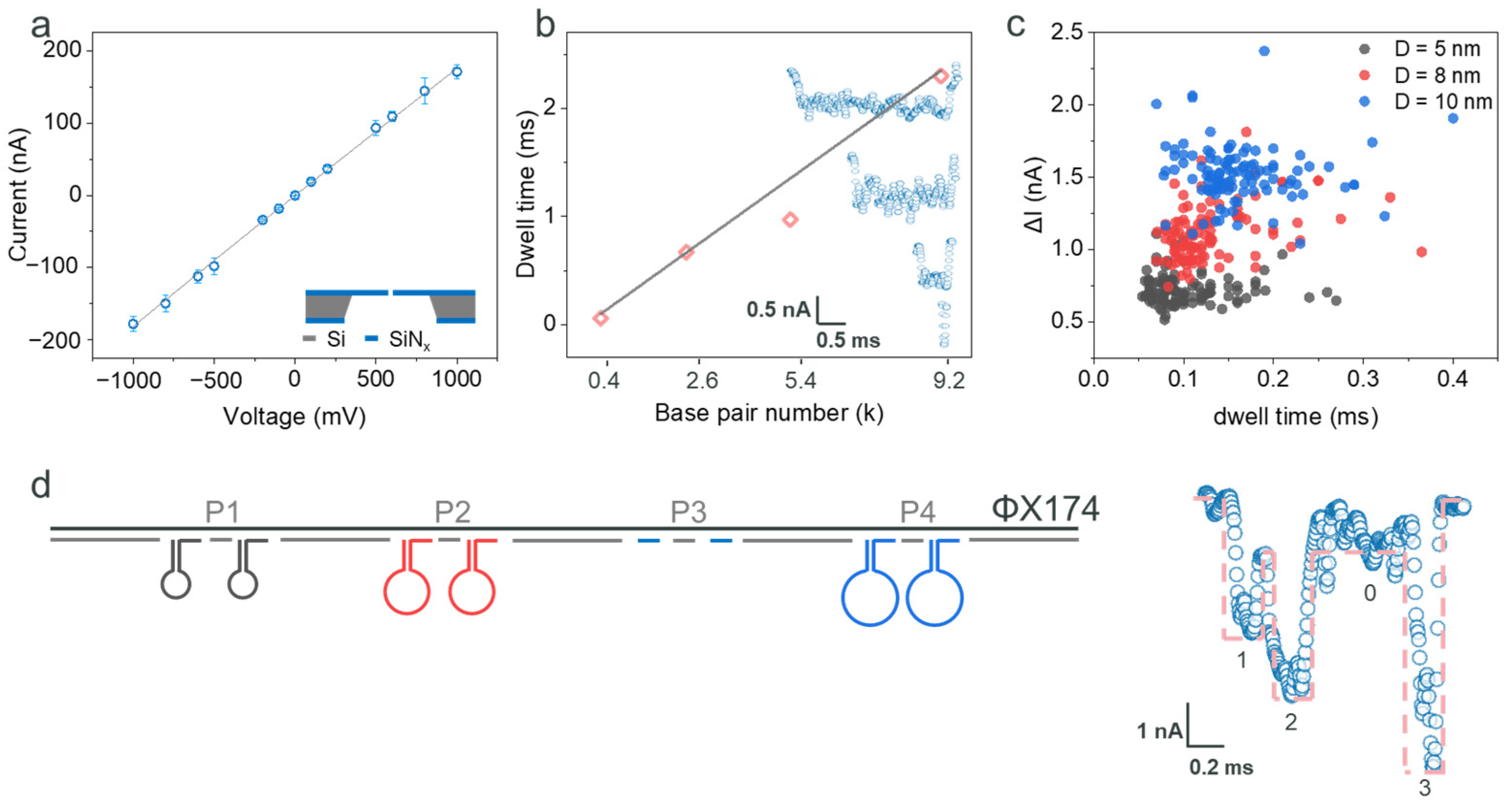
2.1. Detection of the ΦX174 Backbone and DNA Hairpins
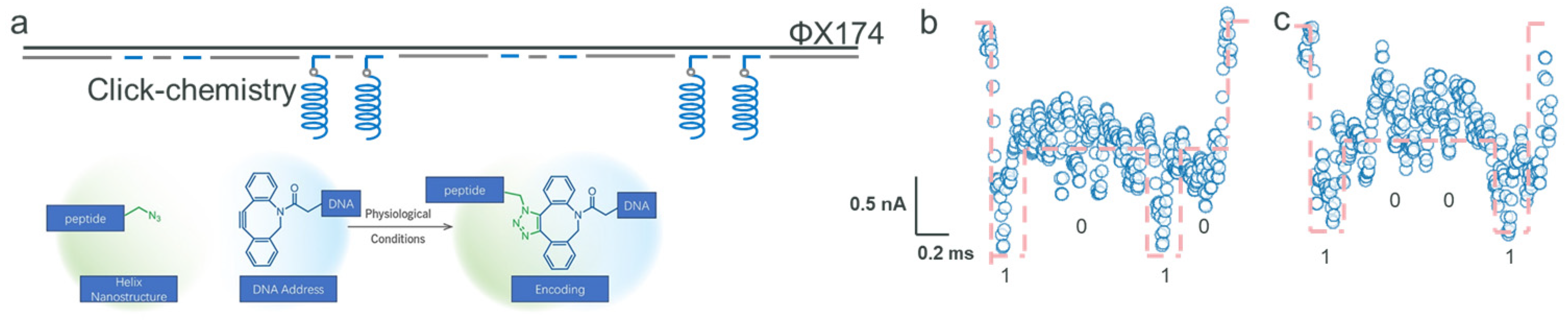
2.2. Encoding with Click Chemistry and Peptides
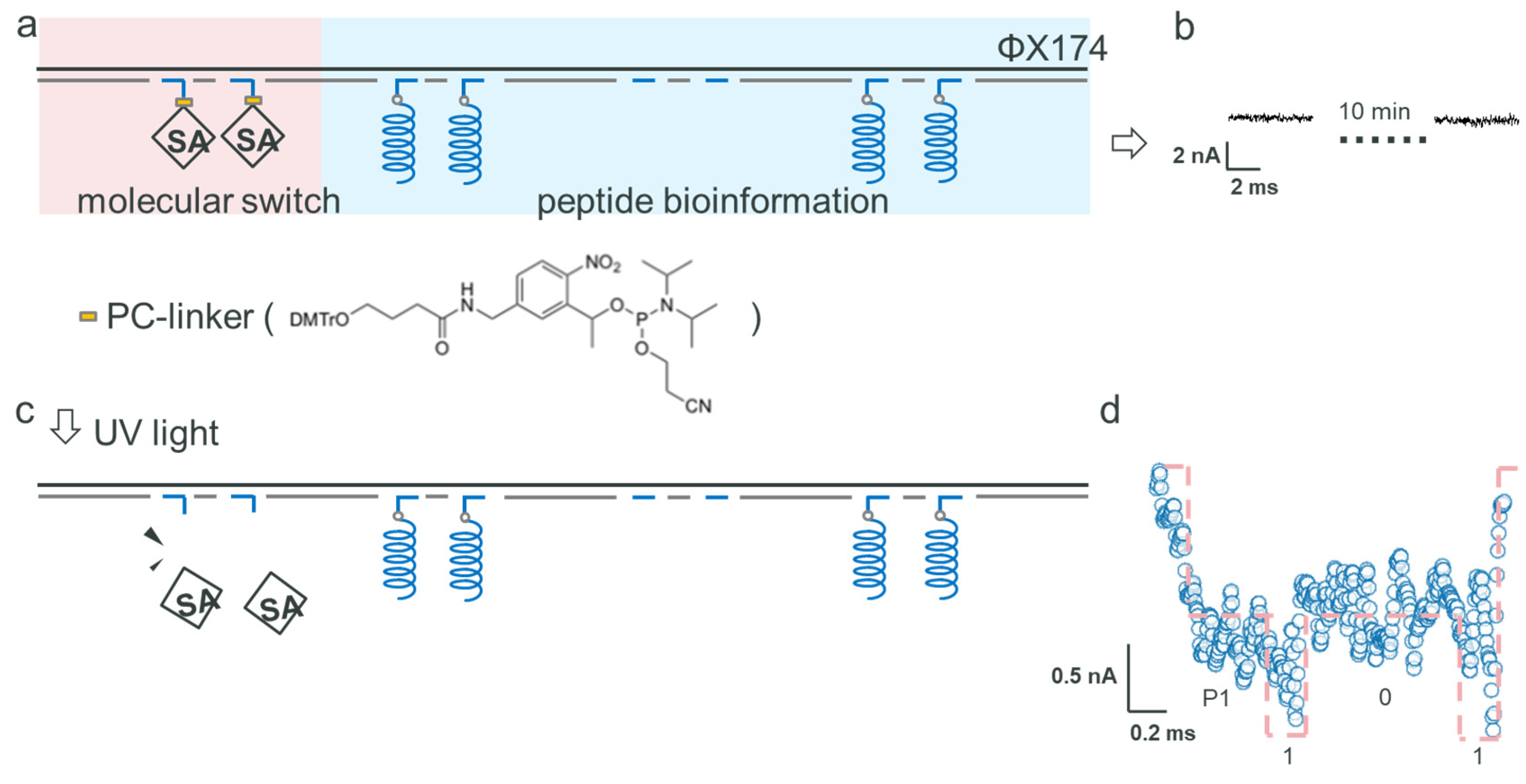
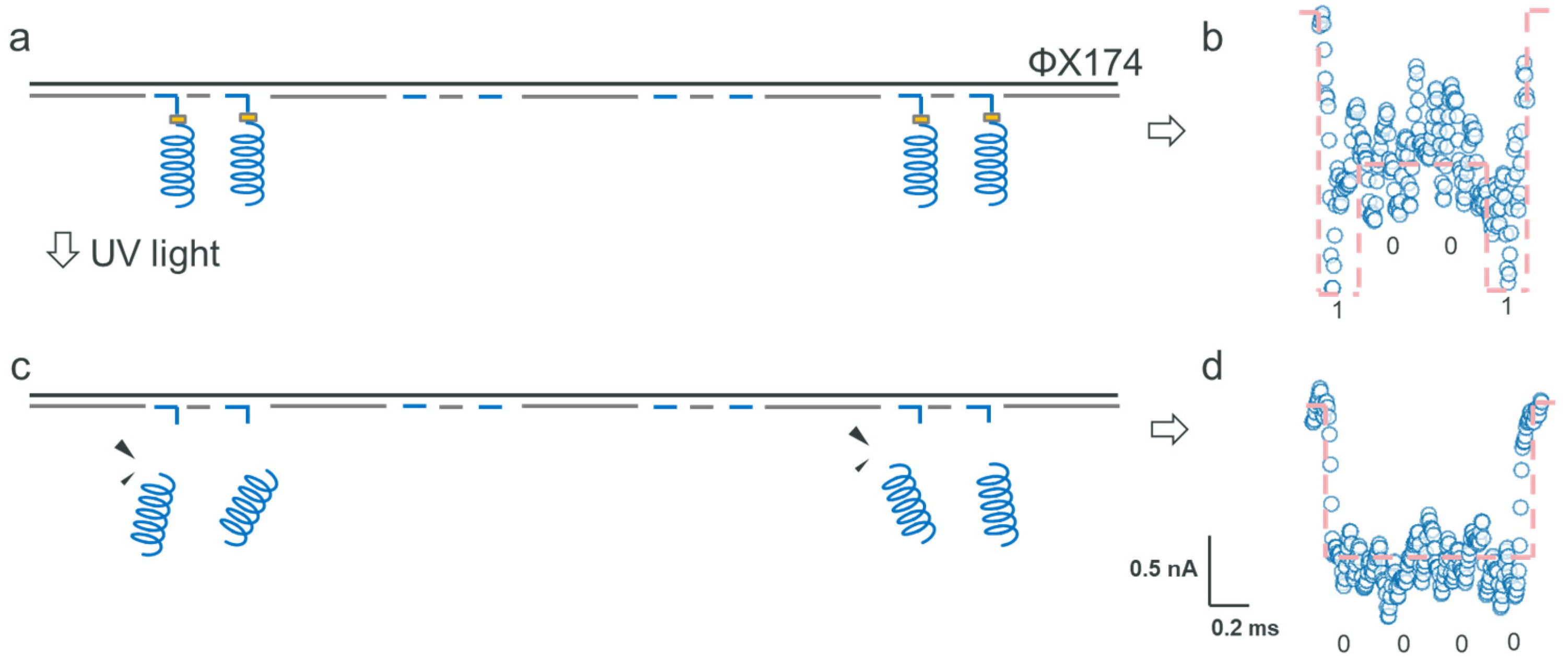
2.3. Data Encryption
2.4. Data Formatting
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Preparation of DNA Backbone Strand
4.3. DNA Hairpins Bind with ΦX174
4.4. Peptides Bind with ΦX174
4.5. SA and Peptides Bind with ΦX174
4.6. Measurement Methods
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lunt, B.M. How long is long-term data storage? In Proceedings of the Archiving Conference, Society for Imaging Science and Technology, Salt Lake City, UT, USA, 16–19 May 2011; pp. 29–33. [Google Scholar]
- Extance, A. How DNA could store all the world’s data. Nature 2016, 537, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Zhirnov, V.; Zadegan, R.M.; Sandhu, G.S.; Church, G.M.; Hughes, W.L. Nucleic acid memory. Nat. Mater. 2016, 15, 366–370. [Google Scholar] [CrossRef] [PubMed]
- Jo, S.; Shin, H.; Joe, S.-y.; Baek, D.; Park, C.; Chun, H. Recent progress in DNA data storage based on high-throughput DNA synthesis. Biomed. Eng. Lett. 2024, 14, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Mao, X.; Wang, F.; Zuo, X.; Fan, C. Data Storage Using DNA. Adv. Mater. 2023, 36, e2307499. [Google Scholar] [CrossRef]
- Organick, L.; Ang, S.D.; Chen, Y.-J.; Lopez, R.; Yekhanin, S.; Makarychev, K.; Racz, M.Z.; Kamath, G.; Gopalan, P.; Nguyen, B.; et al. Scaling up DNA data storage and random access retrieval. bioRxiv 2017. [Google Scholar] [CrossRef]
- Kong, J.; Zhu, J.; Chen, K.; Keyser, U.F. Specific Biosensing Using DNA Aptamers and Nanopores. Adv. Funct. Mater. 2018, 29, 1807555. [Google Scholar] [CrossRef]
- Chen, K.; Kong, J.; Zhu, J.; Ermann, N.; Predki, P.; Keyser, U.F. Digital Data Storage Using DNA Nanostructures and Solid-State Nanopores. Nano Lett. 2018, 19, 1210–1215. [Google Scholar] [CrossRef]
- Bell, N.A.W.; Keyser, U.F. Digitally encoded DNA nanostructures for multiplexed, single-molecule protein sensing with nanopores. Nat. Nanotechnol. 2016, 11, 645–651. [Google Scholar] [CrossRef]
- Feng, N.; Zhang, L.; Shen, J.J.; Hu, Y.L.; Wu, W.B.; Fodjo, E.K.; Chen, S.F.; Huang, W.; Wang, L.H. SERS molecular-ruler based DNA aptamer single-molecule and its application to multi-level optical storage. Chem. Eng. J. 2022, 433, 133666. [Google Scholar] [CrossRef]
- Roelen, Z.; Briggs, K.; Tabard-Cossa, V. Analysis of Nanopore Data: Classification Strategies for an Unbiased Curation of Single-Molecule Events from DNA Nanostructures. Acs. Sens. 2023, 8, 2809–2823. [Google Scholar] [CrossRef]
- Li, X.; Tong, X.; Lu, W.; Yu, D.; Diao, J.; Zhao, Q. Label-free detection of early oligomerization of α-synuclein and its mutants A30P/E46K through solid-state nanopores. Nanoscale 2019, 11, 6480–6488. [Google Scholar] [CrossRef]
- Jiang, W.J.; Li, J.R.; Lin, Z.A.; Guo, J.; Ma, J.X.; Wang, Z.; Zhang, M.Z.; Wu, Y.Z. Recent Advances of DNA Origami Technology and Its Application in Nanomaterial Preparation. Small Struct. 2023, 4, 2200376. [Google Scholar] [CrossRef]
- Poppleton, E.; Mallya, A.; Dey, S.; Joseph, J.; Šulc, P. Nanobase.org: A repository for DNA and RNA nanostructures. Nucleic Acids Res. 2022, 50, D246–D252. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.; Yao, Y.; Guo, X. Single-Molecule Nanotechnologies: An Evolution in Biological Dynamics Detection. ACS Appl. Bio Mater. 2019, 3, 68–85. [Google Scholar] [CrossRef] [PubMed]
- Akkilic, N.; Geschwindner, S.; Höök, F. Single-molecule biosensors: Recent advances and applications. Biosens. Bioelectron. 2020, 151, 111944. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.K.; Choudhary, A.; Sandler, S.E.; Maffeo, C.; Ducati, C.; Aksimentiev, A.; Keyser, U.F. Super-Resolution Detection of DNA Nanostructures Using a Nanopore. Adv. Mater. 2023, 35, 2207434. [Google Scholar] [CrossRef]
- Derrington, I.M.; Craig, J.M.; Stava, E.; Laszlo, A.H.; Ross, B.C.; Brinkerhoff, H.; Nova, I.C.; Doering, K.; Tickman, B.I.; Ronaghi, M.; et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 2015, 33, 1073. [Google Scholar] [CrossRef]
- Zvuloni, E.; Zrehen, A.; Gilboa, T.; Meller, A. Fast and Deterministic Fabrication of Sub-5 Nanometer Solid-State Pores by Feedback-Controlled Laser Processing. ACS Nano 2021, 15, 12189–12200. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, Q.; Wang, W.; Fang, F.; Zhang, J. Solid-State Nanopore Array: Manufacturing and Applications. Small 2022, 19, 2205680. [Google Scholar] [CrossRef]
- Yuan, Z.; Wang, C.; Yi, X.; Ni, Z.; Chen, Y.; Li, T. Solid-State Nanopore. Nanoscale Res. Lett. 2018, 13. [Google Scholar] [CrossRef]
- Wanunu, M. Nanopores: A journey towards DNA sequencing. Phys. Life Rev. 2012, 9, 125–158. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gershow, M.; Stein, D.; Brandin, E.; Golovchenko, J.A. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Mater. 2003, 2, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, K.B.; Kim, H.J.; Yu, J.S.; Chae, H.; Kim, H.M.; Kim, K.B. Recent Progress in Solid-State Nanopores. Adv. Mater. 2018, 30, e1704680. [Google Scholar] [CrossRef]
- Magierowski, S.; Huang, Y.; Wang, C.; Ghafar-Zadeh, E. Nanopore-CMOS Interfaces for DNA Sequencing. Biosensors 2016, 6, 42. [Google Scholar] [CrossRef]
- Uddin, A.; Yemenicioglu, S.; Chen, C.H.; Corigliano, E.; Milaninia, K.; Theogarajan, L. Integration of solid-state nanopores in a 0.5 mum CMOS foundry process. Nanotechnology 2013, 24, 155501. [Google Scholar] [CrossRef]
- Watson, J.D.; Crick, F.H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Awasthi, S.; Ying, C.F.; Li, J.L.; Mayer, M. Simultaneous Determination of the Size and Shape of Single α-Synuclein Oligomers in Solution. Acs. Nano 2023, 17, 12325–12335. [Google Scholar] [CrossRef]
- Schmid, S.; Stömmer, P.; Dietz, H.; Dekker, C. Nanopore electro-osmotic trap for the label-free study of single proteins and their conformations. Nat. Nanotechnol. 2021, 16, 1244–1250. [Google Scholar] [CrossRef]
- Chen, X.Y.; Zhao, X.J.; Ma, R.P.; Hu, Y.; Cui, C.J.; Mi, Z.; Dou, R.F.; Pan, D.; Shan, X.Y.; Wang, L.H.; et al. Ionic Current Fluctuation and Orientation of Tetrahedral DNA Nanostructures in a Solid-State Nanopore. Small 2022, 18, 2107237. [Google Scholar] [CrossRef]
- Mojtabavi, M.; Greive, S.J.; Antson, A.A.; Wanunu, M. High-Voltage Biomolecular Sensing Using a Bacteriophage Portal Protein Covalently Immobilized within a Solid-State Nanopore. J. Am. Chem. Soc. 2022, 144, 22540–22548. [Google Scholar] [CrossRef]
- Osuofa, J.; Husson, S.M. Preparation of Protein A Membrane Adsorbers Using Strain-Promoted, Copper-Free Dibenzocyclooctyne (DBCO)-Azide Click Chemistry. Membranes 2023, 13, 824. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B.; Branson, H.R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. USA 1951, 37, 205–211. [Google Scholar] [CrossRef]
- Wang, V.; Ermann, N.; Keyser, U.F. Current Enhancement in Solid-State Nanopores Depends on Three-Dimensional DNA Structure. Nano Lett. 2019, 19, 5661–5666. [Google Scholar] [CrossRef] [PubMed]
- Markham, N.R.; Zuker, M. UNAFold: Software for nucleic acid folding and hybridization. In Bioinformatics. Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2008. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2010, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Fornace, M.E.; Huang, J.; Newman, C.T.; Porubsky, N.J.; Pierce, M.B.; Pierc, N.A. NUPACK: Analysis and design of nucleic acid structures, devices, and systems. ChemRxiv 2022. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices 1 1Edited by G. Von Heijne. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Wang, J.; Wu, L.-S.; Zhao, X. Solid-State Nanopore Readout of Programmable DNA and Peptide Nanostructures for Scalable Digital Data Storage. Biosensors 2025, 15, 287. https://doi.org/10.3390/bios15050287
Zhao L, Wang J, Wu L-S, Zhao X. Solid-State Nanopore Readout of Programmable DNA and Peptide Nanostructures for Scalable Digital Data Storage. Biosensors. 2025; 15(5):287. https://doi.org/10.3390/bios15050287
Chicago/Turabian StyleZhao, Lihuan, Jiajun Wang, Lin-Sheng Wu, and Xin Zhao. 2025. "Solid-State Nanopore Readout of Programmable DNA and Peptide Nanostructures for Scalable Digital Data Storage" Biosensors 15, no. 5: 287. https://doi.org/10.3390/bios15050287
APA StyleZhao, L., Wang, J., Wu, L.-S., & Zhao, X. (2025). Solid-State Nanopore Readout of Programmable DNA and Peptide Nanostructures for Scalable Digital Data Storage. Biosensors, 15(5), 287. https://doi.org/10.3390/bios15050287




