Rigor & Reproducibility: pH Adjustments of Papain with L-Cysteine Dissociation Solutions and Cell Media Using Phenol Red Spectrophotometry
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Analysis of Published Protocols
3.2. pH Measurements and Calculations
3.3. Culture of Isolated Neurons
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ECM | Extracellular matrix |
| EDTA | Ethylenediaminetetraacetic acid |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| RNA | Ribonucleic acid |
References
- Zhao, F.; Wu, T.; Lau, A.; Jiang, T.; Huang, Z.; Wang, X.J.; Chen, W.; Wong, P.K.; Zhang, D.D. Nrf2 promotes neuronal cell differentiation. Free Radic. Biol. Med. 2009, 47, 867–879. [Google Scholar] [CrossRef]
- Sahu, M.P.; Nikkilä, O.; Lagas, S.; Kolehmainen, S.; Castrén, E. Culturing primary neurons from rat hippocampus and cortex. Neuronal Signal. 2019, 3, 20180207. [Google Scholar] [CrossRef] [PubMed]
- Katzenell, S.; Cabrera, J.R.; North, B.J.; Leib, D.A. Isolation, Purification, and Culture of Primary Murine Sensory Neurons. Methods Mol. Biol. 2017, 1656, 229–251. [Google Scholar]
- Ray, B.; Bailey, J.A.; Sarkar, S.; Lahiri, D.K. Molecular and immunocytochemical characterization of primary neuronal cultures from adult rat brain: Differential expression of neuronal and glial protein markers. J. Neurosci. Methods. 2009, 184, 294–302. [Google Scholar] [CrossRef][Green Version]
- Kaneko, A.; Sankai, Y. Long-Term Culture of Rat Hippocampal Neurons at Low Density in Serum-Free Medium: Combination of the Sandwich Culture Technique with the Three-Dimensional Nanofibrous Hydrogel PuraMatrix. PLoS ONE 2014, 9, e102703. [Google Scholar] [CrossRef]
- Carrodus, N.L.; Teng, K.S.L.; Munro, K.M.; Kennedy, M.J.; Gunnersen, J.M. Differential Labeling of Cell-surface and Internalized Proteins after Antibody Feeding of Live Cultured Neurons. J. Vis. Exp. 2014, 84, e51139. [Google Scholar]
- Badman, R.P.; Moore, S.L.; Killian, J.L.; Feng, T.; Cleland, T.A.; Hu, F.; Wang, M.D. Dextran-coated iron oxide nanoparticle-induced nanotoxicity in neuron cultures. Sci. Rep. 2020, 10, 11239. [Google Scholar] [CrossRef]
- Sun, M.Y.; Taylor, A.; Zorumski, C.F.; Mennerick, S. 24S-hydroxycholesterol and 25-hydroxycholesterol differentially impact hippocampal neuronal survival following oxygen-glucose deprivation. PLoS ONE 2017, 12, e0174416. [Google Scholar] [CrossRef] [PubMed]
- Volpicelli-Daley, L.A.; Luk, K.C.; Lee, V.M.Y. Addition of exogenous α-synuclein preformed fibrils to primary neuronal cultures to seed recruitment of endogenous α-synuclein to Lewy body and Lewy neurite–like aggregates. Nat. Protoc. 2014, 9, 2135–2146. [Google Scholar] [CrossRef] [PubMed]
- Nobuhara, C.K.; DeVos, S.L.; Commins, C.; Wegmann, S.; Moore, B.D.; Roe, A.D.; Costantino, I.; Frosch, M.P.; Pitstick, R.; Carlson, G.A.; et al. Tau Antibody Targeting Pathological Species Blocks Neuronal Uptake and Interneuron Propagation of Tau in vitro. Am. J. Pathol. 2017, 187, 1399–1412. [Google Scholar] [CrossRef]
- Jovičič, A.; Mertens, J.; Boeynaems, S.; Bogaert, E.; Chai, N.; Yamada, S.B.; Paul, J.W.; Sun, S.; Herdy, J.R.; Bieri, G.; et al. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci. 2015, 18, 1226–1229. [Google Scholar] [CrossRef]
- Henderson, M.X.; Sedor, S.; McGeary, I.; Cornblath, E.J.; Peng, C.; Riddle, D.M.; Howard, L.L.; Zhang, G.; Broawn, H.J.; Olufemi, M.F.; et al. Glucocerebrosidase Activity Modulates Neuronal Susceptibility to Pathological α-Synuclein Insult. Neuron 2020, 105, 822–836.e7. [Google Scholar] [CrossRef]
- Kramer, N.J.; Haney, M.S.; Morgens, D.W.; Jovičić, A.; Couthouis, J.; Li, A.; Ousey, J.; Ma, R.; Bieri, G.; Tsui, C.K.; et al. CRISPR–Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Gen. 2018, 50, 603–612. [Google Scholar] [CrossRef]
- Chesi, A.; Staahl, B.T.; Jovičić, A.; Couthouis, J.; Fasolino, M.; Raphael, A.R.; Yamazaki, T.; Elias, L.; Polak, M.; Kelly, C.; et al. Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 2013, 16, 851–855. [Google Scholar] [CrossRef]
- Shin, W.S.; Di, J.; Cao, Q.; Li, B.; Seidler, P.M.; Murray, K.A.; Bitan, G.; Jiang, L. Amyloid β-protein oligomers promote the uptake of tau fibril seeds potentiating intracellular tau aggregation. Alzheimers Res. Ther. 2019, 11, 86. [Google Scholar] [CrossRef] [PubMed]
- Haileselassie, B.; Joshi, A.U.; Minhas, P.S.; Mukherjee, R.; Andreasson, K.I.; Mochly-Rosen, D. Mitochondrial dysfunction mediated through dynamin-related protein 1 (Drp1) propagates impairment in blood brain barrier in septic encephalopathy. J. Neuroinflammation. 2020, 17, 36. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Liu, G.; Jin, S.M.; Parisiadou, L.; Xie, C.; Yu, J.; Sun, L.; Ma, B.; Ding, J.; Vancraenenbroeck, R.; et al. MicroRNA-205 regulates the expression of Parkinson’s disease-related leucine-rich repeat kinase 2 protein. Hum. Mol. Genet. 2013, 22, 608–620. [Google Scholar] [CrossRef]
- Argaw, A.; Duff, G.; Zabouri, N.; Cécyre, B.; Chainé, N.; Cherif, H.; Tea, H.; Lutz, B.; Ptito, M.; Bouchard, J.-F. Concerted Action of CB1 Cannabinoid Receptor and Deleted in Colorectal Cancer in Axon Guidance. J. Neurosci. 2011, 31, 1489–1499. [Google Scholar] [CrossRef]
- Smith, H.L.; Freeman, O.J.; Butcher, A.J.; Holmqvist, S.; Humoud, I.; Schätzl, T.; Hughes, D.T.; Verity, N.C.; Swinden, D.P.; Hayes, J.; et al. Astrocyte Unfolded Protein Response Induces a Specific Reactivity State that Causes Non-Cell-Autonomous Neuronal Degeneration. Neuron 2020, 105, 855–866.e5. [Google Scholar] [CrossRef]
- Ding, X.; Wang, J.; Huang, M.; Chen, Z.; Liu, J.; Zhang, Q.; Zhang, C.; Xiang, Y.; Zen, K.; Li, L. Loss of microglial SIRPα promotes synaptic pruning in preclinical models of neurodegeneration. Nat. Commun. 2021, 12, 2030. [Google Scholar] [CrossRef] [PubMed]
- Abdelmotilib, H.; Maltbie, T.; Delic, V.; Liu, Z.; Hu, X.; Fraser, K.B.; Moehle, M.S.; Stoyka, L.; Anabtawi, H.; Krendelchtchikova, V.; et al. α-Synuclein fibril-induced inclusion spread in rats and mice correlates with dopaminergic Neurodegeneration. Neurobiol. Dis. 2017, 105, 84–98. [Google Scholar] [CrossRef]
- He, Z.; McBride, J.D.; Xu, H.; Changolkar, L.; Kim, S.; Zhang, B.; Narasimhan, S.; Gibbons, G.S.; Guo, J.L.; Kozak, M.; et al. Transmission of tauopathy strains is independent of their isoform composition. Nat. Commun. 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.S.; Farhi, S.L.; Zhao, Y.; Brinks, D.; Zou, P.; Ruangkittisakul, A.; Plastisa, J.; Pieribone, V.A.; Ballanyi, K.; Cohen, A.E.; et al. A Bright and Fast Red Fluorescent Protein Voltage Indicator That Reports Neuronal Activity in Organotypic Brain Slices. J. Neurosci. 2016, 36, 2458–2472. [Google Scholar] [CrossRef] [PubMed]
- Klapoetke, N.C.; Murata, Y.; Kim, S.S.; Pulver, S.R.; Birdsey-Benson, A.; Cho, Y.K.; Morimoto, T.K.; Chuong, A.S.; Carpenter, E.J.; Tian, Z.; et al. Independent optical excitation of distinct neural populations. Nat. Methods. 2014, 11, 338–346. [Google Scholar] [CrossRef]
- Duque, M.; Lee-Kubli, C.A.; Tufail, Y.; Magaram, U.; Patel, J.; Chakraborty, A.; Mendoza, L.J.; Edsinger, E.; Vasan, A.; Weiss, C.; et al. Sonogenetic control of mammalian cells using exogenous Transient Receptor Potential A1 channels. Nat. Commun. 2022, 13, 600. [Google Scholar] [CrossRef]
- Hagihara, H.; Shoji, H.; Otabi, H.; Toyoda, A.; Katoh, K.; Namihira, M.; Myakawa, T. Protein lactylation induced by neural excitation. Cell Rep. 2021, 37, 109820. [Google Scholar] [CrossRef]
- Wani, A.; Gupta, M.; Ahmad, M.; Shah, A.M.; Ahsan, A.U.; Qazi, P.H.; Malik, F.; Singh, G.; Sharma, P.R.; Kaddoumi, A.; et al. Alborixin clears amyloid-β by inducing autophagy through PTEN-mediated inhibition of the AKT pathway. Autophagy 2019, 15, 1810–1828. [Google Scholar] [CrossRef]
- Wall, M.J.; Collins, D.R.; Chery, S.L.; Allen, Z.D.; Pastuzyn, E.D.; George, A.J.; Nikolova, V.D.; Moy, S.S.; Philpot, B.D.; Shepherd, J.D.; et al. The Temporal Dynamics of Arc Expression Regulate Cognitive Flexibility. Neuron 2018, 98, 1124–1132.e7. [Google Scholar] [CrossRef]
- Urrios, A.; Parra-Cabrera, C.; Bhattacharjee, N.; Gonzalez-Suarez, A.M.; Rigat-Brugarolas, L.G.; Nallapatti, U.; Samitier, J.; DeForest, C.A.; Posas, F.; Garcia-Cordero, J.L.; et al. 3D-Printing of Transparent Bio-Microfluidic Devices in PEG-DA. Lab Chip. 2016, 16, 2287. [Google Scholar] [CrossRef]
- Ouyang, Q.; Lizarraga, S.B.; Schmidt, M.; Yang, U.; Gong, J.; Ellisor, D.; Kauer, J.A.; Morrow, E.M. Christianson Syndrome Protein NHE6 Modulates TrkB Endosomal Signaling Required for Neuronal Circuit Development. Neuron 2013, 80, 97–112. [Google Scholar] [CrossRef] [PubMed]
- Piatkevich, K.D.; Jung, E.E.; Straub, C.; Linghu, C.; Park, D.; Suk, H.J.; Hochbaum, D.R.; Goodwin, D.; Pnevmatikakis, E.; Pak, H.; et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters. Nat. Chem. Biol. 2018, 14, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.Y.; Pan, L.; Su, S.C.; Quinn, E.J.; Sasaki, M.; Jimenez, J.C.; Mackenzie, I.R.A.; Huang, E.J.; Tsai, L. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013, 16, 1383–1391. [Google Scholar] [CrossRef]
- Tushev, G.; Glock, C.; Heumüller, M.; Biever, A.; Jovanovic, M.; Schuman, E.M. Alternative 3′ UTRs Modify the Localization, Regulatory Potential, Stability, and Plasticity of mRNAs in Neuronal Compartments. Neuron 2018, 98, 495–511.e6. [Google Scholar] [CrossRef]
- Staahl, B.T.; Benekareddy, M.; Coulon-Bainier, C.; Banfal, A.A.; Floor, S.N.; Sabo, J.K.; Urnes, C.; Nunares, G.A.; Ghosh, A.; Doudna, J.A. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 2017, 35, 431–434. [Google Scholar] [CrossRef]
- Rorabaugh, J.M.; Chalermpalanupap, T.; Botz-Zapp, C.A.; Fu, V.M.; Lembeck, N.A.; Cohen, R.M.; Weinshenker, D. Chemogenetic locus coeruleus activation restores reversal learning in a rat model of Alzheimer’s disease. Brain 2017, 140, 3023–3038. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.; LeVault, K.R.; Barnett, A.J.; Brewer, G.J. A Reversible Early Oxidized Redox State That Precedes Macromolecular ROS Damage in Aging Nontransgenic and 3xTg-AD Mouse Neurons. J. Neurosci. 2012, 32, 5821–5832. [Google Scholar] [CrossRef]
- Wang, Y.J.; Wang, X.; Lu, J.J.; Li, Q.X.; Gao, C.Y.; Liu, X.H.; Sun, Y.; Yang, M.; Lim, Y.; Genevieve, E.; et al. p75NTR Regulates Aβ Deposition by Increasing Aβ Production But Inhibiting Aβ Aggregation with Its Extracellular Domain. J. Neurosci. 2011, 31, 2292–2304. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Liu, S.; Tian, M.; Zhang, W.; Zhu, T.; Li, D.; Wu, J.; Deng, H.; Jia, Y.; X, W.; et al. Activity-induced histone modifications govern Neurexin-1 mRNA splicing and memory preservation. Nat. Neurosci. 2017, 20, 690–699. [Google Scholar] [CrossRef] [PubMed]
- Fares, M.B.; Ait-Bouziad, N.; Dikiy, I.; Mbefo, M.K.; Jovičić, A.; Kiely, A.; Holton, J.L.; Lee, S.J.; Gitler, A.D.; Eliezer, D.; et al. The novel Parkinson’s disease linked mutation G51D attenuates in vitro aggregation and membrane binding of α-synuclein, and enhances its secretion and nuclear localization in cells. Hum. Mol. Genet. 2014, 23, 4491–4509. [Google Scholar] [CrossRef]
- Lee, J.W.; Huang, B.X.; Kwon, H.S.; Rashid, M.A.; Kharebava, G.; Desai, A.; Patnaik, S.; Marugan, J.; Kim, H.Y. Orphan GPR110 (ADGRF1) targeted by N-docosahexaenoylethanolamine in development of neurons and cognitive function. Nat. Commun. 2016, 7, 13123. [Google Scholar] [CrossRef]
- Descloux, A.; Grußmayer, K.S.; Bostan, E.; Lukes, T.; Bouwens, A.; Sharipov, A.; Geissbuehler, S.; Mahul-Mellier, A.-L.; Lashuel, H.A.; Leutenegger, M.; et al. Combined multi-plane phase retrieval and super-resolution optical fluctuation imaging for 4D cell microscopy. Nat. Photonics 2018, 12, 165–172. [Google Scholar] [CrossRef]
- Smith, D.E.; Lipsky, B.P.; Russell, C.; Ketchem, R.R.; Kirchner, J.; Hensley, K.; Huang, Y.; Friedman, W.J.; Boissonneault, V.; Plante, M.-M.; et al. A Central Nervous System-Restricted Isoform of the Interleukin-1 Receptor Accessory Protein Modulates Neuronal Responses to Interleukin-1. Immunity 2009, 30, 817–831. [Google Scholar] [CrossRef]
- Ortega, Z.; Díaz-Hernández, M.; Maynard, C.J.; Hernández, F.; Dantuma, N.P.; Lucas, J.J. Acute Polyglutamine Expression in Inducible Mouse Model Unravels Ubiquitin/Proteasome System Impairment and Permanent Recovery Attributable to Aggregate Formation. J. Neurosci. 2010, 30, 3675–3688. [Google Scholar] [CrossRef]
- Guo, J.L.; Narasimhan, S.; Changolkar, L.; He, Z.; Stieber, A.; Zhang, B.; Stieber, A.; Zhang, B.; Gathagan, R.J.; Iba, M.; et al. Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016, 213, 2635–2654. [Google Scholar] [CrossRef]
- Fang, W.; Zhang, J.; Hong, L.; Huang, W.; Dai, X.; Ye, Q.; Chen, X. Metformin ameliorates stress-induced depression-like behaviors via enhancing the expression of BDNF by activating AMPK/CREB-mediated histone acetylation. J. Affect. Disord. 2020, 260, 302–313. [Google Scholar] [CrossRef]
- Sultan, F.A.; Wang, J.; Tront, J.; Liebermann, D.A.; David Sweatt, J. Genetic Deletion of gadd45b, a Regulator of Active DNA Demethylation, Enhances Long-Term Memory and Synaptic Plasticity. J. Neurosci. 2012, 32, 17059–17066. [Google Scholar] [CrossRef]
- Grozdanov, V.; Müller, A.; Sengottuvel, V.; Leibinger, M.; Fischer, D. A Method for Preparing Primary Retinal Cell Cultures for Evaluating the Neuroprotective and Neuritogenic Effect of Factors on Axotomized Mature CNS Neurons. Curr. Protoc. Neurosci. 2010, 53, 3.22.1–3.22.10. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.Y.; Wu, Y.M.; Ji, Z.; Gao, X.Y.; Pan, S.Y. A Modified Technique for Culturing Primary Fetal Rat Cortical Neurons. Biomed. Res. Int. 2012, 2012, 803930. [Google Scholar] [CrossRef] [PubMed]
- Spaethling, J.M.; Na, Y.J.; Lee, J.; Ulyanova, A.V.; Baltuch, G.H.; Bell, T.J.; Brem, S.; Chen, H.I.; Dueck, H.; Fisher, S.A.; et al. Primary Cell Culture of Live Neurosurgically Resected Aged Adult Human Brain Cells and Single Cell Transcriptomics. Cell Rep. 2017, 18, 791–803. [Google Scholar] [CrossRef]
- Fang, H.; Bygrave, A.M.; Roth, R.H.; Johnson, R.C.; Huganir, R.L. An optimized crispr/cas9 approach for precise genome editing in neurons. Elife 2021, 10, e65202. [Google Scholar] [CrossRef]
- Francisco, D.M.F.; Marchetti, L.; Rodríguez-Lorenzo, S.; Frías-Anaya, E.; Figueiredo, R.M.; Winter, P.; Romero, I.A.; de Vries, H.E.; Engelhardt, B.; Bruggmann, R. Advancing brain barriers RNA sequencing: Guidelines from experimental design to publication. Fluids Barriers CNS. 2020, 17, 51. [Google Scholar] [CrossRef]
- Sünwoldt, J.; Bosche, B.; Meisel, A.; Mergenthaler, P. Neuronal culture microenvironments determine preferences in bioenergetic pathway use. Front. Mol. Neurosci. 2017, 10, 294420. [Google Scholar] [CrossRef]
- Mereu, E.; Lafzi, A.; Moutinho, C.; Ziegenhain, C.; McCarthy, D.J.; Álvarez-Varela, A.; Batlle, E.; Sagar; Grün, D.; Lau, J.K.; et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat. Biotechnol. 2020, 38, 747–755. [Google Scholar] [CrossRef]
- Bykov, Y.; Kim, S.H.; Zamarin, D. Preparation of single cells from tumors for single-cell RNA sequencing. Methods Enzymol. 2020, 632, 295–308. [Google Scholar]
- Rohani, N.; Hao, L.; Alexis, M.S.; Joughin, B.A.; Krismer, K.; Moufarrej, M.N.; Soltis, A.R.; Lauffenberger, D.A.; Yaffe, M.B.; Burge, C.B.; et al. Acidification of tumor at stromal boundaries drives transcriptome alterations associated with aggressive phenotypes. Cancer Res. 2019, 79, 1952–1966. [Google Scholar] [CrossRef]
- Phenolsulfonphthalein|C19H14O5S|CID 4766. PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Phenolsulfonphthalein (accessed on 5 September 2025).
- Schornack, P.A.; Gillies, R.J. Contributions of Cell Metabolism and H + Diffusion to the Acidic pH of Tumors. Neoplasia 2003, 5, 135–145. [Google Scholar] [CrossRef]
- Salgado, L.E.V.; Vargas-Hernández, C. Spectrophotometric Determination of the pKa, Isosbestic Point and Equation of Absorbance vs. pH for a Universal pH Indicator. Am. J. Analyt. Chem. 2014, 5, 1290–1301. [Google Scholar] [CrossRef]
- Gillies, R.J.; Martinez-Zaguilan, R.; Martinez, G.M.; Serranot, R.; Perona, R. Tumorigenic 3T3 cells maintain an alkaline intracellular pH under physiological conditions. Proc. Natl. Acad. Sci. USA 1990, 87, 7414–7418. [Google Scholar] [CrossRef]
- Ikeuchi, T.; Akhi, R.; Cardona Rodriguez, B.; Fraser, D.; Williams, D.; Kim, T.S.; Greenwell-Wild, T.; Overmiller, A.; Morasso, M.; Moutsopoulos, N. Dissociation of murine oral mucosal tissues for single cell applications. J. Immunol. Methods 2024, 525, 113605. [Google Scholar] [CrossRef]
- Panchision, D.M.; Chen, H.L.; Pistollato, F.; Papini, D.; Ni, H.T.; Hawley, T.S. Optimized flow cytometric analysis of central nervous system tissue reveals novel functional relationships among cells expressing CD133, CD15, and CD24. Stem Cells 2007, 25, 1560–1570. [Google Scholar] [CrossRef]
- Khoshbakht, S.; Albayrak, Ö.; Tiryaki, E.; Ağcaoğlu, O.; Öktem, A.; Pınar Sun, G.; Er Gülbezer, E.; Ertekin, S.S.; Boyvat, A.; Vural, A.; et al. A cost-effective protocol for single-cell RNA sequencing of human skin. Front. Immunol. 2024, 15, 1393017. [Google Scholar] [CrossRef]
- Clister, T.; Fey, R.M.; Garrison, Z.R.; Valenzuela, C.D.; Bar, A.; Leitenberger, J.J.; Kulkarni, R.P. Optimization of Tissue Digestion Methods for Characterization of Photoaged Skin by Single Cell RNA Sequencing Reveals Preferential Enrichment of T Cell Subsets. Cells 2024, 13, 266. [Google Scholar] [CrossRef]
- Lee, S.J.; Lee, C.R.; Kim, K.J.; Ryu, Y.H.; Kim, E.; Han, Y.N.; Moon, S.H.; Rhie, J.W. Optimal Condition of Isolation from an Adipose Tissue-Derived Stromal Vascular Fraction for the Development of Automated Systems. Tissue Eng. Regen. Med. 2020, 17, 203–208. [Google Scholar] [CrossRef]
- Brewer, G.J. Isolation and culture of adult rat hippocampal neurons. J. Neurosci. Methods 1997, 71, 143–155. [Google Scholar] [CrossRef]
- Aliaghaei, M.; Haun, J.B. Optimization of Mechanical Tissue Dissociation Using an Integrated Microfluidic Device for Improved Generation of Single Cells Following Digestion. Front. Bioeng. Biotechnol. 2022, 10, 841046. [Google Scholar] [CrossRef]
- Denisenko, E.; Guo, B.B.; Jones, M.; Hou, R.; de Kock, L.; Lassmann, T.; Poppe, D.; Clément, O.; Simmons, R.K.; Lister, R.; et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020, 21, 130. [Google Scholar] [CrossRef]
- Marsh, S.E.; Walker, A.J.; Kamath, T.; Dissing-Olesen, L.; Hammond, T.R.; de Soysa, T.Y.; Young, A.M.H.; Murphy, S.; Abdulraouf, A.; Nadaf, N.; et al. Dissection of artifactual and confounding glial signatures by single-cell sequencing of mouse and human brain. Nat. Neurosci. 2022, 25, 306–316. [Google Scholar] [CrossRef]
- Millet, L.J.; Jain, A.; Gillette, M.U. Less Is More: Oligomer Extraction and Hydrothermal Annealing Increase PDMS Adhesion Forces for Materials Studies and for Biology-Focused Microfluidic Applications. Micromachines 2023, 14, 214. [Google Scholar] [CrossRef]
- Worthington Biochemical Corporation. Tissue Dissociation Guide, 18th ed.; Worthington Biochemical Corporation: Lakewood, NJ, USA, 2019. [Google Scholar]

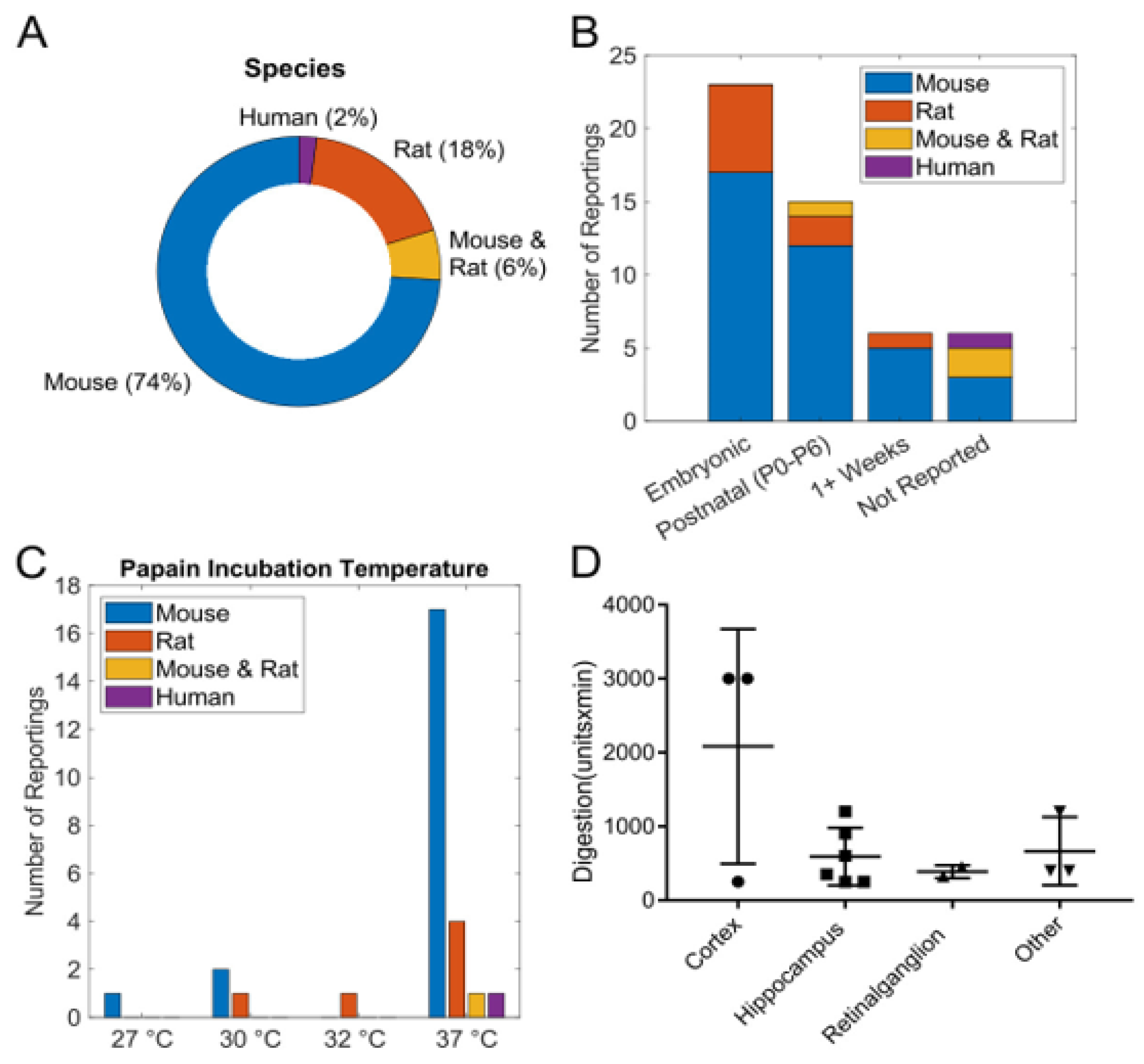

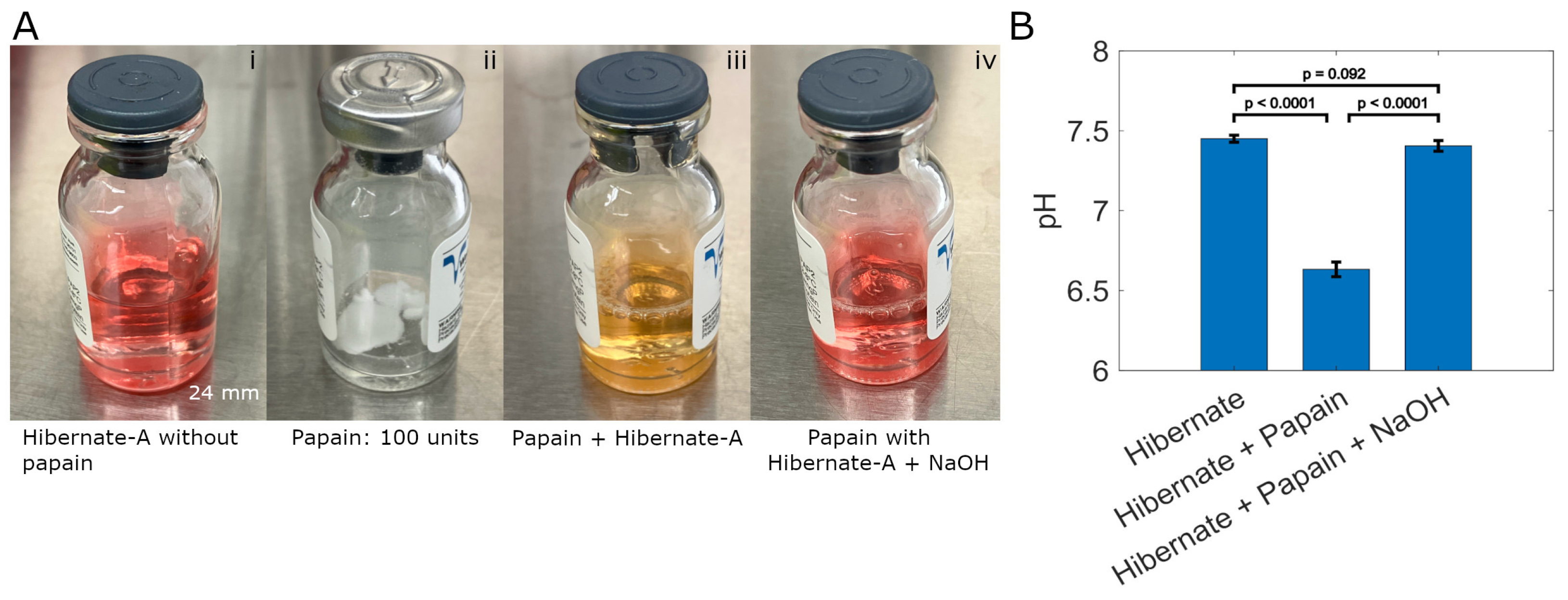
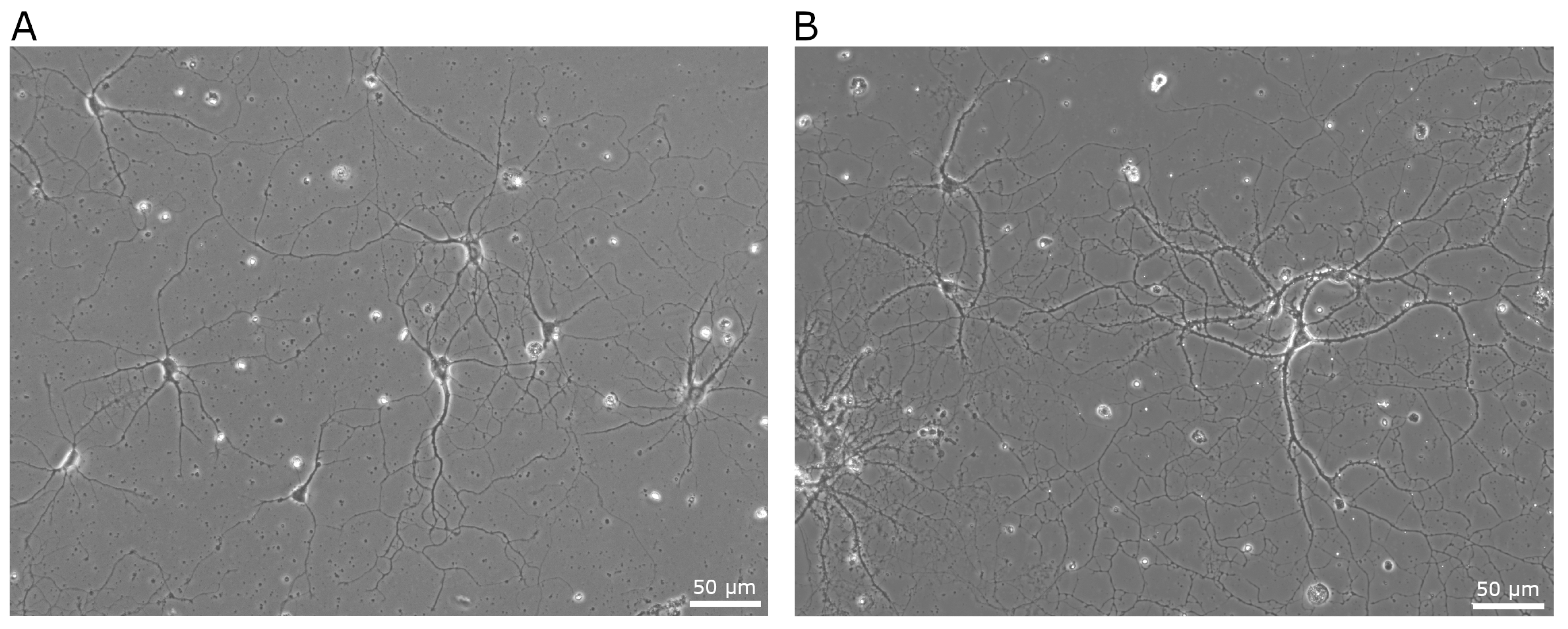
| Domain | Key Considerations | Research Implications |
|---|---|---|
| Enzymatic Variability | Variability in published protocols: enzyme types, storage, activity units, incubation times, and buffer conditions | Underscores the need for harmony and transparency in methods |
| Minimize Mechanical Stress | Gentle dissociation reduces cellular stress and clumping Preservation of fragile cell types | Improve viability and yields for tissue engineering and single-cell analysis |
| Tissue-Specific Enzyme Tuning for Neuroanatomical Regions | ECM composition Glial density, fiber tracts, and vascularity Brain or spine tissue source Region-specific dissociation protocols | Enhances cell recovery Reduces batch variation Demands region-specific adjustments to isolate intact, and representative cell types |
| Avoiding Population Skew | Harsh and weak enzymatic conditions bias robust cell types and deplete fragile populations | Affects conclusions for studies using heterogenous cell populations |
| Age-Dependent Tissue Maturity | Postnatal tissues: more dense cellular connections Embryonic tissues: more effective enzymatic digestion | Requires stage-matched dissociation Shapes cell survival, phenotypic stability, and experimental relevance |
| Oxidative Stress | Neurons are highly susceptible to ROS due to high metabolic rates and low antioxidant defenses | Elevated oxidative stress impairs viability and biases downstream functional assays |
| Ref.# | Year | Citations | Source | Region | Age | mg/mL | Media | U. | Min. | °C | Mfr. | Cat. No. | pH | L-Cys. | DNase |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| [1] | 2009 | 124 | Mouse | C | X | x | Custom | 100 | 30 | 37 | S-A | x | yes (7.4) | Use | x |
| [2] | 2019 | 105 | Rat | H | E17–E18 | 0.1 | PBS | x | 10 | 37 | S-A | P-4762 | x | Use | Use |
| [3] | 2017 | 28 | Mouse | TG | 6–10 weeks | x | HBSS+ | 40 | 30 | 37 | W | x | yes | Use | x |
| [4] | 2009 | 39 | Rat | C | 16 weeks | 2.0 | Hib. A | x | 30 | 30 | W | x | x | x | x |
| [5] | 2014 | 55 | Rat | H | E18 | 3.5 | PBS | x | 12 | 32 | Wako | x | x | x | Use |
| [6] | 2014 | 33 | Rat | H | E18 | x | PDS | 20 | 15–20 | 37 | WBC | PDS | x | Use | Use |
| [7] | 2020 | 28 | Mouse | WB | P0 or P1 | N/A | N/A | N/A | 10–30 | x | BB | N/A | N/A | N/A | N/A |
| [8] | 2017 | 37 | Mouse & Rat | H | P1-3 | 1.0 | L-15 | x | x | x | N/A | N/A | x | Use | x |
| [9] | 2014 | 681 | Mouse | H | E16–E17 | x | HBSS | 20 | 30–60 | 37 | WB | LS 3126 | x | Use | Use |
| [10] | 2017 | 110 | Mouse | C | E14–E15 | x | PDS | 20 | x | x | WBC | x | x | Use | Use |
| [11] | 2015 | 656 | Mouse | C | E17 | x | PDS | 20 | x | x | WB | LK003150 | x | Use | Use |
| [12] | 2020 | 126 | Mouse | C, VM, S | E16–E18; P1 | x | x | 20 | 20 | 37 | WB | LS003126 | x | Use | x |
| [13] | 2018 | 233 | Mouse | C | E16.5 | x | PDS | 20 | x | x | WBC | PDS | x | Use | Use |
| [14] | 2013 | 166 | Mouse | SC | E12.5 | x | PDS | 20 | x | x | W | PDS | x | Use | Use |
| [15] | 2019 | 101 | Mouse | H | P0 or P1 | 0.5 | PGB | x | 20 | 37 | x | x | x | x | Use |
| [16] | 2020 | 136 | Mouse | C | E17 | x | PDS | 20 | x | x | WBC | PDS | x | Use | Use |
| [17] | 2012 | 307 | Mouse | C | P0 | N/A | x | N/A | x | x | W | x | x | x | x |
| [18] | 2011 | 117 | Mouse | RGC | P7–P8 | x | N/A | 15 | 30 | 37 | WB | x | x | Use | Use |
| [19] | 2020 | 188 | Mouse | H | P1 | N/A | x | N/A | 20 | 27 | x | x | x | x | x |
| [20] | 2021 | 101 | Mouse | Ce | N/A | N/A | x | N/A | 30 | x | x | x | x | x | x |
| [21] | 2017 | 180 | Mouse | H | E16–17 | N/A | HBSS+ | N/A | x | x | WB | x | x | x | x |
| [22] | 2020 | 121 | Mouse | H | E16–E18 | N/A | x | N/A | 45 | 37 | WBC | LS003126 | x | Use | x |
| [23] | 2016 | 167 | Rat | H | P0 | x | x | x | x | x | x | x | x | x | x |
| [24] | 2014 | 1275 | Mouse | H | P0–1 | x | x | 50 | 5 | x | WB | x | x | x | x |
| [25] | 2022 | 103 | Rat | C, H, VZ | E18 | 2.0 | x | x | 30 | 37 | BB | PAP | x | x | x |
| [26] | 2021 | 165 | Mouse | C and H | E15.5–16.5 | N/A | HBSS | N/A | 12 | 37 | WBC | LS003127 | x | Use | Use |
| [27] | 2019 | 102 | Mouse | C | E18 | 1.0 | x | x | 10 | 37 | S-A | 76220 | x | x | x |
| [28] | 2018 | 102 | Rat | C | E18 | x | x | N/A | x | x | WB | LS003126 | x | Use | Use |
| [29] | 2016 | 301 | Mouse | H | E18 | x | PDS | 20 | x | x | WB | PDS | x | Use | Use |
| [30] | 2013 | 161 | Mouse | H | P0–P1 | x | EBSS | 20 | 30 | 37 | x | x | x | x | x |
| [31] | 2018 | 344 | Mouse | H | P0–P1 | x | x | 50 | 6–8 | x | WB | x | x | x | x |
| [32] | 2013 | 485 | Mouse | C | E15.5–16.5 | x | HBSS | N/A | x | 37 | W | x | x | x | Use |
| [33] | 2018 | 363 | Rat | H | P0–1 | x | x | x | x | x | x | x | x | x | x |
| [34] | 2017 | 370 | Mouse | C | E13.5 | N/A | x | N/A | 10 | 37 | MB | 130-092-628 | x | x | x |
| [35] | 2017 | 212 | Mouse | LC | P1 | 1.0 | Custom | x | 30 | 37 | S-A | P4762 | x | x | Use |
| [36] | 2012 | 138 | Mouse | H | 2,4,8,11,21 mo. | 2.0 | Hib. A | x | 30 | 30 | W | x | x | x | x |
| [37] | 2011 | 109 | Mouse | C | 1 mo. | x | Custom | x | 30 | 30 | x | x | x | x | x |
| [38] | 2017 | 103 | Mouse | DG/H | 3–4 mo. | x | EBSS | 20 | 60 | 37 | x | x | x | x | Use |
| [39] | 2014 | 278 | Mouse | H | P0 | x | x | x | x | x | x | x | x | x | x |
| [40] | 2016 | 154 | Mouse | C | P0 | x | x | 100 | 30 | 37 | x | x | x | x | x |
| [41] | 2018 | 130 | Mouse | H | P0 | x | x | 20 | x | x | S-A | x | x | x | x |
| [42] | 2009 | 137 | Mouse | C | E17–E18 | x | x | x | 45 | 37 | WB | x | x | x | x |
| [43] | 2010 | 113 | Mouse | C and S | E18 | x | PDS | N/A | x | x | W | PDS | x | Use | Use |
| [44] | 2016 | 443 | Mouse | H and C | E16–18 | x | x | x | x | x | WBC | x | x | x | x |
| [45] | 2020 | 127 | Mouse | WB | N/A | x | x | 20 | 20 | 37 | x | x | yes | x | x |
| [46] | 2012 | 129 | Mouse | H | P0-P2 | N/A | x | N/A | x | x | x | x | x | x | x |
| [47] | 2010 | 34 | Mouse & Rat | RGC | X | x | DMEM | 30-35 | 10 | 37 | W | x | x | Use | x |
| [48] | 2012 | 79 | Rat | C | E18 | N/A | DMEM | x | 30 | 37 | x | x | x | x | Use |
| [49] | 2017 | 53 | Human | C | N/A | x | x | 20 | 10–15 | 37 | WB | x | x | x | x |
| [50] | 2021 | 55 | Mouse & Rat | C | E18 | x | x | x | x | x | x | x | x | x | x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hilner, J.M.; Turner, A.; Vollmar-Zygarlenski, C.; Millet, L.J. Rigor & Reproducibility: pH Adjustments of Papain with L-Cysteine Dissociation Solutions and Cell Media Using Phenol Red Spectrophotometry. Biosensors 2025, 15, 727. https://doi.org/10.3390/bios15110727
Hilner JM, Turner A, Vollmar-Zygarlenski C, Millet LJ. Rigor & Reproducibility: pH Adjustments of Papain with L-Cysteine Dissociation Solutions and Cell Media Using Phenol Red Spectrophotometry. Biosensors. 2025; 15(11):727. https://doi.org/10.3390/bios15110727
Chicago/Turabian StyleHilner, Joshua M., Allison Turner, Calissa Vollmar-Zygarlenski, and Larry J. Millet. 2025. "Rigor & Reproducibility: pH Adjustments of Papain with L-Cysteine Dissociation Solutions and Cell Media Using Phenol Red Spectrophotometry" Biosensors 15, no. 11: 727. https://doi.org/10.3390/bios15110727
APA StyleHilner, J. M., Turner, A., Vollmar-Zygarlenski, C., & Millet, L. J. (2025). Rigor & Reproducibility: pH Adjustments of Papain with L-Cysteine Dissociation Solutions and Cell Media Using Phenol Red Spectrophotometry. Biosensors, 15(11), 727. https://doi.org/10.3390/bios15110727






