Abstract
Colorectal cancer (CRC) remains a leading cause of cancer-related morbidity and mortality worldwide, with patient outcomes highly dependent on early and accurate diagnosis. However, existing diagnostic methods, such as colonoscopy, fecal occult blood testing, and imaging, are often invasive, costly, or lack sufficient sensitivity and specificity, particularly in early-stage disease. In this context, aptamers, which are synthetic single-stranded oligonucleotides capable of binding to specific targets with high affinity, have emerged as a powerful alternative to antibodies for biosensing applications. This review provides a comprehensive overview of aptamer-based strategies for CRC detection, spanning from biomarker discovery to clinical translation. We first examine established and emerging CRC biomarkers, including those approved by regulatory agencies, described in patents, and shared across multiple cancer types. We then discuss recent advances in aptamer selection and design, with a focus on SELEX variants and in silico optimization approaches tailored to CRC-relevant targets. The integration of aptamers into cutting-edge sensing platforms, such as electrochemical, optical, and nanomaterial-enhanced aptasensors, is highlighted, with emphasis on recent innovations that enhance sensitivity, portability, and multiplexing capabilities. Furthermore, we explore the convergence of aptasensing with microfluidics, and wearable technologies to enable intelligent, miniaturized diagnostic systems. Finally, we consider the clinical and regulatory pathways for point-of-care implementation, as well as current challenges and opportunities for advancing the field. By outlining the technological and translational trajectory of aptamer-based CRC diagnostics, this review aims to provide a roadmap for future research and interdisciplinary collaboration in precision oncology.
1. Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed malignancy worldwide and the second leading cause of cancer-related mortality. Its incidence is steadily increasing, driven not only by aging populations but also by modified lifestyle factors such as diet, obesity, and physical inactivity, which underscores the need for effective strategies for prevention, early detection, and treatment [1]. Despite advances in therapeutic approaches, prognosis remains strongly stage-dependent, making timely and accurate diagnosis the cornerstone of improved patient outcomes [2].
Population-based screening programs have been implemented in many countries, generally targeting individuals over 45 years of age. These initiatives have successfully reduced mortality where participation is high, yet adherence rates remain highly variable across countries and socioeconomic groups [2]. Current screening relies predominantly on stool-based assays. The fecal immunochemical test (FIT), which detects hidden blood, is widely used due to its non-invasiveness and affordability. More recently, DNA/RNA-based assays have been developed to detect tumor-associated genetic alterations shed into stool samples [3]. U.S. Food and Drug Administration (FDA)-approved test such as Cologuard, which detects multiple DNA markers [4], and ColoSense, which targets RNA signatures, exemplify this progress. Nevertheless, colonoscopy remains the gold standard for CRC diagnosis, as it allows direct visualization of the colon, detection of early lesions, and removal of precancerous polyps [5].
Despite these advances, the sensitivity and specificity of current non-invasive tests remain imperfect. FIT achieves approximately 70–75% sensitivity and 90–95% specificity, while multitarget stool DNA assays such as Cologuard reach ~92% sensitivity but lower specificity (~87%) [3,4,5]. This performance, although clinically valuable, leaves room for improvement, particularly for early-stage CRC and precancerous lesions. These limitations have spurred increasing interest in alternative blood-based diagnostic strategies that could offer comparable or superior accuracy with higher patient compliance.
Current CRC screening approaches, including stool-based tests and colonoscopy, still face major challenges that limit their effectiveness in real-world settings. Participation in stool-based screening remains suboptimal, often hindered by lack of awareness, socioeconomic barriers, or perceived inconvenience [6]. FIT lacks sensitivity for advanced adenomas and early-stage cancers, while molecular stool tests, although more sensitive, are costly and not universally accessible. Colonoscopy, despite its comprehensiveness, is invasive, expensive, and associated with low patient compliance [7]. These limitations highlight a persistent gap between technical capability and public health impact. In this context, liquid biopsy approaches, based on the detection of circulating tumor-derived material in biological fluids (e.g., blood, saliva, urine) are emerging as promising alternatives that may enhance compliance and broaden screening coverage [8]. By enabling minimally invasive detection of circulating tumor DNA, RNA, proteins, or extracellular vesicles, liquid biopsy holds potential to overcome many of the challenges faced by conventional screening modalities.
Within this landscape, aptamers have gained considerable attention as versatile recognition elements that could accelerate the development of next-generation liquid biopsy strategies [9]. These short, single-stranded nucleic acids fold into defined three-dimensional structures, enabling them to bind a wide spectrum of molecular targets with high affinity and specificity. Compared with antibodies, aptamers offer several practical advantages: they can be generated entirely in vitro through iterative selection (SELEX), are chemically stable under a broad range of conditions, and can be synthesized at scale with high reproducibility and relatively low cost [10]. Their modular nature also facilitates conjugation with nanomaterials, fluorophores, or electroactive compounds, expanding their functionality in biosensing applications.
Importantly, the adaptability of aptamers makes them well suited for integration into liquid biopsy platforms. They can be tailored to capture circulating tumor biomarkers, thus directly addressing the biomarker diversity highlighted in current CRC research. Combined with advances in nanotechnology, microfluidics, and biosensor engineering, aptamer-based systems are emerging as promising tools for highly sensitive, rapid, and minimally invasive CRC diagnostics. While challenges related to clinical validation, standardization, and regulatory approval remain [10], the unique features of aptamers position them as strong candidates to bridge the gap between laboratory innovation and point-of-care implementation.
Previously, comprehensive and high-quality reviews have focused on the use of aptamers for therapeutic applications and their selection through the cell-SELEX strategy [11,12,13]. However, these works predate several important developments in the field. Since 2019, significant progress has been made in expanding the classes of colorectal cancer (CRC) biomarkers recognized by aptamers, including circulating tumor cells, exosomal and stool-derived nucleic acids, and inflammation-associated proteins, as well as in the engineering of miniaturized, multiplexed, and wearable aptasensors. Furthermore, recent studies have introduced new SELEX variants integrating magnetic, microfluidic, and in silico optimization strategies, alongside improved clinical benchmarking for diagnostic validation.
In contrast to earlier reviews, this work provides a comprehensive and up-to-date overview of aptamer-based strategies for CRC detection, emphasizing advances in biosensing platforms, materials engineering, and translational readiness. We first examine CRC-relevant biomarkers, particularly protein targets that serve as foundations for aptamer selection. We then highlight recent advances in SELEX methodologies, aptamer optimization, and their integration into electrochemical and other biosensing platforms. Finally, we discuss the challenges and opportunities for clinical translation and commercialization, emphasizing the potential of aptamer-based diagnostics to complement or even replace current screening tools.
2. Colorectal Cancer Biomarkers
Molecular markers detectable in biological fluids are central to improving the sensitivity and specificity of early cancer detection. They constitute the foundation of liquid biopsy–based strategies, which enable minimally invasive diagnostic testing. In colorectal cancer (CRC), a wide spectrum of circulating biomarkers has been reported [14], including genetic and epigenetic alterations, nucleic acids (DNA and RNA) proteins and circulating tumor cells (Figure 1). In this review, we highlight both clinically validated and emerging CRC biomarkers, as the latter represent promising targets whose translation into practice strongly depends on the development of reliable analytical tools, such as aptamer-based assays. While each class offers valuable diagnostic and prognostic information, proteins remain the most widely implemented in clinical practice and are particularly attractive as targets for aptamer-based recognition due to their abundance, structural diversity, and direct association with tumor biology. The following section provides an overview of the principal biomarker categories, with special emphasis on protein markers as the cornerstone for emerging aptamer-driven diagnostic platforms.
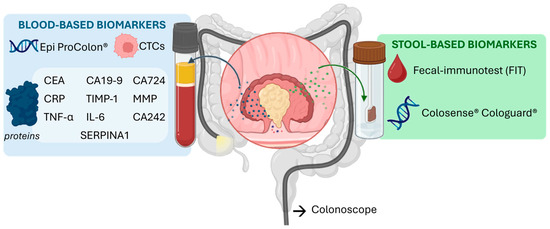
Figure 1.
Different types of biomarkers for CRC in stool and blood.
2.1. Genetic and Epigenetic Biomarkers
Genetic alterations play a central role in CRC pathogenesis and progression, with recurrent mutations in APC, KRAS, TP53, and BRAF among the most extensively studied [15]. APC mutations are detected in up to 90% of CRC cases and serve as early events in tumorigenesis [16], whereas KRAS and TP53 alterations are more frequently associated with prognosis and therapy response [17]. Additional recurrent mutations involve PIK3CA, SMAD4, NRAS, and others, though their utility for population screening remains limited [18,19]. Recently, transcriptomic panels such as ColonSentry, based on expression profiling of six genes, have been approved for clinical use in high-risk patients to support adherence to colonoscopy with a sensitivity of 78% and a specificity of 66% [20].
Epigenetic changes, particularly DNA methylation, also provide valuable biomarkers [21]. The most established is SEPT9 [22], approved for clinical testing (Epi proColon®), which demonstrates high specificity (86–99%) but modest sensitivity (56–79%) for early detection [23]. Other methylation targets, such as NDRG4 and BMP3, are under investigation and incorporated into multi-marker assays.
In addition, microsatellite instability (MSI), arising from defects in mismatch repair genes, represents both a diagnostic marker and a predictor of response to immunotherapy. MSI-high CRCs account for ~15% of cases, particularly in hereditary syndromes, and are increasingly guiding treatment decisions [24].
Taken together, genetic and epigenetic biomarkers provide valuable insights into CRC biology, prognosis, and therapy stratification, but their sensitivity and specificity remain insufficient for widespread use in early, non-invasive screening.
2.2. Circulating Nucleic Acids
Circulating tumor DNA (ctDNA), derived from apoptotic or necrotic tumor cells, is increasingly recognized as a promising biomarker in CRC [25]. ctDNA levels correlate with tumor burden, prognosis, and recurrence risk, and are particularly valuable for the detection of molecular residual disease (MRD) after surgery, enabling more precise, stage-independent treatment decisions [26]. However, ctDNA lacks specificity, as elevated levels may also occur in inflammatory or benign conditions, and its clinical performance improves mainly when combined with other biomarkers [27].
Circulating RNAs, including microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), also show potential as minimally invasive markers [28]. Several miRNAs, such as miR-21 [29], miR-29a [30], miR-92a [31], and miR-223 [32,33], are consistently overexpressed in CRC patients, while others (e.g., miR-145 [34], miR-18a [35]) are downregulated. Similarly, CRC-associated lncRNAs such as CCAT1 [36] and CCAT2 [37] demonstrate diagnostic and prognostic value, whereas others (e.g., HOTAIR, PVT1, H19) are shared across multiple cancers [38]. Despite encouraging results, both miRNAs and lncRNAs suffer from limited reproducibility and lack of standardization across studies, restricting their current clinical utility [39].
2.3. Circulating Tumor Cells
Circulating tumor cells (CTCs) are malignant cells shed from primary tumors or metastatic sites into the bloodstream. They have been identified across virtually all cancer types. In CRC, elevated CTC counts are generally associated with poorer prognosis and increased metastatic potential, highlighting their potential as prognostic biomarkers. Nevertheless, the diagnostic and prognostic value of CTCs remains a subject of debate, as studies to date have produced inconsistent or contradictory findings. Furthermore, false-positive results may arise due to the presence of benign cells that mimic CTCs, complicating their clinical interpretation [40].
2.4. Protein Biomarkers
Several serum proteins have emerged as potential biomarkers for CRC, most studied for diagnostic purposes and a few for prognostic use, aiding in predicting tumor progression and guiding treatment (Table 1). Currently, carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) are the main protein biomarkers used clinically. However, both lack the specificity and sensitivity required for definitive CRC diagnosis, as elevated levels may also occur in conditions such as inflammatory bowel disease or pancreatic cancer. CA19-9, primarily a pancreatic cancer marker, exhibits low sensitivity for CRC (26–48%), whereas CEA is more useful for post-treatment monitoring and detecting disease recurrence [41].
To overcome these limitations, other members of the CA family, including CA72-4 and CA242, have been investigated. While their combined detection can improve diagnostic accuracy and aid prognosis [42], CA72-4 shows only 50% sensitivity due to elevations in other cancers and benign conditions, limiting its utility for CRC screening [43]. CA242 is typically used alongside CEA to enhance sensitivity and serve as a prognostic indicator [44].
In search of more robust alternatives, several promising protein candidates are under evaluation. For instance, tissue inhibitor of metalloproteinases-1 (TIMP-1) [45] and matrix metalloproteinases (MMPs) [46] have shown encouraging diagnostic accuracy in population studies, reflecting tumor invasion and tissue remodelling processes [47]. Notably, Gimeno-Garcia et al. evaluated MMP-9 as a diagnostic biomarker in a cohort of 25 CRC patients, 50 individuals with adenomas, and 75 healthy controls that were previously referred for colonoscopy. Using a cut-off value of 204 ng mL−1, MMP-9 achieved 67% specificity, 80% sensitivity, and an area under the curve (AUC) of 0.8, demonstrating considerable diagnostic potential. Likewise, members of the serpin family (e.g., SERPINA1, SERPINA3, and SERPINC1), exhibit altered expression patterns in CRC, highlighting their involvement in disease progression. For instance, elevated SERPINA1 levels are associated with poorer patient outcomes and promote CRC cell proliferation and migration. Moreover, the use of SERPINA1 has been statistically studied in a cohort of 15 CRC, 15 adenomas, 15 healthy controls with a validation test of 19 CRC patients and 21 healthy controls, obtaining 95% of sensitivity and specificity, an AUC of 0.97 with a cu-off value un serum of 817 µg mL−1. Additionally, some serpins, such as SERPINB5, may interact with established CRC biomarkers like CEA and are linked to more advanced disease stages [48].
To enhance the specificity of colorectal cancer (CRC) detection, systemic inflammatory markers have been extensively studied, observing a correlation between systemic inflammatory indicators and CRC susceptibility. Key indicators include C-reactive protein (CRP), interleukin-6 (IL-6), and tumor necrosis factor-alpha (TNF-α). CRP, an acute-phase protein, shows elevated serum levels in CRC patients; in a study of 164 CRC patients, 34 with adenomas, and 119 healthy controls, CRP achieved an AUC of 0.64, with 17% sensitivity and 90% specificity, indicating limited diagnostic utility alone [49]. IL-6, a pro-inflammatory cytokine, demonstrated improved diagnostic performance with an AUC of 0.794, 61% sensitivity, and 84% specificity [50]. TNF-α, a key signalling molecule, serves primarily as a prognostic marker: elevated levels correlate with advanced disease and poorer outcomes. In a study including 35 CRC patients, 20 with benign colorectal lesions, and 51 healthy controls, TNF-α reached an AUC of 0.9, with 80% sensitivity and 95% specificity [51].
To improve diagnostic performance, multi-analyte panels combining conventional and novel markers (e.g., CEA, CA19-9, CA24-2, and MIC-1, or CEA plus STK4) [52,53,54] have yielded higher accuracy in pilot studies, though validation in larger screening cohorts is still required [52,53,54]. Finally, HER-2/neu overexpression, detected in 2–5% of metastatic CRC cases [55], has emerged as a potential predictive marker, as it correlates with resistance to anti-EGFR therapies [56].

Table 1.
List of serum proteins biomarkers for CRC diagnosis and their clinical parameters in terms of specificity, sensitivity, AUC and cut-off value.
Table 1.
List of serum proteins biomarkers for CRC diagnosis and their clinical parameters in terms of specificity, sensitivity, AUC and cut-off value.
| Name | Clinically Relevant Parameters | Ref. | |||
|---|---|---|---|---|---|
| Specificity | Sensitivity | AUC | Cut-Off Value | ||
| CEA | 89.2% | 64.5% | 0.789 | 3.36 ng mL−1 | [57] |
| CA19-9 | 90.1% | 47.8% | 0.690 | 23.9 U mL−1 | [57] |
| CA72-4 | 86% | 50% | 0.73 | - | [43] |
| CA242 | 59.7% | 66% | 0.651 | 20 U mL−1 | [58] |
| TIMP-1 | 95% | 42–65% | 0.83 | - | [45] |
| CRP | 90% | 17% | 0.64 | 14.6 ng mL−1 | [49] |
| IL6 | 61% | 84% | 0.794 | 4.2 pg mL−1 | [50] |
| TNF-α | 85% | 80% | 0.90 | - | [51] |
| STK4 | 92.3% | 100% | - | - | [59] |
| SerpinA1 | 95% | 95% | 0.97 | 817 µg mL−1 | [48] |
| MMP-9 | 67% | 80% | 0.8 | 204 ng mL−1 | [60] |
Taken together, these findings show that while a limited number of serum proteins (CEA, CA19-9) are currently validated for clinical use, several others (e.g., TIMP-1, MMP-9, SERPINA1) exhibit strong diagnostic potential and molecular characteristics compatible with aptamer recognition. Therefore, continued exploration of both established and emerging biomarkers is essential to expand the repertoire of aptamer targets suitable for minimally processed clinical samples.
3. SELEX for CRC Biomarkers
Aptamers have become invaluable tools for targeting disease-specific proteins, offering high precision in biomarker detection. The Systematic Evolution of Ligands by EXponential enrichment (SELEX) process enables the selection of high affinity aptamers through iterative binding and amplification cycles. Over time, SELEX methodologies have evolved from traditional separation techniques to modern, integrated platforms that combine automation, nanotechnology, and computational tools. In this section, we summarize recent advances in aptamer selection and optimization strategies, focusing on their progressive refinement toward CRC-relevant protein biomarkers (Figure 2).
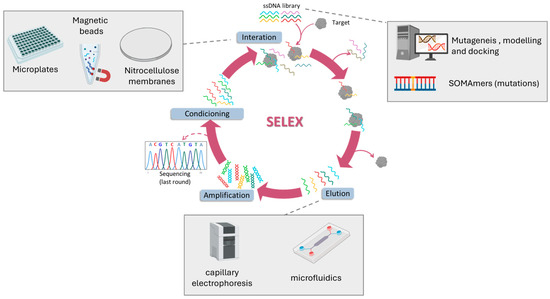
Figure 2.
General scheme of the SELEX method and the different strategies employed to obtained aptamers against CRC biomarkers.
Early SELEX methods relied on simple physical separation systems, such as nitrocellulose membranes (m-SELEX) or microplate immobilization, to separate bound from unbound sequences. Using these classical techniques, aptamers have been isolated against canonical CRC markers including CEA, CA72-4, and CA19-9, typically nanomolar affinities after multiple rounds of selection [61,62,63,64]. Despite their early successes, conventional SELEX was limited by labor-intensive workflows, lengthy selection cycles, and poor translation to physiological conditions. RNA aptamers, although exhibiting promising affinities, suffer from instability in biological fluids, and immobilization on microplate followed by proteolysis can distort target epitopes [63]. Furthermore, specificity against homologous proteins and robustness in complex biological matrices remain largely untested. Collectively, these limitations underscore why classical SELEX-derived aptamers have not yet bridged the gap to clinical application in CRC diagnostics.
To overcome these challenges, SELEX has progressively incorporated magnetic materials and microfluidic tools to improve efficiency and reduce selection bias. Magnetic beads for target immobilization (MBs-SELEX) markedly improved efficiency by enabling small sample volumes, rapid magnetic separation, stringent washing with minimal target loss, and direct polymerase chain reaction (PCR) amplification on bead surfaces, streamlining the selection process [64]. Using this strategy, Wang et al. obtained DNA aptamers against TIMP-1 in seven rounds with a dissociation constant (KD) of 0.41 nM, demonstrating recognition in serum [65]. Similarly, Minagawa et al. selected sub-nanomolar affinity aptamers against CRP [66] and Mashayekhi et al. for TNF-α [67]. A further refinement, Magnetic-Assisted Rapid Aptamer Selection (MARAS), introduced an externally applied rotating magnetic field to apply dynamic selection pressure, enabling the isolation of aptamers with distinct affinities to the target. Using this approach, an aptamer against C-reactive protein with an affinity of 25 nM was obtained [68]. MBs-SELEX has also been integrated with microfluidic or capillary electrophoresis platforms to reduce selection rounds. For example, Nagano et al. achieved aptamer selection against TNF-α in only three rounds using microbead-assisted capillary electrophoresis (MACE) SELEX [69], while Huang et al. selected combined incubation, magnetic extraction, and PCR amplification in a fully automated microfluidic MB-SELEX system [70].
Although these variants significantly increased efficiency and recovery of high-affinity candidates, several limitations remain. First, immobilization of target proteins on magnetic surfaces may induce structural alterations that bias aptamer selection, potentially limiting recognition of the native biomarker in physiological fluids. Second, the high affinities reported in controlled laboratory settings are often not reproduced in complex clinical matrices, raising concerns about real-world applicability. Finally, most reported aptamers, including those against TIMP-1, CRP, and TNF-α, have been validated only in small cohorts, and there is still little evidence that MBs-SELEX consistently yields aptamers ready for translational use. Thus, while magnetic bead-based strategies accelerate the SELEX process, further optimization and rigorous validation are required to confirm their utility in CRC diagnostics.
Parallel to these methods, chemical modification of nucleotides has given rise to Slow Off-rate Modified Aptamers (SOMAmers), representing a major leap in aptamer engineering. By incorporating side chain into DNA nucleotides, SOMAmers achieve enhanced molecular recognition, higher binding affinity and slower dissociation rates. These modifications also enable targeted post-SELEX optimization, improving functional activity and metabolic stability. Using this strategy, Gupta et al. successfully developed SOMAmers against IL-6 with sub-nanomolar affinity and with a great stability in human serum for more than 48 h [71]. This is particularly relevant for CRC, as reliable biomarker detection in complex fluids is a prerequisite for clinical translation.
The latest frontier in aptamer development lies in computational refinement. In silico tools such as mutagenesis, secondary structure modelling, and molecular docking simulations are increasingly used to streamline SELEX outcomes and reduce experimental cycles. A notable example is the affinity improvement of a DNA aptamer targeting CEA by generating mutant sequences and predicting their 3D RNA structures using Mfold and RNA Composer. The RNA models were converted to 3D DNA structures with Write, and their interactions with CEA were evaluated via ZDOCK. This approach successfully enhanced aptamer affinity to the low nanomolar range [72]. Although still in early stages, these approaches promise to streamline aptamer design and reduce experimental cycles.
Overall, SELEX technologies have evolved from classical separation methods to highly integrated, automated, and computationally guided pipelines. These innovations are beginning to deliver high-affinity aptamers against clinically relevant CRC biomarkers, setting the stage for their application in diagnostic assays and biosensing platforms.
4. Aptasensors for CRC Diagnosis
Aptamer-based biosensors (aptasensors) have rapidly evolved into diverse analytical formats for CRC detection. Beyond demonstrating the binding potential of selected aptamers, these platforms translate molecular recognition into measurable analytical signals with the sensitivity and reproducibility required for clinical application. Reported designs range from traditional formats, such as sandwich and direct assays, to advanced architectures integrating nanomaterials or nucleic acid-driven amplification strategies.
A clear trend can be observed among reported CRC aptasensors, which predominantly employ a direct assay configuration where the aptamer is immobilized on gold or nanomaterial-modified electrode surfaces. This arrangement allows for rapid detection and straightforward fabrication, but it is also more prone to matrix interference in complex biological fluids such as serum. Consequently, many studies rely on highly diluted serum samples to minimize nonspecific adsorption, which in turn reduces analytical sensitivity and limits clinical applicability. Although sandwich-type formats improve selectivity through dual recognition, they are generally more complex, involving additional preparation steps and longer assay times. Hybrid and nanomaterial-integrated designs (e.g., those employing AuNPs, graphene, or carbon nanotubes) have emerged as intermediate solutions that balance sensitivity and operational simplicity.
Most reported systems target protein biomarkers detectable in blood, and while a number of studies include recovery tests in spiked serum, only a few have evaluated real patient samples, restricting their translational relevance. Furthermore, despite achieving impressive limits of detection, some platforms remain over-engineered, making them less suitable for reproducible and point-of-care implementation.
Overall, a comparison of these approaches reveals key design rules for CRC aptasensor: direct formats favor speed and miniaturization, sandwich assays enhance selectivity, and hybrid nanomaterial platforms offer tunable performance but require simplified, standardized fabrication for clinical translation. Notably, most reported CRC aptasensors rely on electrochemical transduction (77%), followed by optical (20%) and, to a much lesser extent, mass-based approaches (3%), reflecting the clear advantages of electrochemical sensing in terms of sensitivity, portability, and cost-effectiveness, as already emphasized in previous reviews [11,73].
In addition to these design considerations, methodological rigor varies substantially among primary studies. Many CRC aptasensor reports lack appropriate negative controls to assess nonspecific binding, or rely on limited replicates, hindering statistical validation of results. Furthermore, essential analytical parameters such as the limit of detection, dynamic range, and inter-assay reproducibility are not always reported in a standardized format, complicating comparison between studies. Greater adherence to reporting standards, including the systematic use of negative and blank controls, independent replicates, and transparent analytical validation, will be critical to ensure reproducibility and accelerate clinical translation of aptamer-based biosensors.
4.1. Traditional Approaches
Traditional aptasensor designs for CRC biomarkers (e.g., TIMP-1, IL-6, MMP-9, TNF-α, CRP, and CEA) rely on either sandwich or direct assay formats (Table 2 summarizes their analytical performance, including limit of detection, selectivity, detection time, and sample matrix; Figure 3).
Sandwich assays use an aptamer as the capture element and either a second aptamer or an antibody for detection, thereby improving selectivity through dual recognition. Representative examples include chemiluminescent detection of TIMP-1 [65], piezoelectric aptasensors for MMP-9 [74] and photoelectric strategies for split-type assays [75]. Although these designs can enhance analytical performance, they generally involve more complex fabrication steps, longer assay times, and, in hybrid formats, partially forfeit the advantages of fully aptamer-based systems.
Direct assays are more common due to their simpler construction and shorter analysis time. These rely on label-free techniques such as surface plasmon resonance (SPR) [76], or label-based techniques such as photoelectrochemical [77], fluorescence [78] and most predominantly electrochemical detection. In electrochemical formats, aptamers are typically immobilized on gold electrodes, and target binding is transduced as a change in signal. Strategies include methylene blue-labeled aptamers, which brings the reporter closer to the electrode upon target capture (demonstrated for IL-6 [79], TNF-α [80] and CRP [81,82]) (Figure 3A), or unmodified aptamers combined with solution-phase redox mediators, such as ferrocene, to monitor binding events (applied to CRP [83] and CEA [84,85]). Label-free capacitive approaches, including non-faradic impedance spectroscopy [86] (Figure 3B) and interdigitated electrode systems [87], have also been developed, particularly for IL-6 detection, with the advantage of eliminating the need for chemical labeling.
While sandwich and direct aptasensors have successfully demonstrated the feasibility of aptamer-based recognition for CRC biomarkers, key limitations persist. Sandwich assays achieve higher selectivity but at the cost of complexity, extended assay times, and, in some cases, antibody dependency. Direct assays offer speed and simplicity but are more vulnerable to nonspecific binding and matrix effects in clinical samples. Electrochemical approaches dominate the field owing to their sensitivity, portability, and compatibility with point-of-care miniaturization; however, challenges remain regarding long-term stability of the aptamer–electrode interface, reproducibility between fabrication batches, and robustness under physiological conditions (e.g., blood, variable temperatures). These issues highlight why, despite encouraging analytical performance in controlled studies, translation to clinical practice and large-scale validation remains limited.
To overcome these shortcomings, particularly in terms of sensitivity, stability, and real-world applicability, researchers are increasingly integrating nanomaterials into aptasensor designs. These materials enable signal amplification, improved robustness, and novel detection formats, representing the next major step in aptasensor development for CRC detection.
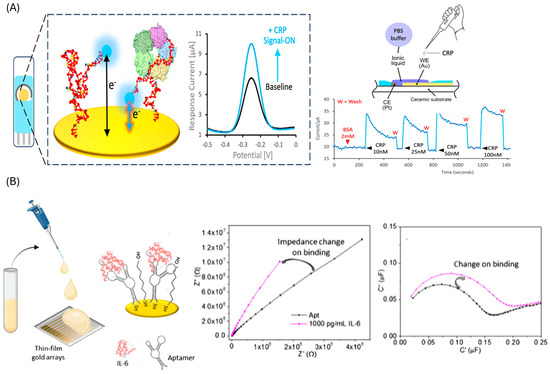
Figure 3.
Traditional aptasensors. (A) Direct approach employing methylene blue labelled aptamer and electrochemical detection for CRP. Reprinted with permission from Ref. [81] (B) Direct approach employing aptamer modified gold electrodes and capacitive EIS for IL-6 detection. Reprinted with permission from Ref. [86].

Table 2.
Summary of aptasensors employing a traditional design.
Table 2.
Summary of aptasensors employing a traditional design.
| Protein | Sensing Platform/ Assay Format | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CEA | ITO/Fe3O4@SiO2@CdS/Apt1/SiO2–Au-Apt2 Sandwich assay | Photo- electrochemical | 1–6000 pg mL−1/ 0.3 pg mL−1 | AFP, PSA, BSA, HCG | 2 h | - | 4 weeks | [75] |
| PtμEs/Au/aptamer Direct assay | Electrochemistry (SWV) | 10 pg mL−1–100 ng mL−1/ 7.7 pg mL−1 | AFP, BSA, pancreatin, IgG | 1 h | 5 Blood samples | - | [84] | |
| IDE/Apt/MCH Direct assay | Electrochemistry (EIS) | 2 pg mL−1–2 ng mL−1/ 2.4 pg mL−1 | HSA, IL-6, SARS-CoV-2-S | 20 min | 1% spiked serum | - | [85] | |
| Apt + 1,1′-biphenyl]-4,4′-diyldiboronic acid Direct assay | Fluorescence | 0.003–10 ng mL−1/ 1 pg mL−1 | AFP, IgG, VEGF, BSA, CA125, IgE, PSA, PTK7 | 55 min | 3 cancer serum | - | [78] | |
| CRP | Au/Apt-MB/MCH Direct assay | Electrochemistry (SWV) | 1–200 nM/ 20 nM | pfLDH, PCT | 1 min | - | 1 week | [81] |
| Au/apt-MB/MCH Direct assay | Electrochemistry (SWV) | 1 pM to 100 pM/ 1 pM | BSA, IgE | 30 min | 10% spiked serum | - | [82] | |
| Au/aptamer/MCH/BSA Direct assay | Electrochemistry (DPV) | 0–1000 ng mL−1/ 1.84 ng mL−1 | cTnT, cTnI | - | 60 blood samples | - | [83] | |
| Fiber/Au/aptamer/MCH Direct assay | SPR | 0–100 nM/ 1.7 nM | Hemoglobin, fibrinogen, cTn-I | - | - | - | [76] | |
| PTB7-Th/GCE-Au/Apt/HT Direct assay | Photo- electrochemical | 1 pM–1000 nM/ 0.33 pM | BSA, IgG, Thrombin | 30 min | - | - | [77] | |
| IL-6 | Au/apt-MB/MCH Direct assay | Electrochemistry (SWV) | 50–200 nM/ 50 nM | TNF-α, HSA, H-calprotectin | - | - | 7 days | [79] |
| TFGA-Au/Apt/MCH Direct assay | Electrochemistry (non-faradaic EIS) | 10–10,000 pg mL−1/ 10 pg mL−1 | MMP3 | 30 min | Spiked serum | 2 weeks | [86] | |
| IDE-strep/btn-apt Direct assay | Electrochemistry (current changes) | 1 fM–100 pM/ 10 fM | - | 5 min | 1% spiked serum | - | [87] | |
| MMP-9 | Apt1/apt2-btn/strep Sandwich assay | Piezoelectric (QZM) | 8.3–2075 ng mL−1/ 100 pg mL −1 | HSA, IgG | 1 h | - | - | [74] |
| TIMP-1 | MBs-strep/btn-apt/Ab-AE Sandwich assay | Chemi- luminescence | 1–500 ng mL−1/ 1 ng mL−1 | AFP, CEA, CA199, BSA, 8HIS peptide | - | 40 serum samples | - | [65] |
| TNF-α | Au/apt-MB/MCH | Electrochemistry (SWV) | 10–100 ng mL−1/ 10 ng ml−1 | - | 15 min | - | 10 h (blood) | [80] |
Ab: antibody; AE: acridinium ester; AFP: alpha feto protein; Apt: aptamer; Au: gold; BSA: bovine serum albumin; Btn: biotin; HCG: human chorionic gonadotropin; CA125; cancer antigen 125; CA199: cancer antigen 199, CEA: Carcinoembryonic antigen; CRP: C-reactive protein; cTnT: cardiac troponin; TcTn-I: troponin I; DPV: differential pulse voltammetry; EIS: electrochemical impedance spectroscopy; GCE: glassy carbon electrode; HCG: human chorionic gonadotropin; HSA: Human serum albumin; HT: hexanothiol; IDE: interdigitated electrodes; IgE: immunoglobulin E; IgG: immunoglobulin G; IL-6: interleukin 6; ITO: indium tin oxide; MB: methyleneblue; MBs: Magnetic beads; MCH: mercaptohexanol; MMP3: Matrix Metallopeptidase 3; PCT: procalcitonin; pfLDH: plasmodium lactate dehydrogenase; PTB7-Th: poly{4,8-bis[5-(2-ethylhexyl) thiophen-2-yl]benzo[1,2-b:4,5-b′]dithiophene-2,6-diyl-alt-3-fluoro-2-[(2-ethylhexyl)carbonyl] thieno[3,4-b]-thiophene-4,6-diyl}; PSA: prostate specific antigen; PTK7: Tyrosine-protein kinase-like 7; SPR: surface plasmon resonance; strep: streptavidin; SWV: square wave voltammetry; PtμEs: platinum microelectrode: TFGA: surface flexible thin film gold arrays chips; VEGF: vascular endothelial growth factor.
4.2. Nanomaterial-Based Sensors
Nanomaterials have emerged as a transformative component in the development of aptamer-based sensors, primarily due to their ability to improve sensitivity and robustness. Their high surface-to-volume ratio, tuneable optical and electronic properties, and versatile surface chemistry enhance aptamer immobilization, stabilize aptamer-target complexes, and enable efficient signal transduction. These features are particularly advantageous for detecting CRC biomarkers, where low analyte concentrations in complex biological samples demand exceptional analytical performance. In this section, we review the main classes of nanomaterials integrated into CRC biomarkers aptasensors, highlighting their specific roles in signal amplification and assay design.
4.2.1. Gold Nanoparticles
Gold nanoparticles (AuNPs) are among the most widely used nanomaterials in aptasensor design due to their high surface area and versatile binding molecules, which enable highly sensitive target detection [88]. Depending on the sensing strategy, AuNPs can serve either as the sensing phase or as signal labels (Table 3). As a sensing phase, AuNPs increase the available surface for aptamer immobilization (Figure 4A), thereby improving sensitivity in electrochemical [89,90,91,92,93], electrochemiluniniscence (ECL) [94], quartz crystal-microbalance (QCM) [95] and surface-enhanced Raman scattering (SERS) [96] platforms. When used as labels, AuNPs have been employed in multiple ways: conjugated to secondary aptamers and coupled with enzymes (e.g., horseradish peroxidase, HRP) for electrochemical detection of enzymatic products [97], as quenchers of quantum dot photoluminescence [98], and in aggregation-based assays (Figure 4B), where inhibition of nanoparticle clustering produces measurable fluorescence changes [99]. Additionally, AuNPs have been integrated with other nanomaterials like ZrO2 nanoparticles that improve the signal and stability of the sensor [100] and with Pt, mimicking the peroxidase activity [101].
Despite their versatility, AuNP-based sensors also face important challenges. Colloidal stability and aggregation under physiological conditions can compromise reproducibility, while variability in nanoparticle size, shape, and surface chemistry often introduces batch-to-batch inconsistencies that affect analytical performance. Moreover, although AuNPs enhance sensitivity, their incremental benefit over other nanomaterials is sometimes limited once baseline signal amplification is achieved. Most reported AuNP-aptasensors remain at the proof-of-concept stage, with scarce validation in real clinical samples, underscoring the need for further work to confirm their robustness in complex biological matrices.
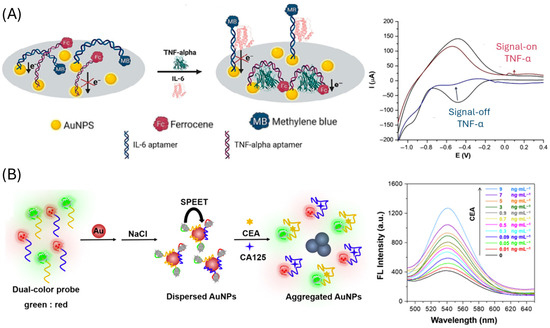
Figure 4.
Gold nanoparticles-based CRC aptasensors. (A) approach employing AuNPs modified electrodes for the immobilization of the aptamer. Reprinted with permission from Ref. [89]. (B) Aptasensor based on salt-induced gold nanoparticles aggregation. Reprinted with permission from Ref. [99].

Table 3.
Summary of aptamer-based sensors employing gold nanoparticles.
Table 3.
Summary of aptamer-based sensors employing gold nanoparticles.
| Protein | Sensing Platform/ Assay Format | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CA125 | DNA-AgNCs-apt+ AuNPs Direct assay | Fluorescence | 0.01–2.0 U mL−1/ 0.015 U mL−1 | DP, BSA, OVA, GOD, Tb | 25 min | serum (n = 4) | 2 h | [99] |
| CEA | Au/4-mercaptophenyl/AuNPs/apt Direct assay | QCM | 0.1–25 ng mL−1/ 102 pg mL−1 | AFP, CA-125, VEGF | 50 min | Spiked serum | - | [95] |
| GrSPE/AuNPs/apt/MCH Direct assay | Electrochemistry (EIS) | 0.2–15.0 ng mL−1/ 0.085 ng mL−1 | DHEA, AFP, Leptin, AA, BSA | 1 h | - | - | [93] | |
| Au@AgNPs/4-MBA/apt Direct assay | SERS | 0.01–200 ng mL−1/ 3.24 pg mL−1 | AFP, DP, IgG, BSA | 20 min | serum (n = 3) | - | [96] | |
| ITO/E-Si/ZrO2/AuNPS/apt Direct assay | Electrochemistry (DPV) | 0.01 pg mL−1–100 ng mL−1 42.504 fg mL−1 | cTnI, CA199, NSE, l-cys | 30 min | serum (n = 43) | - | [100] | |
| AuE/apt/MCH/CEA/apt2-Au@PtNPs Sandwich assay | Electrochemistry (amperometry) | 0.1–100 ng mL−1/ 0.02–0.31 ng mL−1 | AFP, AA, BSA, CA125, | 2 h 30 min | spiked 20% serum | 2 weeks | [101] | |
| DNA-AgNCs-apta+ AuNPs Direct assay | Fluorescence | 0.01–0.9 ng mL−1/ 0.03–7.5 pg mL−1 | DP, BSA, OVA, GOD, Tb | 25 min | Serum (n = 4) | 2 h | [99] | |
| CRP | ePAD/EGaIn-PPD/AuNPs/Apt/ MCH/AuNPs Direct assay | Electrochemistry (DPV) | 1–100 ng mL−1/ 0.039 ng mL−1 | Cys, Glu, urea, Hcy | 1 h | Saliva (n = 38) | - | [92] |
| SPCE/AuNPs/Apt/casein/JNP-AuNPs/HRP Direct assay | Electrochemistry (amperometry) | 10 pg mL−1–1 ng mL−1/ 3.1 pg mL−1 | cTnT, cTnI, TB, HigG, HSA, Myo | 1 h | serum (n = 3) | 32 days | [97] | |
| IL-6 | Electrode/AuNPs/apt/MB Direct assay | Electrochemistry (CV) | 5–5000 pg mL−1/ 1.6 pg mL−1 | IL-1, IL-18, BSA, HSA | 2 h | 15 saliva, 15 sweat | 3 days | [89] |
| TNF-α | Electrode/AuNPs/apt/ferrocene Direct assay | Electrochemistry (CV) | 5–5000 pg mL−1/ 1.6 pg mL−1 | IL-1, IL-18, BSA, HSA | 2 h | 15 saliva, 15 sweat | 3 days | [89] |
| GrSPECoHCF-AuNps/Apt/1-HT/Ab-HRP Direct assay | Electrochemistry (DPV) | 1–100 pg mL−1/ 0.52 pg mL−1 | APC, HSA, IgG | 80 min | Serum (n = 3) | - | [90] | |
| GCE/AuNPS/Apt/MCH/TNF-α apt2-Ru(phen)2+/GO Sandwich assay | ECL | 0.05–50 ng mL−1/ 36 pg mL−1 | MMP-2, IL-2/6, BSA, IFN-γ | 1h 45 min | - | - | [94] | |
| Fe3O4/AuNPs/Apt1/cApt-MB AuNPs Direct-displacement assay | Electrochemistry (SWV) | 10 pg mL−1–100 ng mL−1/ 10 pg mL−1 | IL-1, IL-2, IL-6, IL-12, IFN-γ | 30 min | Spiked serum | - | [91] | |
| Quantum dot/apt/AuNPs Direct assay | FRET | 0–22.3 nM/ 97.2 pM | HSA, CRP, transferrin, Tb | 1 h | spiked serum (n = 5) | - | [98] |
AA: ascorbic acid; AFP: alpha-feto protein; APC: activated protein-C; AgNPS: silver nanoparticles; Apt: aptamer; Au: gold electrode, AuNPs: gold nanoparticles; BSA: bovine serum albumin; CA: cancer antigen; cAPT: aptamer complementary; CEA: Carcinoembryonic antigen; CRP: C reactive protein; cTnI: troponin I; cTnT cardiac troponin; CV: cyclic voltammetry; Cys: cysteine; DHEA: dehydroepiandrosterone; DNA: deoxyribonucleic acid; DP: dopamine; DPV: differential pulse voltamety; EGaIn: eutectic gallium indium; EIS: electrochemical impedance spectroscopy; ePAD: electrochemical paper-based analytical devices; Glu: glucose; GO: graphene oxide; GOD: glucose oxidase; GrSPE: graphene screen printed electrodes; Hcy: homocysteine; HRP: horse radish peroxidase HSA: human serum albumin; HT: hexanothiol; IFN-γ: interferon-γ; IgG: immunoglobulin-g; IL: interleukin; ITO: indium tin oxide; JNP: janus particles; MB: methylene blue; MCH: mercaptohexanol; MMP: matrix metalloproteinases; Myo: myoglobin; NSE: neuron-specific enolase OVA: ovalbumin QCM: quartz microbalance; SERS: Surface-Enhanced Raman Scattering; SPCE: screen printed carbon electrodes; SWV: square wave voltammetry; TB: thrombin; VEGF: vascular endothelial growth factor; 4-MBA: 4-mercaptobenzoic acid.
4.2.2. Carbon-Based Nanomaterials
Carbon-based nanomaterials have been extensively employed as sensing elements in biosensors owing to their large surface area, chemical stability, mechanical strength, biocompatibility, and facile surface functionalization [102]. They can be categorized by dimensionality into 0D, 1D, 2D, and 3D structures, a classification that helps highlight their different modes of action in aptasensor design. In the context of CRC, these nanomaterials have been applied both to canonical CRC biomarkers such as CEA and to inflammation-related proteins (e.g., CRP, IL6, TNF-α), which are increasingly recognized as contributors to CRC onset and progression.
Although several examples target non-CRC-specific inflammatory biomarkers, these studies demonstrate the broad adaptability of carbon nanomaterials for protein detection, and their underlying sensing principles are directly translatable to CRC biomarker assays (Table 4).
0D materials such as carbon quantum dots (CQDs) doped with nitrogen and AuNPS were employed in Förster resonance energy transfer (FRET)-based aptasensors [103]. Similarly, graphene quantum dots combined with AuNPs have enabled the development of fluorescence aptasensors (Figure 5A) [104]. These optical strategies have been applied to markers associated with CRC-related inflammation, underscoring their potential for integration into CEA or CA19-9 detection formats.
1D materials, particularly carbon nanotubes (CNTs), both multi-walled and single-walled, are frequently used to modify electrodes in electrochemical aptasensors and as scaffolds in chemiluminescent sensors, enabling aptamer immobilization and direct target sensing. Multi-walled CNTs (MWCNTs) have been integrated with graphitic carbon nitride/zirconium dioxide (Figure 5B) [105], cobalt hexacyanoferrate [106], and flower-shaped Ag@ZIF-67 [107]. Single-walled CNTs (SWCNTs) have been used in combination with gold nanoparticles [108] and silver nanoparticle–mesoporous carbon composites [109]. Compared to quantum dots, CNTs are particularly effective in enhancing electrical conductivity and facilitating electron transfer, features that have been leveraged in electrochemical aptasensors for CRC-relevant biomarkers, including IL-6, TNF-α, and CEA.
Graphene and its derivatives, as 2D materials are the most widely exploited carbon-based nanomaterials for CRC biomarker detection. They are typically employed to modify electrode surfaces for aptamer immobilization [110,111,112,113]. Graphene can also form hybrid systems with Ag@Pt core–shell nanoparticles [114], AuNPs [115,116] (Figure 5C), silver nanoparticles [117], PdNPs [118], Cu3(PO4)2 Hybrid Nanoflowers [119], and even MWCNTs and AuNPs [120]. Additionally, graphene has been used as a carrier for generating electrochemical labels in combination with polymers [121]. This versatility stems from graphene’s exceptionally high surface area and ease of functionalization, which allow synergistic effects when integrated with metallic nanostructures, enhancing both conductivity and catalytic activity. Graphene-based systems have been among the most successful platforms for CEA detection, serving as proof-of-concept for CRC diagnostic biosensors.
Beyond planar systems, 3D carbon nanostructures introduce intrinsic cavities and hierarchical porosity that are advantageous for aptasensor applications. Carbon nanoboxes and carbon nanofibers exemplified this category [122]. Rahmati et al. exploited the hollow cavities of N-doped carbon nanoboxes to load thioine, the sensing molecule, while using AuNPs to immobilize the aptamer for CEA detection [123] (Figure 5D) Such designs enable the loading of both recognition elements and redox mediators within a single structure, facilitating high-performance sensing.
Carbon-based nanomaterials provide robust, versatile, and highly functional platforms for biosensor development. However, their intrinsic heterogeneity, potential cytotoxicity, and challenges in large-scale, reproducible synthesis remain limitations for clinical translation. In addition, while hybrid composites with metals or polymers enhance sensitivity, they also complicate fabrication protocols, potentially increasing variability between sensors. Future efforts should focus on standardizing these nanomaterial-based aptasensors for clinically validated CRC biomarkers, thereby accelerating their translation into diagnostic practice.
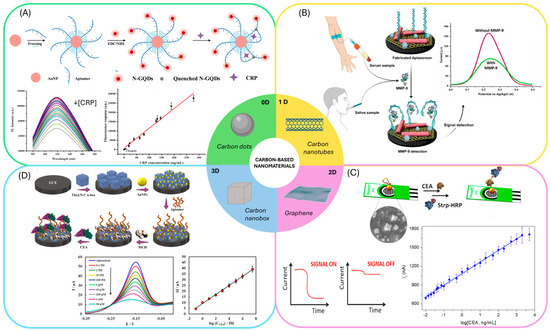
Figure 5.
Aptasensors employing carbon-based nanomaterials: (A) Example of an aptasensors employing carbon dots and gold nanoparticles. Reprinted with permission from Ref. [104]. (B) Aptasensors employing carbon nanotubes for the immobilization of the aptamer. Reprinted with permission from Ref. [105]. (C) Sandwich aptasensors employing graphene modified electrodes. Reprinted with permission from Ref. [116]. (D) Aptasensor employing carbon nanoboxes to modify the electrode surface and gold nanoparticles for the immobilization of the aptamer. Reprinted with permission from Ref. [123].

Table 4.
Summary of the aptasensors for CRC biomarkers based on carbon nanomaterials.
Table 4.
Summary of the aptasensors for CRC biomarkers based on carbon nanomaterials.
| Protein | Sensing Platform/ Assay Format | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CA19-9 | GCE/GO-cMWCNTs/AuNPs cDNA-AQ/BSA/Apt-Fc Displacement assay | Electrochemistry (ACV) | 1.0–1.0 × 106 mUmL−1/ 0.65 mU mL−1 | CA242, AFP, PSA | 1 h | 1% serum | 15 days | [120] |
| CEA | MACNTs/apt/DNA-Ag@ZIF Displacement assay | Chemiluminescence | 0.05–500 ng mL−1/ 4.53 pg mL−1 | AFP, glu, adrenaline, Na+, BSA, Carbamide, | 20 min | - | 21 days | [107] |
| GCE/AgNPs/SWCNT/MCF/Apt/BSA Direct assay | Electrochemistry (DPV) | 1 fg mL−1–20 ng mL−1/ 0.30 fg mL−1 | Glu, insulin, urea, gly, AA, HSA, Hb | 1 h | Serum (n = 5) | 16 days | [109] | |
| GO/AgNPS/aptamer Direct assay | Electrochemistry (DPV) | 0.001–10 pg mL−1/ 0.5 fg mL-1 | HER2, VEGF, IgG, MUC1 CFP10 | - | Serum (n = 2) | - | [117] | |
| IDE/GO/apt1/BSA/CEA/apt2 Sandwich assay | Electrochemistry (LSW) | 0.5–500 ng mL−1/ 0.96 ng mL−1 | - | - | 1% serum | - | [112] | |
| SPCE/rGO/AuNPs/casein/apt-btn/strep-HRP Direct assay | Electrochemistry (amperometry) | 20 pg mL−1–2 μg mL−1/ 16 pg mL−1 | cTnI, CRP, HSA, TBA, IgG | 35 min | - | 60 days | [116] | |
| PEDOT:PSS/GO/APTES/apt Direct assay | Electrochemistry (EIS) | 0.77–14 ng mL−1/ 0.45 ngmL−1 | BSA, PSA, insulin | - | Spiked serum | - | [113] | |
| Cu3(PO4)2HNF/GO/strep/btn-apt/Cu2+ Direct assay | Electrochemistry (SWV) | 10 fg mL−1–500 ng mL−1/2.4 fg mL−1 | PSA, TB, Hb | 1 h | Serum | 15 days | [119] | |
| GCE/Thi@N-C n-box/AuNPs /apt/MCH Direct assay | Electrochemistry (SWV) | 0.1 fM–30 nM/ 0.03 fM | HCG, AFP, PSA | 25 min | Spiked 0.1% serum | 15 days | [123] | |
| GNSPE/PdNPs/apt/BSA Direct assay | Electrochemistry (DPV) | 0.002–200 ng mL−1/ 1.0 pg mL−1 | insulin, cys, glu, arg, gly, BSA, cholesterol | 30 min | 1% serum | 5 weeks | [122] | |
| Au/apt/BSA/Apt2-CNTs- PFcGE Sandwich assay | Electrochemistry (SWV) | 1 fg mL−1–10 ng mL−1/ 0.28 fg mL−1 | DA, AA, UA, l-cys, | 1 h | 5% spiked serum | 2 weeks | [121] | |
| CRP | IDE/SWCNTs/Apt-AuNPs/ PEG-COOH Direct assay | Electrochemistry (SWV) | 1 pM–10 nM/ 10 pM | Troponin I | 30 min | - | - | [108] |
| GCE/PDES/GO/AuNPs/Apt Direct assay | Electrochemistry (EIS) | 0.001–50 ng mL−1/ 0.0003 ng mL−1 | AFP, ly, UA | 80 min | serum (n = 3) | 10 days | [115] | |
| CSPE/CNFs-CHIT/RNA-apt/BSA/MB Direct assay | Electrochemistry (SWV) | 1–150 pM/ 0.37 pM | HSA, IgG | 1 h | Spiked 2.5% serum | 15 days | [122] | |
| AuNPs/apt/N-GQDs Direct assay | FRET | 0.2–300 ng mL−1/ 0.2 ng mL−1 | BSA, cTNI, MYO | 40 min | Serum | - | [104] | |
| IL-6 | GCE/MWCNTs/CoHCF/ /AuNPs/Apt/MCH Direct assay | Electrochemistry (DPV) | 0.5–1000 pg mL−1/ 0.17 pg mL−1 | BSA, IgG, CEA, MUC1 | 1 h | serum (n = 5) | 15 days | [106] |
| AuNPS-apt/NCD Direct assay | FRET | 1.5–5.9 pg mL−1/ 0.84 pg mL−1 | BSA, TNF-α | 1 h | 5% serum | - | [103] | |
| MMP-9 | GCE/MWCNTS-Gc3n4/OZrO2NPs/apt/BSA Direct assay | Electrochemistry (DPV) | 50–1250 pg mL−1 10.51 pg mL-1 | leptin, TB, VEGF, MUC-1, IL-6, Apo-A1, NGAL | 45 min | 1% serum, 10% saliva | 14 days | [105] |
| IDE/Graphene/apt/6-FHH Direct assay | Electrochemistry (DPV) | 10 fM–1000 nM/ 0.1 pM | - | - | Wound fluids | 1 month | [110] | |
| TNF-α | GO/strep/btn-PEG-Apt-Fc Direct assay | Electrochemistry (SWV) | 5 –200 pg mL−1/ 5 pg mL−1 | BSA, IL-6, IL-1β | 1 h | Serum, sweat | 30 days | [111] |
| AuSPE/GO/Ag@PtNPs/Apt Direct assay | Electrochemistry (DPV) | 0–60 pg mL−1/ 2.07 pg mL−1 | BSA, Hb, cys, l-arg | 2 h | serum (n = 3) | 15 days | [124] | |
| SPCE/Au-GO/CS/apt/MCH/Apt-Ag@Pt-GRs Sandwich assay | Electrochemistry (DPV) | 5–70 pg mL−1 1.64 pg mL−1 | BSA, Hb, L-cys, L-arg | 2 h+ overnight | serum (n = 3) | 14 days | [114] |
AA: ascorbic acid; ACV: alternating current voltammetry; AFP: alpha-feto protein; AgNPS: silver nanoparticles; apt: aptamer; APTES: 3-Aminopropyl)triethoxysilane; Apo: apolipoprotein; AuNPs: gold nanoparticles; AuSPE: gold screen printed electrode; Arg: arginine; BSA: bovine serum albumin; btn: biotin; cDNA: complementary DNA; CEA: carcinoembryonic antigen; CNTs: carbon nanotubes; CRP: C-reactive protein; CSPE: carbon screen printed electrodes; cTnI: troponin I; Cys: cysteine; DPV: differential pulse voltammetry; EIS: electrochemical impedance spectroscopy; Fc: ferrocene; FH: ferrocenylhexanethiol; FRET: fluorescence resonance energy transfer; GCE: glassy carbon electrode; Glu: glucose; Gly; glycine; GO: graphene oxide; Hb: hemoglobulin; HCG: human chorionic gonadotropin HER-2: Human Epidermal growth factor Receptor 2; HNF: Hepatocyte nuclear factors; HRP; horse radish peroxidase; HSA: human serum albumin; IDE: interdigitated electrodes IgG: immunoglobulin G; IL: interleukin; LSW: linear sweep voltammetry; MACNTs: Magnetic Carbon nanotubes; MB: methylene blue; MCF: mesoporous carbon foam; MMP: matrix metalloprotease; MUC: mucin; MWCNTs: multi well carbon nanotubes; Myo: myoglobin; NCD: nitrogen dopped carbon dots; NGAL: lipocalin-2; N-GQDSNPs: nitrogen-graphene quantum dots nanoparticles; PEDOT:PSS: 3,4-ethylenedioxythiophene):poly(styrenesulfonate; PFcGE: poly(ferrocenyl glycidyl ether)-grafted; PEG: polyethylene glycol; PSA: prostate specific antigen; strep: streptavidin; SWCNTs: single well carbon nanotubes; SWV: square wave voltammetry; TNF-alpha: TB: thrombin; UA: uric acid; VEGF: Vascular Endothelial Growth Factor.
4.2.3. Metal–Organic and Covalent Organic Frameworks
Metal–organic frameworks (MOFs) and covalent organic frameworks (COFs) are emerging as versatile porous nanomaterials for biosensing due to their high sorption capacity, tunable structures, and excellent electrocatalytic and electrochemical properties [125]. MOFs are commonly used to modify sensor substrates, enhancing surface area for aptamer immobilization and facilitating signal labelling. Different types of MOFs have been employed for developing aptasensors for CRC biomarkers (Table 5), such as ZIF-8@Au NPs@S QDs-MOFs enabling electrochemical, fluorescence and colorimetric detection [126]; nanoporous carbon-AuNP nanocomposites (Figure 6) [127]; Cu-MOF with intrinsic fluorescence emission and peroxidase-like activity [128]; zirconium–cobalt metal–MOFs [129], and zeolitic imidazolate MOFs [130]. Integration with other nanomaterials further enhances performance, such as Fe-MOFs combined with high-entropy alloy nanoparticles [131], AuNPs [132,133] and CdS quantum dots [134].
Similarly, covalent organic frameworks (COFs) have been employed as fluorescent quenchers [135] and integrated with 2D porphyrinic structures for photoelectrochemical transduction [136].
Both MOFs and COFs stand out as highly tunable platforms for aptasensor development, offering modular chemistry, high porosity, and multifunctional integration with other nanomaterials. However, despite their promising performance in proof-of-concept studies, challenges remain for their translation to robust CRC diagnostics. First, their structural and chemical stability in complex biological media is often limited, leading to partial degradation or loss of function. Second, fabrication protocols tend to be complex, with variability in crystallinity and particle size that may compromise reproducibility between sensor batches. Moreover, while their capacity for multimodal detection is attractive, this feature is not always aligned with clinical needs, where assay simplicity and robustness are usually prioritized. Taken together, MOFs and COFs provide a powerful toolkit for aptasensors, but their practical impact will likely depend on simplifying synthesis, improving biostability, and validating their performance in clinically relevant settings.
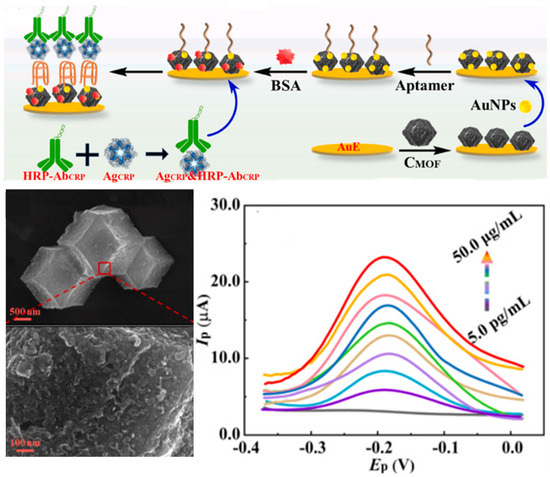
Figure 6.
Sandwich aptasensors employing metal–organic frameworks (MOFs) in combination with gold nanoparticles for the electrochemical detection of CRP. Reprinted with permission from Ref. [127].

Table 5.
Summary of aptasensors employing MOFs and COFs.
Table 5.
Summary of aptasensors employing MOFs and COFs.
| Protein | Sensing Platform/ Assay Format | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CEA | 6FAM-apt/COF Displacement assay | Fluorescence | 0.01–15 ng mL−1/ 5 pg mL−1 | CA125, BSA, HSA, PSA, AGP, VEGF | 55 min | serum | - | [135] |
| GCE/HEANPs-Fe-MOF/apt/BSA Direct assay | Electrochemistry (DPV) | 1 fg mL−1–10 μg mL−1 0.115 fg mL−1 | AFP, PSA, CP | 35 min | 10% plasma | 12 days | [131] | |
| GE/rGO/AuNPS-MOF/apt Direct assay | Electrochemistry (EIS) | 0.0025–0.25 ng L−1/ 0.8 pg L−1 | MUC-1, HSA, IgG, HGB, BSA | 1 h | Spiked serum | - | [132] | |
| AuE/ZrCo-MOF/apt/BSA Direct assay | Electrochemistry (DPV) | 0.001–100 pg mL−1/ 0.35 fgmL−1 | BSA, PSA, VEGF, IgG, MUC1, HER2, HSA, Tb, HGB | 80 min | Spiked serum | 15 days | [129] | |
| GCE/CdS QDs@MOF/TEOA@ AuNPs/apt/b-ME Direct assay | ECL | 0.0001–10 ng mL−1 0.085 pg mL−1 | BSA, thrombin, lysozyme, trypsin | 90 min | 0.1% serum | - | [134] | |
| GCE/Fe-MOFAu@PDA/apt/BSA Direct assay | Electrochemistry (DPV) | 1 fg mL−1–1 μg mL−1/ 0.33 fg mL−1 | AFP, PSA | 35 min | serum (n = 3; SA) | 12 days | [133] | |
| CRP | PCB/AuE/CMOF/AuNPs/aptamer/BSA/Ag-CRP+Ab-HRP Sandwich Assay | Electrochemistry (DPV) | 5 pgmL−1–50 µgmL−1 0.3 pg mL−1 | IGF-I, GHBP, IL-6, IL-10, CA125, BSA | 2 h | serum (n = 3) | 21 days | [127] |
| Cu-MOF/apt Displacement assay | Colorimetric/ Fluorometry | 0.1–50 ng mL−1 40 pg mL−1 | Glu, GSH, AA, Fe, Cr, Ca, albumin, | 15 min /8 h | 10% serum | - | [128] | |
| ITO/NiS/pCOFs/AgNP/apt/MCH Direct assay | Photo-electrochemical | 0.5–100 ng mL−1/ 0.1 ng mL−1 | PSA, BSA, HCG, MC-LR | 50 min | 10% serum | 20 days | [136] | |
| MMP-9 | GCE/rGO-AuNRs/cDNA/BSA/ apt-ZIF-8@Au NPs@S QDs Nanorod displacement | DPV Colorimetric Fluorescence | 1 ng mL−1–10 μg mL−1 10 fg mL−1–10 μg mL−1 0.01–5 μM | annexin CAVIN2-, FEN1-, RAB26 | 40 min | - | 30 days | [126] |
AA: ascorbic acid; Ab: antibody; AFP: alpha-feto protein; apt: aptamer; AGP: Arabinogalactan protein; AuE: gold electrode; AuNPS: gold nanoparticles; AuNRs: gold nanorods; b-ME:β-mercaptoethanol; BSA: bovine serum albumin; CA: cancer antigen; CAVIN-2: caveolae-associated protein 2; cDNA: complementary DNA; CEA: carcinoembryonic antigen; COF: covalent organic framework; CP: calprotectin; CR; creatinine; CRP: c-reactive protein; DPV: differential pulse voltammetry; EIS: electrochemical impedance spectroscopy; FEN1: flap endonuclease 1; GCE: glassy carbon electrode; GE: graphene electrode; GHBP: growth hormone receptor; Glu: glucose; GSH: glutathione; HCG: human chorionic gonadotropin; HER2: Human Epidermal Growth Factor Receptor 2; HGB: hemoglobin; HRP: horse radish peroxidase; HSA: human serum albumin; IGF: Insulin-like growth factor; IgG: immunoglobulin G; ITO: indium tin oxide; MCH: mercaptohexanol; MC-LR: microcystin; MMP: matrix metaloprotease; MOF: metal–organic framework; MUC: mucin; NPs: nanoparticles; PCB: printed circuit board; PSA: prostate specific antigen; QDs: quantum dots; RAB26: ras-related protein; rGO: reduce graphene oxide; SA: standard addition; Tb: thrombin; VEGF: Vascular endothelial growth factor; 6-FAM: fluorescein.
4.2.4. Other Nanomaterials
In the literature, other nanomaterials, including nanochannels, nanocomposites, nanocubes, nanosheets and quantum dots (QDs), have also been explored for the development of CRC biomarker aptasensors (Table 6).
Vertically oriented ultrasmall nanochannels provide a high surface area, enhancing adsorption, reaction kinetics, and small-molecule transport. They also exhibit strong anti-interference and anti-fouling properties. These features make them highly suitable for ECL-based aptasensors, where the nanochannel architecture offers space to immobilize aptamers and localize ECL emitters near the electrode. In such systems, target binding to the aptamer typically leads to a decrease in the ECL signal [137,138,139,140] (Figure 7A).
Among nanocomposites, Ti3C2Tx has emerged as a dominant platform. It has been integrated with AuNPs [141] (Figure 7B) and MoS2 [142] to functionalize electrode surfaces, improve conductivity, and facilitate aptamer immobilization. Composites of Ti3C2Tx with silver/gold nanoparticles have also been used as electrochemical signal amplification strategies [143]. Another example is PdPt@PCN-224, a nanocomposite with excellent catalytic activity for the reduction of H2O2, enabling sensitive dual-signal output aptasensing [144].
Other nanostructures have also been investigated. CdS nanocubes have been applied in photoelectrochemical aptasensors as signal generators, combined with NiCo2O4-Au acting as a signal quencher [145]. Mn3(PO4)2/aptamer nanosheets have been used as labels in sandwich aptasensors [146]. Additionally, mercaptosuccinic acid capped nickel selenide quantum dots (MSA-NiSe2 QDs) have demonstrated promising electrochemical sensing capabilities [147].
While these other nanomaterials illustrate the breadth of strategies pursued in CRC aptasensor research, their application remains largely exploratory. Most studies report strong analytical sensitivity, yet reproducibility, biocompatibility, and long-term stability are seldom addressed. For instance, nanochannel-based systems offer elegant control over analyte transport but require highly controlled fabrication, limiting scalability. Similarly, advanced nanocomposites and QDs show signal amplification potential, but their complex synthesis routes and possible cytotoxicity may hinder translational progress. In this sense, these materials represent valuable platforms for proof-of-concept innovation, but their real impact will depend on balancing performance with cost-effectiveness, ease of fabrication, and clinical robustness.
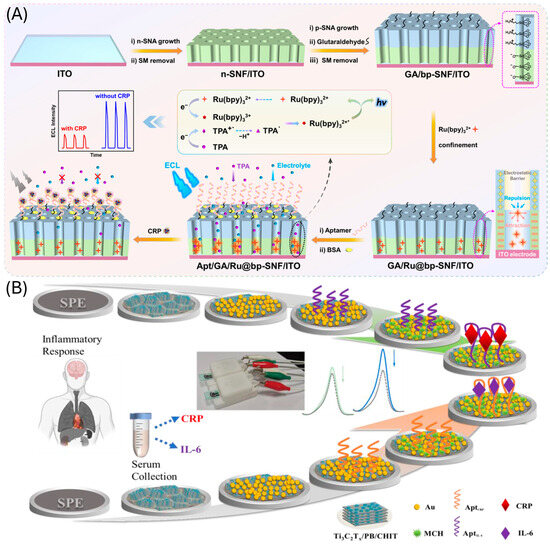
Figure 7.
CRC aptasensors based on the use of (A) nanochannel for the immobilization of the aptamer and ECL detection. Reprinted with permission from Ref. [137] and (B) Ti3C2Tx and gold nanoparticles composites. Reprinted with permission from Ref. [141].

Table 6.
Summary of aptasensors employing different types of nanomaterials.
Table 6.
Summary of aptasensors employing different types of nanomaterials.
| Protein | Sensing Platform/ Assay Format | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CEA | ITO/SNF-ngqdS/apt/BSA Direct Assay | ECL | 0.001–10 ng mL−1/ 0.4 pg mL−1 | Glucosa, NGAL, S100, AFP | - | 2% FBS (SA) | - | [139] |
| ITO/VMSF/PtNPs/Apt/BSA Direct assay | ECL | 0 fg mL−1–100 ngmL−1/ 0.4 fg mL−1 | AFP, CA19-9, S100, Hb, HSA, IgG, AA, UA, Glu, DA | 90 min | 2% spiked serum | - | [140] | |
| AuSPE/cDA-Fc/apt/PdPt@PCN-224 Displacement assay | Electrochemistry (DPV) | 1 pg mL−1–100 ng mL−1/ 0.98 pg mL−1 | Thrombin, PSA, HSA, AFP, IgG | 30 min | Serum (n = 2) | - | [144] | |
| CRP | ITO/bp-SNF/Ru(bpy)32+/apt Direct assay | ECL | 0.01–1000 ng mL−1/ 8.5 pg mL−1 | CEA, SAA, CA-15-3, CA19-9, Glu, TRP | 1 h | Spiked FBS | 11 days | [137] |
| ITO/SNF/AuNPs/Apt/BSA Direct assay | ECL | 10 pgmL−1–1 μgmL−1 4 pg mL−1 | AFP, CA15-3, glu, l-serines, Na, K, Ca | 30 min | 2% spiked serum | 15 days | [138] | |
| SPCE/Ti3C2Tx/PBA/Chit/AuNP/Apt/MCH Direct assay | Electrochemistry (DPV) | 0.1–15 μg mL−1/ 0.075 μg mL−1 | Hb, cholesterol, cys, LPS, BSA, Glu | 30 min | serum (n = 3) | 30 days | [141] | |
| SPCE/MSA-NiSe2/QD/apt/BSA Direct assay | Electrochemistry (CC) | 10–110 pg mL−1/ 2.80 pg mL−1 | l-cys, BSA, cTnI, | - | 10% serum (n = 3) | 15 days | [147] | |
| SPCE/Ti3C2Tx-AgNO3/AuNPS/apt/MCH Direct assay | Electrochemistry (DPV) | 0.1–200 ng mL−1/ 41 pg mL−1 | IL-6, BSA, Hb, Glu, Cys | 40 min | Spiked serum (n = 2) | 10 days | [143] | |
| GCE/ZIF67-C@AuNPs/apt/BSA/CRP/ Ab-HRP Sandwich assay | Electrochemistry (DPV) | 10 pg mL−1–10 μg mL−1 0.44 pg mL−1 | IL-6/10, GHBP, IFN-γ, CA-125, BSA | 1 h | Plasma (n = 4) | 15 days | [130] | |
| GCE/PDA/AuNPs/Ab/BSA/CRP/aptamer@ Mn3(PO4)2 Sandiwch assay | Electrochemistry (DPV) | 1 pg mL−1–1 ng mL−1 0.37 pg mL−1 | CEA, cTnI, NMP22, PSA | 2 h 40 min | Serum (n = 8) | - | [146] | |
| IL-6 | SPCE/Ti3C2Tx/PBA/Chit/AuNP/apt/MCH Direct assay | Electrochemistry (DPV) | 0.01–100 ng mL−1/ 7 pg mL−1 | Hb, cholesterol, cys, LPS, BSA, Glu | 40 min | serum (n = 3) | 30 days | [141] |
| SPE/Ti3C2Tx/MoS2/Au/NPs/apt/MCH Direct assay | Electrochemistry (CA) | 5 pg mL−1–100 ng mL−1/ 2.9 pg mL−1 | CRP, bovine l-cys, AA, BSA, UA | 30 min | 1% serum | - | [142] | |
| TNF-α | ITO/CdS/Apt/MCH/TNF-α/NiCo3O4-Au-Apt2 Sandwich assay | Photoelectrochemistry | 1 fg mL−1–1 ng mL−1/ 0.63 fg mL−1 | h-FABP, CEA, l-cys | 2 h 30 min | Spiked 10% serum | 20 days | [145] |
AA: ascorbic acid; Ab: antibody; AFP: alpha-feto protein; apt: aptamer; AuNPS: gold nanoparticles; AuSPE: gold screen printed, electrodes; BSA: bovine serum albumin; bp-SNF: bipolar silica nanochannel film; CA: cancer antigen; CEA: carcinoembryonic antigen; cDNA: complementary DNA: DA: dopamine; CHIT; Chitosan; CRP: C-reactive protein; cTnI: troponin I Cys: cysteine; DPV: differential pulse voltammetry; ECL: electrochemiluminescence; Fc: ferrocene; FBS: fetal bovine serum; GCE: glassy carbon electrode; GHBP; Growth Hormone Binding Protein; Glu: glucose; Hb: hemoglobin; hFBPA: heart-type fatty acid-binding protein; HSA: human serum albumin; HRP: horse radish peroxidase; IFN-γ: interferon-γ; IgG: immunoglobulin G; IL: interleukin; ITO: indium tin oxide; LPS: lipopolysaccharide; MCH: mercaptohexanol; MSA; mercaptosuccinic acid; NGAL: lipocalin-2; NGQDs: nitrogen-graphene quantum dots; NMP: Nuclear Matrix Protein NPs: nanoparticles; PBA: Phenylboronic acid; PDA: polymerized dopamine; PSA: prostate specific antigen; PtNPs: platinum nanoparticles; QDs: quantum dots; SA: standard additions; SAA: Serum Amyloid A; SPCE: screen printed carbon electrodes; S100: calcium related proteins; SNF: TNF-alpha: TRP: tryptophan; UA: uric acid; VMSF: vertically ordered mesoporous silica film.
4.3. DNA-Based Amplifications
To improve the sensitivity of aptamer-based sensors, DNA amplification strategies have emerged as effective alternatives. Since aptamers are typically single-stranded DNA (ssDNA), amplification methods such as the hybridization chain reaction (HCR) and exonuclease-assisted reactions can be directly integrated into aptasensors. In addition, DNAzymes provide another powerful means of enhancing sensitivity, as they can be readily incorporated into oligonucleotide-based signal amplification processes (Table 7).
One example is an RNase H-assisted DNA recycling strategy applied to a fluorescence assay for C-reactive protein (CRP). Binding of CRP to its aptamer releases the DNA strand P1, which hybridizes with a fluorescently labelled RNA. RNase H then selectively cleaves the RNA strand, generating fluorescent fragments that do not adsorb on graphene oxide, evading quenching and therefore producing fluorescence. This cycle repeats until enzyme depletion, resulting in strong signal amplification (Figure 8A) [148].
Endonuclease-assisted amplification has also been explored. A representative strategy employed magnetic beads functionalized with an aptamer and its complementary strand. Upon recognition of CEA the complementary DNA activated the endonuclease to cleave detection probes labelled with fluorophore–quencher pairs. Continuous cleavage resulted in the accumulation of fluorescent fragments in the supernatant, markedly enhancing the signal after magnetic separation [149].
Exonuclease-based amplification is among the most widely applied strategies, particularly Exonuclease III (Exo III) [150,151,152,153] and Exonucelase I [154,155,156]. For instance, Huang et al. combined Exo III-assisted target recycling with HCR amplification to achieve ultrasensitive electrochemical detection of CEA. The process involved multiple hairpin probes and enzymatic cleavage steps, producing abundant dsDNA polymers that interacted with a triblock polyadenine probe (TPP) immobilized on a gold electrode. The resulting steric hindrance and electrostatic repulsion effects enhanced the electrochemical impedance signal. This strategy achieves an impressive detection limit though at the cost of a long (>4 h) and complex experimental workflow (Figure 8B) [157].
Beyond enzymatic recycling, the structural features of aptamers have been exploited through DNAenzymes. Hemin intercalates into G-quadruplex (G4) motifs, forming a G4/hemin DNAzyme with horseradish peroxidase (HRP)-like activity [158]. This approach has been applied to CEA detection via photoelectrochemical [159] and fluorescence methods. However, the experimental protocol is excessively long (>8 h), and analyzing real samples requires a high dilution (1:1000), which complicates its use in hospitals and limits practical sample analysis. More sophisticated systems combine DNAzyme amplification with structural DNA nanodesigns: for example, a dendrimer-like DNA nanoassembly carrying multiple G-quadruplex motifs and a dangling aptamer. In the presence of CEA, the aptamer dissociates from the electrode, exposing the complementary DNA (cpDNA) to capture the nanoassembly. This arrangement harnesses the high density of G-quadruplex DNAzyme motifs to catalyze BQ efficiently, resulting in a significantly amplified electrochemical signal and a remarkably low detection limit of 0.24 ng·mL−1. The system demonstrates a synergistic strategy that combines target-triggered aptamer release with DNAzyme-based signal amplification for highly sensitive CEA detection (Figure 8C) [160].
Recent approaches increasingly combine multiple amplification mechanisms to maximize sensitivity. Examples include integrating rolling circle amplification (RCA) with exonuclease-assisted target cycling and G4/hemin catalysis [161] or coupling Exonuclease I-assisted recycling with ECL readout for CEA detection [162]. Hybrid systems combining Fe3O4@Au nanocomposites with Exo III and G-quadruplex/hemin amplification further highlight the trend toward synergistic amplification cascades [163].
Although DNA-based amplification strategies have achieved ultrasensitive detection limits, their complexity poses barriers to translation. Long incubation times, reliance on multiple enzymes and oligonucleotide constructs, and susceptibility to experimental variability limit their reproducibility and compatibility with point-of-care (POC) diagnostics. In particular, protocols often require hours of incubation and specialized expertise, which contrasts with the rapid and robust workflows needed in clinical settings. These strategies therefore remain highly innovative proof-of-concept approaches that demonstrate the versatility of aptamer chemistry but still face significant hurdles before practical implementation.
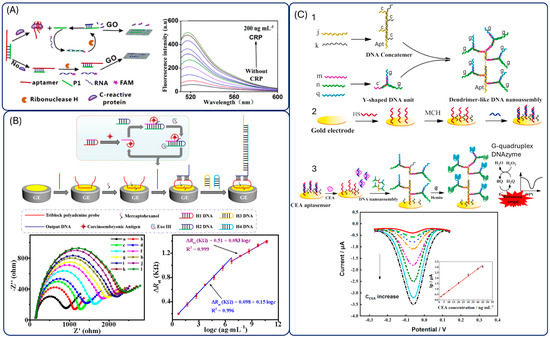
Figure 8.
Aptasensors employing DNA-based amplification strategies. (A) Fluorescence sensor employing ribonuclease H reaction and graphene oxide. Reprinted with permission from Ref. [148]. (B) Electrochemical aptasensors employing exonuclease III reaction. Reprinted with permission from Ref. [157]. (C) Electrochemical sensor employing G-quadruplex formation and hemin catalysis. Reprinted with permission from Ref. [160].

Table 7.
Summary of aptasensors employing DNA-based amplifications.
Table 7.
Summary of aptasensors employing DNA-based amplifications.
| Protein | Sensing Platform | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CEA | AuE/E1/MCH/T1-L2-L1/CEA ExoIII | Electrochemistry (DPV) | 10 fg mL−1–50 ng mL−1/ 4.88 fg mL−1 | TB, Hb, BSA | 2 h 40 min | Serum (n = 3) | 5 days | [153] |
| AuE/CTF-ssDNA/MCH/rHp/ CuxMn3-x(HITP)2+ Exo-III | ECL | 1 pg mL−1–50 ng mL−1/ 2.91 fg mL−1 | AFP, CD63, NCL, CA-50 | 1 h 40 min | Serum (SA) | 15 days | [150] | |
| AuE/AuNPs/Fc-HP2/MCH/exoIII/T4-DNA ligase+ Phi29-DNA polymerase+ dNTPs/MB | Ratiometric | 1 pg mL−1–100 ng mL−1/ 0.59 pg mL−1 | AFP, PSA, HSA | 4 h 30 min | 10% serum (n= 5) | - | [151] | |
| GCE/Au/cDNA/MCH/apt/ CEA+ExoI+ Ag+ | ECL | 100 ag mL−1–10 ng mL1/ 38.86 ag mL−1 | HSA, PSA, AFP, IgG | 1 h 30 min | 1% serum (n = 3) | - | [154] | |
| AuE/AuNPs/hpDNA/MCH/Fc-apt/exoI+CEA | Ratiometric | 10 pg mL−1–100 ng mL−1/ 1.9 pg mL−1 | AFP, PSA, HSA | 2 h | 10% serum (SA, n = 3) | 1 week | [155] | |
| AuE/AuNP/cpDNA-MCH/fcDNA ExoI+ CEA/tDNA/H1+H2+MB | Ratiometric | 1 pg mL−1–100 ng mL−1/ 0.479 pg mL−1 | AFP, PSA, HSA | 1 h 30 min | 10% serum (n =4) | 21 days | [156] | |
| AuE/H2/apt/Exo-III+CEA/S1/S2/MB | Electrochemistry (DPV) | 10 pg mL−1–100 ng mL−1/ 0.84 pg.mL−1 | AFP, CA125, CA199,CA153 | 1 h 15 min | 10% serum (n = 3) | 7 days | [152] | |
| Photoanode: ITO/BVO/ZIS Photocathode: AuNP/CuBi2O4/ ITO/Apt1/CEA/Apt2/G4/hemin | Photo-electrochemistry | 0.1 pg mL−1–10 ng mL−1/ 0.021 pg mL−1 | AFP, BSA, IgG, PSA | 2 h 20 min | Serum (n = 4) | 600 s | [159] | |
| MCH/cpDNA/Au/Apt/MCH /cpDNA/Au | Electrochemistry (DPV) | 2–45 ng mL−1/ 0.24 ng mL−1 | BSA, HSA, IgG, CRP, CA125, AFP | 3 h 24 min | Serum (n = 9) | 31 days | [160] | |
| Aptamer/Harpins/DNA/hemin/G4+ HCR | Fluorescence | 0.25–1.5 nM/ 0.2 nM | IgG, AFP, PSA | 8 h 20 min | 0.1% serum | - | [164] | |
| ExoI+ExoIII on MBs/AuE | Electrochemistry (DPV) | 10 fg mL−1–100 ng mL−1/1.26 fg mL−1 | BSA, AFP, UA CA125, Hb, AA | 3 h | Spiked serum | 30 days | [161] | |
| GCE/rGO-IL/PtNPs/Apt-G4/ hemin + exoI | Ratiometric- ECL | 10 fg mL−1–10 ng mL−1/0.85 fg mL−1 | HSA, AFP, PSA, IgG, | 30 min | 1% serum | 10 days | [162] | |
| Fe3O4@AuNPs-S1-S2-S3 + exo III+ G4. | Electrochemistry (DPV) | 0.1–200 ng mL−1/ 0.4 pg mL−1 | HSA, Hb, l-cys, BSA | 2 h 35 min | serum | - | [163] | |
| MNPs/strep/btn-cDNA/apt | Fluorescence | 1–500 ng mL−1/ 0.7 ng mL−1 | SCD146, IgG, BSA | 3 h | plasma | - | [149] | |
| AuSPE/TPP+ exoIII | Electrochemistry (EIS) | Up to 1010 ag mL−1/ 0.39 ag mL−1 | HSA, Hb, AFP, lysz ,BSAVEGF | 4h 30 min | 2% serum (SA) | 20 days | [157] | |
| CRP | GO+ aptamer+RNA-FAM+ ribonuclease H | Fluorescence | 50 pg mL−1–100 ng mL−1/ 0.01 ng mL−1 | d-dimer, MB, MUC1 | 50 min | 2% serum, urine, saliva | - | [148] |
AA: ascorbic acid; AFP: alpha-feto protein; AuE: gold electrode; AuNPs: gold nanoparticles; apt: aptamer; AuSPE: gold screen printed electrodes; BSA: bovine serum albumin; btn: biotin; BVO: BiVO4; CA: cancer antigen; cDNA: complementary DNA; CD63: Granulophysin; CEA: carcinoembryonic antigen; CRP: C-reactive protein; CTF: covalent triazine framework; dNTPS: nucleotides; DPV: differential pulse voltammetry; ECL: electrochemiluminescence; EIS: electrochemical impedance spectroscopy; Exo: exonuclease; FAM: fluorescein; Fc: ferrocene; Fc-HP: ferrocene-hairpin DNA; GCE: glassy carbon electrode; GO: graphene oxide; G4: g-quadruplex; Hb: hemoglobin; HCR: hybridization chain reaction; HITP: 2,3,6,7,10,11-hexaiminotriphenylene; HSA: human serum albumin; IgG: immunoglobulin G; ITO: indium tin oxide; l-cys: l-cysteines; Lyz: Lysozymes; MB: methylene blue; MBs: magnetic beads; MCH: mercaptohexanol; MNTPS: magnetic nanoparticles; MUC: mucin; NCL: nucleolin; PSA: prostate specific antigen; PtNPS: platinum nanoparticles; rGO/IL: reduced graphene oxide ionic liquid; rHp: residual hairpins strands; SCD146: soluble melanoma cell adhesion molecule; ssDNA: single stranded DNA; TB: thrombin; tDNA: trigger DNA; TTP: triblock polyadenine probe; UA: uric acid; VEGF: Vascular endothelial growth factor; ZIS: ZnIn2S4.
5. Adaptations to POC (Point-of-Care) Formats
Translating aptamer-based diagnostic tools into Point-of-Care (POC) applications requires balancing accuracy, speed, and usability. POC settings demand rapid results, streamlined workflows, and seamless integration into clinical environments, often operated by non-specialist staff. While highly sensitive, multistep amplification protocols can deliver outstanding sensitivity, they must demonstrate consistent reliability to be practical in real-world POC settings. Consequently, simplified assay formats, such as direct electrochemical or colorimetric platforms, are preferred as they reduce operational errors and enable reliable, real-time decision-making [165]. This section explores the strategies and innovations implemented to optimize sensing devices and protocols for effective and reliable POC deployment (Table 8).
Microfluidic chips are gained traction as compact and automated systems that minimize reagent use while integrating sample pretreatment, separation, mixing, reactions, and detection steps [166]. These platforms enable fully integrated assays, combining sample handling, target enrichment, and sensitive detection on a single device. For example, Zhao et al. developed a microfluidic platform incorporating an electrochemical aptasensor for CEA. The device consists of a herringbone-embedded microfluidic chip above a sensor constructed from a MXene–carbon nanotube composite, providing strong signal amplification. The herringbone structures generate localized mixing, enhancing interactions between target molecules and the sensing interface. This design allows automated sample injection, efficient target capture, and sensitive electrochemical detection on a single, user-friendly platform [167] (Figure 9A). Similarly, Liu et al. integrated an electrode sensor array for TNF-alpha into a microfluidic device, enabling controlled fluid flow over multiple sensing elements [168]. Extending this principle to optical detection, Huang et al. coupled a SERS-based aptasensor with a microfluidic system integrating mixing chambers, serpentine channels, and magnetic separation. The system utilizes two nanomaterials with excellent SERS properties: gold-coated iron oxide particles (Fe3O4@AuNPs) and gold nanocages (AuNCs). In the assay, ssDNA1 and aptamers are immobilized on Fe3O4@AuNPs, while ssDNA2 and Raman tags are attached to AuNCs. Upon target binding, the aptamer detaches from Fe3O4@AuNPs, allowing ssDNA2 to hybridize with ssDNA1 and form the Fe3O4@AuNPs@AuNCs@SERS tag complex. This platform has been successfully applied to distinguish healthy individuals from cancer patients in just 15 min [169]. Microfluidic paper-based analytical devices (µPADs) represent another step toward affordable and disposable POC systems. The optical properties of gold nanoparticles (GNPs) functionalized with aptamers enable straightforward colorimetric readouts. Upon target binding, nanoparticle aggregation induces a visible color change, supporting rapid detection without the need for specialized equipment. For instance, Malekmohamadi et al. developed a µPAD aptasensor for CRP and IL-6, providing a low-cost approach to monitoring sepsis biomarkers [170].
Microneedles (MNs) have recently emerged as promising platforms for minimally invasive wearable and POC sensing technologies [171]. Using MNs, Yuan et al. developed an electrochemical aptamer-based sensor for real-time monitoring of C-reactive protein (CRP) in interstitial fluid (ISF) (Figure 9B). Integrated with a smartphone interface, the platform enabled real-time in vivo monitoring in animal models, demonstrating its potential for wearable healthcare technologies [172].
Collectively, these strategies highlight how aptasensor technology is moving closer to POC implementation by prioritizing automation, miniaturization, and user-friendliness. Microfluidic and paper-based systems reduce complexity while maintaining sensitivity, and wearable devices such as microneedles pave the way for continuous monitoring in outpatient or home settings. Nevertheless, widespread adoption still depends on demonstrating robustness in diverse clinical environments, scaling up fabrication, and validating performance across large patient cohorts.
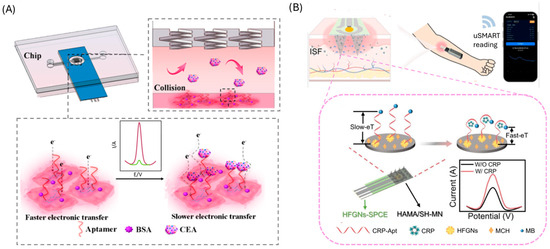
Figure 9.
Adaptations of aptasensors for colorectal cancer diagnosis to a point-of-care format. (A) Aptasensor employing a microfluidic chip and electrochemical detection. Reprinted with permission from Ref. [167]. (B) Microneedle-based electrochemical aptasensors for the real time and continuous determination of CRP in interstitial fluid. Reprinted with permission from Ref. [172].

Table 8.
Summary of CRC aptasensors adapted to a point-of-care format.
Table 8.
Summary of CRC aptasensors adapted to a point-of-care format.
| Protein | Sensing Platform/POC Device | Detection Method | Linear Range/ LOD | Selectivity | Time | Samples | Stability | Ref. |
|---|---|---|---|---|---|---|---|---|
| CEA | Ti3C2-He@CCNT/Apt/BSA/CEA microfluidic chip | Electrochemistry (DPV) | 10 pg mL−1–1 µg mL−1/ 2.88 pg mL−1 | HSA, IgG, glu | 1 h | Serum (n = 3) | - | [167] |
| CRP | uPAD/AuNPs/apt/salt paper-based + microfluidic | Colorimetric | 0 -1000 mg L−1 1.9 mg L−1/ | BSA, IL-6, TNF-α | 40 min | plasma (spiked) | 1 year | [170] |
| HFGN-SPCE/MB-apt Microneedles | Electrochemistry (SWV) | 1 ngmL−1–100 μgmL−1/ 0.85 ng mL−1 | UA, AA, Hb BSA, cTnI | seconds | In vivo (rats) | 16 days | [172] | |
| IL-6 | Fe3O4AuNPs/apt/AuNC-RTG-cDNA Microfluidic chip | SERS | 1 pg mL−1–1 µg mL−1 /0.178 pg mL−1 | OPN, AFP | 15 min | Serum (n = 30) | - | [169] |
| uPAD/AuNPs/apt/salt paper-based + microfluidic | Colorimetric | 1–25 ng L−1/ 0.07 ng L−1 | BSA, CRP, TNF-α | 40 min | plasma (spiked) | 1 year | [170] | |
| MMP9 | Fe3O4AuNPs/apt/AuNC-RTG-cDNA Microfluidic chip | SERS | 1 pg mL−1–1 µg mL−1/ 0.178 pg mL−1 | OPN, AFP | 15 min | Serum (n = 30) | [169] | |
| TNF-α | Glass/Au/apt-MB/MCH + PDMS microfluidic device | Electrochemistry (SWV) | 9–88ng mL−1/ 5.46 ng mL−1 | IL-12, IL-6, IL-10, BSA, IFN-γ, | 1 h | - | - | [168] |
AA: ascorbic acid; AFP: alpha-feto protein; Apt: aptamer; Au: gold; AuNCs: gold nanocages; AuNPS: gold nanoparticles; BSA: bovine serum albumin; CCNT: carboxylic carbon nanotube; cDNA: complementary DNA; CEA: Carcinoembryonic antigen; CRP: C-reactive protein; cTnI: troponin I; DPV: differential pulse voltammetry; Glu: glucose; Hb: hemoglobin; He: hemin; HFGN: hierarchical flower-like gold nanostructure; HSA: human serum albumin; IFN-α: interferon alpha; IgG: Immunoglobulin G, IL: interleukin; MB: methylene blue; MCH: mercaptohexanol; MMP: matrix metalloprotease; OPN: Osteopontin; POC: point-of-care; µPADs: paper-based analytical devices; PDMS: Polydimethylsiloxane; RTG: Raman-tag; SPCE: screen printed carbon electrodes; SERS: Surface-Enhanced Raman Scattering; SWV: square wave voltammetry; UA: uric acid.
6. Market Transfer
For aptamer-based diagnostic tools to transition from research laboratories to clinical practice, several translational challenges must first be addressed. Stability in complex biological matrices, such as blood and stool, remains a major concern due to nuclease degradation and the presence of interfering biomolecules. This limitation can be mitigated by converting aptamers into peptide nucleic acids (PNAs) or by introducing chemical backbone modifications, which enhance enzymatic resistance while preserving binding specificity. Equally important is the need for validation using real patient samples and statistically robust cohorts, as many current studies rely on artificial matrices or spiked samples. Reproducibility across laboratories, standardisation of reporting practices, and evaluation under clinically relevant conditions are also essential prerequisites for regulatory acceptance.
Regulatory approval represents a critical milestone. In the U.S., the Food and Drug Administration (FDA) regulates in vitro diagnostic devices (IVDs) under the Center for Devices and Radiological Health (CDRH). Aptamer-based tests fall into this category and must demonstrate analytical validity (accuracy, precision, reproducibility), clinical validity (correlation with disease), and clinical utility (impact on patient outcomes) [173]. Depending on risk classification, regulatory clearance may proceed via the 510(k) pathway (when a predicate device exists) or via the more rigorous Premarket Approval (PMA) process [174].
In Europe, the In Vitro Diagnostic Regulation (IVDR) imposes comparable requirements, emphasizing clinical performance evaluation, risk stratification, and conformity assessment by notified bodies. For global commercialization, compliance with both FDA and IVDR standards, along with local regulatory frameworks, will be indispensable [175].
To date, only a few nucleic acid-based diagnostics (e.g., Cologuard, Guardant360) have successfully met these standards, providing a precedent for future aptamer-based tests. However, challenges remain, including the need for large-scale clinical validation, reproducibility across heterogeneous populations, and demonstration of cost-effectiveness.
Looking forward, the convergence of advanced sensor designs, nanomaterials, microfluidic integration, and wearable formats positions aptasensors as strong candidates for next-generation colorectal cancer diagnostics. Achieving market transfer will require not only technological refinement but also close collaboration between researchers, clinicians, regulatory bodies, and industry partners. If these hurdles are addressed, aptamer-based diagnostics have the potential to complement or even reshape existing CRC screening and monitoring strategies, bridging the gap between laboratory innovation and patient-centred healthcare.
Author Contributions
M.J.L.-C.: conceptualization, writing—review and editing, project administration, funding acquisition. A.D.-F.: conceptualization, writing—original draft preparation, visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Spanish Government (Project PID-2021-123183OB-I00) and Principado de Asturias (IDE/2024/000677). A. Díaz Fernández was supported by a senior postdoctoral contract from Instituto de Investigación del Principado de Asturias (ISPA).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| AA | Ascorbic acid |
| Ab | Antibody |
| ACV | Alternating Current Voltammetry |
| AE | Acridinium ester |
| AFP | Alpha-feto protein |
| AgNPS | Silver nanoparticles |
| AGP | Arabinogalactan protein |
| APC | Activated protein-C |
| Apo | Apolipoprotein |
| Apt | Aptamer |
| APTES | 3-Aminopropyltriethoxysilane |
| Arg | Arginine |
| Au | Gold |
| AUC | Area under the curve |
| AuE | Gold electrode |
| AuNCs | Gold nanocages |
| AuNPs | Gold nanoparticles |
| AuNRs | Gold nanorods |
| AuSPE | Gold screen printed electrode |
| b-ME | β-mercaptoethanol |
| Bp-SNF | Bipolar silica nanochannel film |
| BSA | Bovine serum albumin |
| btn | Biotin |
| BVO | BiVO4 |
| CA | Carbohydrate antigen |
| cAPT | Aptamer complementary strand |
| CAVIN | Caveolae-associated protein |
| CCNT | Carboxylic carbon nanotube |
| cDNA | Complementary DNA |
| CDRH | Center for Devices and Radiological Health |
| CD63 | Granulophysin |
| CEA | Carcinoembryonic antigen |
| CHIT | Chitosan |
| CNTs | Carbon nanotubes |
| COFs | Covalent organic frameworks |
| CP | Calprotectin |
| cpDNA | Complementry DNA |
| CQDs | Carbon quantum dots |
| CR | Creatinine |
| CRC | Colorectal cancer |
| CRP | C-reactive protein |
| CSPE | Carbon screen printed electrode |
| CTCs | Ciculating tumor cells |
| ctDNA | Circulating tumor DNA |
| CTF | Covalent triazine framework |
| cTnI | Troponin I |
| cTnT | Cardiac troponin |
| CV | Cyclic voltammetry |
| cys | Cysteine |
| DA | Dopamine |
| DHEA | Dehydroepiandrosterone |
| DNA | Deoxyribonucleic acid |
| dNTPs | Nucleotides |
| DP | Dopamine |
| DPV | Differential pulse voltammetry |
| ECL | Electrochemiluminescence |
| EGaIn | Eutectic gallium indium |
| EIS | Electrochemical impedance spectroscopy |
| ePAD | Electrochemical paper-based analytical devices |
| Exo | Exonuclease |
| FBS | Fetal bovine serum |
| Fc | Ferrocene |
| Fc-HP | Ferrocene-hairpin DNA |
| FDA | Food and drug administration |
| FEN | Flap endonuclease |
| FH | Ferrocenylhexanethiol |
| FIT | Fecal immunotest |
| FRET | Förster resonance energy transfer |
| GCE | Glassy carbon electrode |
| GE | Graphene electrode |
| GHBP | Growth hormone receptor |
| Glu | Glucose |
| Gly | Glycine |
| GNPs | gold nanoparticles |
| GO | Graphene oxide |
| GOD | Glucose oxidase |
| GrSPE | Graphene screen printed electrodes |
| GSH | Glutathione |
| G4 | G-quadruplex |
| Hb | Hemoglobulin |
| HCG | Human chorionic gonadotropin |
| HCR | Hybridization chain reaction |
| Hcy | Homocysteine |
| He | Hemin |
| HER-2 | Human Epidermal growth factor Receptor 2 |
| hFBPA | Heart-type fatty acid-binding protein |
| HFGN | Hierarchical flower-like gold nanostructure |
| HITP | 2,3,6,7,10,11-hexaiminotriphenylene |
| HRP | Horse radish peroxidase |
| HSA | Human serum albumin |
| HT | Hexanothiol |
| IDE | Interdigitated electrodes |
| IFN | Interferon |
| Ig | Immunoglobulin |
| IL | Interleukin |
| ISF | Interstitial fluid |
| ITO | Indium tin oxide |
| IVDR | In vitro diagnostic regulation |
| IVDs | In vitro diagnostic devices |
| JNP | janus particles |
| KD | Dissociation constant |
| lncRNAs | Long non-coding RNA |
| LPS | Lipopolysaccharide |
| LSW | Linear sweep voltammetry |
| Lyz | Lysozymes |
| m-SELEX | Membrane-SELEX |
| MACE | Microbead-assisted capillary electrophoresis |
| MACNTs | Magnetic Carbon nanotubes |
| MARAS | Magnetic-Assisted Rapid Aptamer Selection |
| MB | Methylene blue |
| MBs | Magnetic beads |
| MCF | Mesoporous carbon foam |
| MCH | Mercaptohexanol |
| MC-LR | Microcystin |
| miRNAs | microRNAs |
| MMPs | Matrix metalloproteinases |
| MNs | Microneedles |
| MNTPS | Magneitc nanoparticles |
| MOFs | Metal–organic frameworks |
| MRD | Molecular residual disease |
| MSA | Mercaptosuccinic acid |
| MSI | Microsatellite instability |
| MUC | Mucin |
| MWCNTs | Multi-walled carbon nanotubes |
| Myo | Myoglobin |
| NCD | nitrogen dopped carbon dots |
| NCL | Nucleolin |
| NGAL | Lipocalin-2 |
| N-GQDSNPs | Nitrogen-graphene quantum dots nanoparticles |
| NMP | Nuclear matrix protein |
| NPs | Nanoparticles |
| NSE | Neuron-specific enolase |
| OPN | Osteopotin |
| OVA | Ovalbumin |
| PBA | Phenylboronic acid |
| PCB | Printed circuit board |
| PCR | Polymerase chain reaction |
| PCT | Procalcitonin |
| PDA | Polymerized dopamine |
| PDMS | Polydimethylsiloxane |
| PEDOT:PSS | 3,4-(ethylenedioxythiophene):poly(styrenesulfonate) |
| PEG | Polyethylene glycol |
| PFcGE | Poly(ferrocenyl glycidyl ether)-grafted |
| pfLDH | Plasmodium lactate dehydrogenase |
| PMA | Premarket Approval |
| PNAs | Peptide nucleic acids |
| PSA | Prostate specific antigen |
| PTB7-Th | poly{4,8-bis[5-(2-ethylhexyl) thiophen-2-yl]benzo[1,2-b:4,5-b′]dithiophene-2,6-diyl-alt-3-fluoro-2-[(2-ethylhexyl)carbonyl] thieno[3,4-b]-thiophene-4,6-diyl} |
| PTK7 | Tyrosine-protein kinase-like 7 |
| PtNPs | Platinum nanoparticles |
| QCM | Quartz crystal microbalance |
| QDs | Quantum dots |
| RAB26 | Ras-related protein |
| RCA | Rolling circle amplification |
| rGO | Reduce graphene oxide |
| rGO/IL | Reduced graphene oxide ionic liquid |
| rHP | Residual hairpins strands |
| RTG | Rammna-tag |
| SA | Standard addition |
| SAA | Serum Amyloid A |
| SCD146 | Soluble melanoma cell adhesion molecule |
| SELEX | Systematic Evolution of Ligands by Exponential Enrichment |
| SERS | Surface-Enhanced Raman Scattering |
| SOMAMERS | Slow Off-rate Modified Aptamers |
| SPCE | Screen printed carbon electrodes |
| SPR | Surface plasmon resonance |
| ssDNA | Single stranded DNA |
| strep | Streptavidin |
| SWCNTs | Single well carbon nanotubes |
| SWV | Square wave voltammetry |
| S100 | Calcium related proteins |
| POC | Point-of-care |
| PtμEs: | Platinum microelectrode |
| TB | Thrombin |
| TcTn-I | Troponin I |
| tDNA | Trigger DNA |
| TFGA | Surface flexible thin film gold arrays chips |
| TIMP-1 | Metalloproteinases-1 |
| TNF-α | Tumor necrosis factor-alpha |
| TPP | Triblock polyadenine probe |
| TRP | Tryptophan |
| UA | Uric acid |
| VEGF | Vascular endothelial growth factor |
| VMSF | Vertically ordered mesoporous silica film |
| ZIS | ZnIn2S4 |
| 4-MBA | 4-mercaptobenzoic acid |
| 6-FAM | Fluorescein |
| µPADs | Microfluidic paper-based analytical devices |
References
- Disc Klimeck, L.; Heisser, T.; Hoffmeister, M.; Brenner, H. Colorectal cancer: A health and economic problem. Best Pract. Res. Clin. Gastroenterol. 2023, 66, 101839. [Google Scholar] [CrossRef]
- Mannucci, A.; Zuppardo, R.A.; Rosati, R.; Di Leo, M.; Perea, J.; Cavestro, G.M. Colorectal cancer screening from 45 years of age: Thesis, antithesis and synthesis. World J. Gastroenterol. 2019, 25, 2565. [Google Scholar] [CrossRef]
- McCabe, M.A.; Mauro, A.J.; Schoen, R.E. Novel colorectal cancer screening methods—Opportunities and challenges. Nat. Rev. Clin. Oncol. 2025, 22, 581–591. [Google Scholar] [CrossRef]
- Zamanian, M.Y.; Darmadi, D.; Darabi, R.; Al-Aouadi, R.F.A.; Ivraghi, M.S.; Akkol, E.K. Biomarkers for colorectal cancer detection: An insight into colorectal cancer and FDA-approved biomarkers. Bioimpacts 2025, 15, 31211. [Google Scholar] [CrossRef]
- Shaukat, A.; Levin, T.R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 521–531. [Google Scholar] [CrossRef]
- López Salas, M.; De Haro Gázquez, D.; Fernández Sánchez, B.; Amador Muñoz, M.L. Knowledge, Compliance, and Inequities in Colon Cancer Screening in Spain: An Exploratory Study. Healthcare 2023, 11, 2475. [Google Scholar] [CrossRef] [PubMed]
- Berg-Beckhoff, G.; Leppin, A.; Nielsen, J.B. Reasons for participation and non-participation in colorectal cancer screening. Public Health 2022, 205, 83–89. [Google Scholar] [CrossRef]
- Ma, L.; Guo, H.; Zhao, Y.; Liu, Z.; Wang, C.; Bu, J.; Sun, T.; Wei, J. Liquid biopsy in cancer current: Status, challenges and future prospects. Signal Transduct. Target. Ther. 2024, 9, 336. [Google Scholar] [CrossRef] [PubMed]
- Lin, B.; Jiang, J.T.; Zhou, X. Recent advances in design strategies of aptamer-based liquid biopsy. J. Polym. Sci. 2024, 62, 2848–2870. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Ahmadyousefi, Y.; Malih, S.; Mirzaee, Y.; Saidijam, M. Nucleic acid aptamers in diagnosis of colorectal cancer. Biochimie 2019, 156, 1–11. [Google Scholar] [CrossRef]
- Goh, K.W.; Stephen, A.; Wu, Y.S.; Sim, M.S.; Batumalaie, K.; Gopinath, S.C.B.; Guad, R.M.; Kumar, A.; Sekar, M.; Subramaniyan, V.; et al. Molecular Targets of Aptamers in Gastrointestinal Cancers: Cancer Detection, Therapeutic Applications, and Associated Mechanisms. J. Cancer 2023, 14, 2491–2516. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, S.; Cai, Y.; Tang, F. Nucleic acid aptamer application in diagnosis and therapy of colorectal cancer based on cell-SELEX technology. npj Precis. Oncol. 2017, 1, 37. [Google Scholar] [CrossRef]
- Zhang, J.; Zhu, H.; Liu, W.; Miao, J.; Mao, Y.; Li, Q. Prognostic and predictive molecular biomarkers in colorectal cancer. Front. Oncol. 2025, 15, 1532924. [Google Scholar] [CrossRef]
- Ding, Y.; Li, W.; Wang, K.; Xu, C.; Hao, M.; Ding, L. Perspectives of the Application of Liquid Biopsy in Colorectal Cancer. BioMed Res. Int. 2020, 2020, 6843180. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and quantification of mutations in the plasma of patients with colorectal tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed]
- Nikolouzakis, T.K.; Vassilopoulou, L.; Fragkiadaki, P.; Sapsakos, T.M.; Papadakis, G.Z.; Spandidos, D.A.; Tsatsakis, A.M.; Tsiaoussis, J. Improving diagnosis, prognosis and prediction by using biomarkers in CRC patients (Review). Oncol. Rep. 2018, 39, 2455–2472. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Zhang, B.; Li, P.; Zhao, Y. Methods and biomarkers for early detection, prediction, and diagnosis of colorectal cancer. Biomed. Pharmacother. 2023, 163, 114786. [Google Scholar] [CrossRef]
- Łukaszewicz-zając, M.; Mroczko, B. Circulating Biomarkers of Colorectal Cancer (CRC)—Their Utility in Diagnosis and Prognosis. J. Clin. Med. 2021, 10, 2391. [Google Scholar] [CrossRef]
- Marshall, K.W.; Mohr, S.; El Khettabi, F.; Nossova, N.; Chao, S.; Bao, W.; Ma, J.; Li, X.J.; Liew, C.C. A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int. J. Cancer 2010, 126, 1177–1186. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; De Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef]
- DeVos, T.; Tetzner, R.; Model, F.; Weiss, G.; Schuster, M.; Distler, J.; Steiger, K.V.; Grützmann, R.; Pilarsky, C.; Habermann, J.K.; et al. Circulating Methylated SEPT9 DNA in Plasma Is a Biomarker for Colorectal Cancer. Clin. Chem. 2009, 55, 1337–1346. [Google Scholar] [CrossRef]
- Shirley, M. Epi proColon® for Colorectal Cancer Screening: A Profile of Its Use in the USA. Mol. Diagn. Ther. 2020, 24, 497–503. [Google Scholar] [CrossRef]
- Han, Y.J.; Shao, C.Y.; Yao, Y.; Zhang, Z.; Fang, M.Z.; Gong, T.; Zhang, Y.J.; Li, M. Immunotherapy of microsatellite stable colorectal cancer: Resistance mechanisms and treatment strategies. Postgrad. Med. J. 2024, 100, 373–381. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients with Stages i to III Colorectal Cancer. JAMA Oncol. 2019, 5, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B.; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal–Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020, 17, 757. [Google Scholar] [CrossRef]
- Ma, D.; Gao, X.; Wang, L.; Yin, H.; Feng, L.; Zhu, Y. Circulating tumor DNA for MRD detection in colorectal cancer: Recent advances and clinical implications. Biomark. Res. 2025, 13, 89. [Google Scholar] [CrossRef]
- Link, A.; Balaguer, F.; Shen, Y.; Nagasaka, T.; Lozano, J.J.; Boland, C.R.; Goel, A. Fecal microRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1766–1774. [Google Scholar] [CrossRef]
- Bao, Z.; Gao, S.; Zhang, B.; Shi, W.; Li, A.; Tian, Q. The critical role of the miR-21–MEG2 axis in colorectal cancer. Crit. Rev. Eukaryot. Gene Expr. 2020, 30, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Yamada, A.; Horimatsu, T.; Okugawa, Y.; Nishida, N.; Honjo, H.; Ida, H.; Kou, T.; Kusaka, T.; Sasaki, Y.; Yagi, M.; et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin. Cancer Res. 2015, 21, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.; Fawzy, A.; Akel, S.Y.; Gamal, H.; Elshimy, R.A.A. Evaluation of microRNA 92a Expression and Its Target Protein Bim in Colorectal Cancer. Asian Pac. J. Cancer Prev. 2022, 23, 723–730. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, T.; Duan, J.; Liu, X.; Liu, L. MicroRNA-223-induced inhibition of the FBXW7 gene affects the proliferation and apoptosis of colorectal cancer cells via the Notch and Akt/mTOR pathways. Mol. Med. Rep. 2020, 23, 154. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, H.A.; El Amin, H.A.; Ahmed, E.S.M.; Kenawy, A.G.; El-Ebidi, A.M.; Elnakeeb, I.; Kholef, E.F.M.; Elsewify, W.A.E. Role of MicroRNA-223 and MicroRNA-182 as Novel Biomarkers in Early Detection of Colorectal Cancer. Int. J. Gen. Med. 2022, 15, 3281–3291. [Google Scholar] [CrossRef]
- Eslamizadeh, S.; Zare, A.-A.; Talebi, A.; Tabaeian, S.P.; Eshkiki, Z.S.; Heydari-Zarnagh, H.; Akbari, A. Differential Expression of miR-20a and miR-145 in Colorectal Tumors as Potential Location-specific miRNAs. MicroRNA 2021, 10, 66–73. [Google Scholar] [CrossRef]
- Humphreys, K.J.; McKinnon, R.A.; Michael, M.Z. miR-18a Inhibits CDC42 and Plays a Tumour Suppressor Role in Colorectal Cancer Cells. PLoS ONE 2014, 9, e112288. [Google Scholar] [CrossRef]
- Alaiyan, B.; Ilyayev, N.; Stojadinovic, A.; Izadjoo, M.; Roistacher, M.; Pavlov, V.; Tzivin, V.; Halle, D.; Pan, H.; Trink, B.; et al. Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer 2013, 13, 196. [Google Scholar] [CrossRef]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed]
- Badowski, C.; He, B.; Garmire, L.X. Blood-derived lncRNAs as biomarkers for cancer diagnosis: The Good, the Bad and the Beauty. Npj Precis. Oncol. 2022, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Lippi, G.; Betsou, F.; Cadamuro, J.; Cornes, M.; Fleischhacker, M.; Fruekilde, P.; Neumaier, M.; Nybo, M.; Padoan, A.; Plebani, M.; et al. Preanalytical challenges-time for solutions. Clin. Chem. Lab. Med. 2019, 57, 974–981. [Google Scholar] [CrossRef]
- Hauptman, N.; Glavač, D. Colorectal Cancer Blood-Based Biomarkers. Gastroenterol. Res. Pract. 2017, 2017, 2195361. [Google Scholar] [CrossRef]
- Lakemeyer, L.; Sander, S.; Wittau, M.; Henne-Bruns, D.; Kornmann, M.; Lemke, J. Diagnostic and Prognostic Value of CEA and CA19-9 in Colorectal Cancer. Diseases 2021, 9, 21. [Google Scholar] [CrossRef]
- Carpelan-Holmström, M.; Louhimo, J.; Stenman, U.H.; Alfthan, H.; Järvinen, H.; Haglund, C. CEA, CA 242, CA 19-9, CA 72-4 and hCGbeta in the diagnosis of recurrent colorectal cancer. Tumour Biol. 2004, 25, 228–234. [Google Scholar] [CrossRef]
- Yanqing, H.; Cheng, D.; Ling, X. Serum CA72-4 as a Biomarker in the Diagnosis of Colorectal Cancer: A Meta-analysis. Open Med. 2018, 13, 164. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yu, H.; Shao, Y.; Han, X.; Zhang, Y. Significance of cea and CA242 in the diagnosis of colorectal carcinoma. Chin. J. Cancer Res. 1996, 8, 272–275. [Google Scholar] [CrossRef]
- Holten-Andersen, M.; Christensen, I.; Nielsen, H.; Stephens, R.; Jensen, V.; Nielsen, O.; Sørensen, S.; Overgaard, J.; Lilja, H.; Harris, A.; et al. Total levels of tissue inhibitor of metalloproteinases 1 in plasma yield high diagnostic sensitivity and specificity in patients with colon cancer. Clin. Cancer Res. 2002, 8, 156–164. [Google Scholar] [PubMed]
- Hadler-Olsen, E.; Winberg, J.O.; Uhlin-Hansen, L. Matrix metalloproteinases in cancer: Their value as diagnostic and prognostic markers and therapeutic targets. Tumour Biol. 2013, 34, 2041–2051. [Google Scholar] [CrossRef]
- Otero-Estévez, O.; De Chiara, L.; Rodríguez-Girondo, M.; Rodríguez-Berrocal, F.J.; Cubiella, J.; Castro, I.; Hernández, V.; Martínez-Zorzano, V.S. Serum matrix metalloproteinase-9 in colorectal cancer family-risk population screening. Sci. Rep. 2015, 5, 13030. [Google Scholar] [CrossRef]
- Peltier, J.; Roperch, J.P.; Audebert, S.; Borg, J.P.; Camoin, L. Quantitative proteomic analysis exploring progression of colorectal cancer: Modulation of the serpin family. J. Proteom. 2016, 148, 139–148. [Google Scholar] [CrossRef]
- Bünger, S.; Haug, U.; Kelly, M.; Posorski, N.; Klempt-Giessing, K.; Cartwright, A.; Fitzgerald, S.P.; Toner, V.; McAleer, D.; Gemoll, T.; et al. A novel multiplex-protein array for serum diagnostics of colon cancer: A case-control study. BMC Cancer 2012, 12, 393. [Google Scholar] [CrossRef]
- Xu, J.; Ye, Y.; Zhang, H.; Szmitkowski, M.; Mäkinen, M.J.; Li, P.; Xia, D.; Yang, J.; Wu, Y.; Wu, H. Diagnostic and Prognostic Value of Serum Interleukin-6 in Colorectal Cancer. Medicine 2016, 95, e2502. [Google Scholar] [CrossRef]
- Dressen, K.; Hermann, N.; Manekeller, S.; Walgenbach-Bruenagel, G.; Schildberg, F.A.; Hettwer, K.; Uhlig, S.; Kalff, J.C.; Hartmann, G.; Holdenrieder, S. Diagnostic Performance of a Novel Multiplex Immunoassay in Colorectal Cancer. Anticancer Res. 2017, 37, 2477–2486. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.; Yu, F.; Chen, J.; Zhao, S.; Zhang, D.; Yu, Y.; Liu, X.; Tang, H.; Peng, Z. Combined detection of preoperative serum CEA, CA19-9 and CA242 improve prognostic prediction of surgically treated colorectal cancer patients. Int. J. Clin. Exp. Pathol. 2015, 8, 14853. [Google Scholar] [PubMed]
- Alves Martins, B.A.; de Bulhões, G.F.; Cavalcanti, I.N.; Martins, M.M.; de Oliveira, P.G.; Martins, A.M.A. Biomarkers in Colorectal Cancer: The Role of Translational Proteomics Research. Front. Oncol. 2019, 9, 1284. [Google Scholar] [CrossRef]
- Yu, J.; Zhai, X.; Li, X.; Zhong, C.; Guo, C.; Yang, F.; Yuan, Y.; Zheng, S. Identification of MST1 as a potential early detection biomarker for colorectal cancer through a proteomic approach. Sci. Rep. 2017, 7, 14265. [Google Scholar] [CrossRef] [PubMed]
- Djaballah, S.A.; Daniel, F.; Milani, A.; Ricagno, G.; Lonardi, S. HER2 in C olorectal Cancer: The Long and Winding Road From Negative Predictive Factor to Positive Actionable Target. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 219–232. [Google Scholar] [CrossRef]
- Benli, Y.; Arıkan, H.; Akbulut-Çalışkan, Ö. HER2-targeted therapy in colorectal cancer: A comprehensive review. Clin. Transl. Oncol. 2025, 27, 3607–3624. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Lin, M.; Zhang, H.B. Diagnostic value of carcinoembryonic antigen and carcinoma antigen 19-9 for colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 9404. [Google Scholar]
- Luo, H.; Shen, K.; Li, B.; Li, R.; Wang, Z.; Xie, Z. Clinical significance and diagnostic value of serum NSE, CEA, CA19-9, CA125 and CA242 levels in colorectal cancer. Oncol. Lett. 2020, 20, 742. [Google Scholar] [CrossRef]
- Babel, I.; Barderas, R.; Diaz-Uriarte, R.; Moreno, V.; Suarez, A.; Fernandez-Aceñero, M.J.; Salazar, R.; Capellá, G.; Casal, J.I. Identification of MST1/STK4 and SULF1 Proteins as Autoantibody Targets for the Diagnosis of Colorectal Cancer by Using Phage Microarrays. Mol. Cell. Proteom. 2011, 10, M110.001784. [Google Scholar] [CrossRef]
- Janer, A.; Warsinggih, W.; Uwuratuw, J.A.; Ladju, R.B.; Sampetoding, S.; Syarifuddin, E. Accuracy of Faecal Matrix-Metalloproteinase-9 (MMP-9) for Colorectal Cancer Detection in Makassar, Indonesia. Asian Pac. J. Cancer Prev. 2025, 26, 603. [Google Scholar] [CrossRef] [PubMed]
- Yunussova, N.; Sypabekova, M.; Zhumabekova, Z.; Matkarimov, B.; Kanayeva, D. A Novel ssDNA Aptamer Targeting Carcinoembryonic Antigen: Selection and Characterization. Biology 2022, 11, 1540. [Google Scholar] [CrossRef]
- Pan, Q.; Law, C.O.K.; Yung, M.M.H.; Han, K.C.; Pon, Y.L.; Lau, T.C.K. Novel RNA aptamers targeting gastrointestinal cancer biomarkers CEA, CA50 and CA72-4 with superior affinity and specificity. PLoS ONE 2018, 13, e0198980. [Google Scholar] [CrossRef]
- Gu, L.; Yan, W.; Liu, S.; Ren, W.; Lyu, M.; Wang, S. Trypsin enhances aptamer screening: A novel method for targeting proteins. Anal. Biochem. 2018, 561–562, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Manea, I.; Casian, M.; Hosu-Stancioiu, O.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Cristea, C. A review on magnetic beads-based SELEX technologies: Applications from small to large target molecules. Anal. Chim. Acta 2024, 1297, 342325. [Google Scholar] [CrossRef]
- Wang, Q.; Li, Y.; Yao, L.; Li, H.; Zhang, L.; Wang, Y.; Li, J.; Chen, T.; Chai, K.; Gao, J.; et al. High-affinity ssDNA aptamer and chemiluminescent aptasensor for TIMP-1 detection in human serum. Anal. Sci. 2025, 41, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, H.; Kataoka, Y.; Fujita, H.; Kuwahara, M.; Horii, K.; Shiratori, I.; Waga, I. Modified DNA Aptamers for C-Reactive Protein and Lactate Dehydrogenase-5 with Sub-Nanomolar Affinities. Int. J. Mol. Sci. 2020, 21, 2683. [Google Scholar] [CrossRef] [PubMed]
- Mashayekhi, K.; Ganji, A.; Sankian, M. Designing a new dimerized anti human TNF-α aptamer with blocking activity. Biotechnol. Prog. 2020, 36, e2969. [Google Scholar] [CrossRef]
- Tsao, S.M.; Lai, J.C.; Horng, H.E.; Liu, T.C.; Hong, C.Y. Generation of aptamers from A primer-free randomized ssDNA library using magnetic-assisted rapid aptamer selection. Sci. Rep. 2017, 7, 45478. [Google Scholar] [CrossRef]
- Nagano, M.; Oguro, T.; Sawada, R.; Yoshitomi, T.; Yoshimoto, K. Accelerated Discovery of Potent Bioactive anti-TNFα Aptamers by Microbead-Assisted Capillary Electrophoresis (MACE)-SELEX. ChemBioChem 2021, 22, 3341–3347. [Google Scholar] [CrossRef]
- Huang, C.J.; Lin, H.I.; Shiesh, S.C.; Lee, G. Bin Integrated microfluidic system for rapid screening of CRP aptamers utilizing systematic evolution of ligands by exponential enrichment (SELEX). Biosens. Bioelectron. 2010, 25, 1761–1766. [Google Scholar] [CrossRef]
- Gupta, S.; Hirota, M.; Waugh, S.M.; Murakami, I.; Suzuki, T.; Muraguchi, M.; Shibamori, M.; Ishikawa, Y.; Jarvis, T.C.; Carter, J.D.; et al. Chemically Modified DNA Aptamers Bind Interleukin-6 with High Affinity and Inhibit Signaling by Blocking Its Interaction with Interleukin-6 Receptor. J. Biol. Chem. 2014, 289, 8706–8719. [Google Scholar] [CrossRef]
- Wang, Q.L.; Cui, H.F.; Du, J.F.; Lv, Q.Y.; Song, X. In silico post-SELEX screening and experimental characterizations for acquisition of high affinity DNA aptamers against carcinoembryonic antigen. RSC Adv. 2019, 9, 6328–6334. [Google Scholar] [CrossRef]
- Zhang, W.; Xiao, G.; Chen, J.; Wang, L.; Hu, Q.; Wu, J.; Zhang, W.; Song, M.; Qiao, J.; Xu, C. Electrochemical biosensors for measurement of colorectal cancer biomarkers. Anal. Bioanal. Chem. 2021, 413, 2407–2428. [Google Scholar] [CrossRef]
- Scarano, S.; Dausse, E.; Crispo, F.; Toulmé, J.J.; Minunni, M. Design of a dual aptamer-based recognition strategy for human matrix metalloproteinase 9 protein by piezoelectric biosensors. Anal. Chim. Acta 2015, 897, 1–9. [Google Scholar] [CrossRef]
- Li, J.; Xu, L.; Shen, Y.; Guo, L.; Yin, H.; Fang, X.; Yang, Z.; Xu, Q.; Li, H. Superparamagnetic Nanostructures for Split-Type and Competitive-Mode Photoelectrochemical Aptasensing. Anal. Chem. 2020, 92, 8607–8613. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Wang, R.; Li, D.A.; Li, X.; Wang, Y.; Hua, Y.; Wang, R.; Li, D. A Fiber-Based SPR Aptasensor for the In Vitro Detection of Inflammation Biomarkers. Micromachines 2022, 13, 1036. [Google Scholar] [CrossRef]
- Li, M.J.; Wang, H.J.; Yuan, R.; Chai, Y.Q. A sensitive label-free photoelectrochemical aptasensor based on a novel PTB7-Th/H2O2 system with unexpected photoelectric performance for C-reactive protein analysis. Biosens. Bioelectron. 2021, 181, 113162. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, M.; Yang, Y.; Tang, Q.; Zeng, Y.; Wang, H.; Zhang, L.; Pan, C.; Hu, C.; Fu, Z.; et al. Detecting carcinoembryonic antigen based on the aggregation-induced emission enhancement effect. Chem. Commun. 2024, 60, 13570–13573. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Duan, W.; Deng, F.; Tang, W.; Payne, S.C.; Guo, T.; Goldys, E.M.; Lovell, N.H.; Shivdasani, M.N. Towards an Implantable Aptamer Biosensor for Monitoring in Inflammatory Bowel Disease. Biosensors 2025, 15, 546. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhou, Q.; Revzin, A. An aptasensor for electrochemical detection of tumor necrosis factor in human blood. Analyst 2013, 138, 4321–4326. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, W.L.; Lo, L.H.Y.; Kinghorn, A.B.; Shiu, S.C.C.; Tanner, J.A. Structure-Switching Electrochemical Aptasensor for Rapid, Reagentless, and Single-Step Nanomolar Detection of C-Reactive Protein. ACS Appl. Bio Mater. 2024, 7, 3721–3730. [Google Scholar] [CrossRef] [PubMed]
- Jarczewska, M.; Rębiś, J.; Górski, Ł.; Malinowska, E. Development of DNA aptamer-based sensor for electrochemical detection of C-reactive protein. Talanta 2018, 189, 45–54. [Google Scholar] [CrossRef]
- Beduk, D.; Beduk, T.; Ait Lahcen, A.; Mani, V.; Guler Celik, E.; Iskenderoglu, G.; Demirci, F.; Turhan, S.; Ozdogan, O.; Ozgur, S.; et al. Multiplexed aptasensor for detection of acute myocardial infraction (AMI) biomarkers. Sens. Diagn. 2024, 3, 1020–1027. [Google Scholar] [CrossRef]
- Zhai, J.; Ji, P.; Xin, Y.; Liu, Y.; Qu, Q.; Han, W.; Zhao, G. Development of Carcinoembryonic Antigen Rapid Detection System Based on Platinum Microelectrode. Front. Chem. 2022, 10, 899276. [Google Scholar] [CrossRef]
- Yunussova, N.; Tilegen, M.; Pham, T.T.; Kanayeva, D. Rapid detection of carcinoembryonic antigen by means of an electrochemical aptasensor. iScience 2024, 27, 109637. [Google Scholar] [CrossRef]
- Sánchez-Salcedo, R.; Miranda-Castro, R.; De-los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Corrigan, D.K. Comparing nanobody and aptamer-based capacitive sensing for detection of interleukin-6 (IL-6) at physiologically relevant levels. Anal. Bioanal. Chem. 2023, 415, 7035–7045. [Google Scholar] [CrossRef]
- Chen, N.; Yang, H.; Li, Q.; Song, L.; Gopinath, S.C.B.; Wu, D. An interdigitated aptasensor to detect interleukin-6 for diagnosing rheumatoid arthritis in serum. Biotechnol. Appl. Biochem. 2021, 68, 1479–1485. [Google Scholar] [CrossRef] [PubMed]
- Kumalasari, M.R.; Alfanaar, R.; Andreani, A.S. Gold nanoparticles (AuNPs): A versatile material for biosensor application. Talanta Open 2024, 9, 100327. [Google Scholar] [CrossRef]
- Irimes, M.B.; Pusta, A.; Coker, D.; Martian, P.C.; Suciu, M.; Pandrea, S.L.; Tertis, M.; Cristea, C.; Oprean, R. Dual-target electrochemical aptasensor for the simultaneous detection of interleukin-6 and tumor necrosis factor-α in biological fluids. Sens. Bio-Sens. Res. 2025, 49, 100866. [Google Scholar] [CrossRef]
- Hosseini Ghalehno, M.; Mirzaei, M.; Torkzadeh-Mahani, M. Electrochemical aptasensor for tumor necrosis factor α using aptamer–antibody sandwich structure and cobalt hexacyanoferrate for signal amplification. J. Iran. Chem. Soc. 2019, 16, 1783–1791. [Google Scholar] [CrossRef]
- Miao, P.; Yang, D.; Chen, X.; Guo, Z.; Tang, Y. Voltammetric determination of tumor necrosis factor-α based on the use of an aptamer and magnetic nanoparticles loaded with gold nanoparticles. Microchim. Acta 2017, 184, 3901–3907. [Google Scholar] [CrossRef]
- Diao, W.; Zhou, C.; Zhang, Z.; Cao, Y.; Li, Y.; Tang, J.; Liu, G. EGaIn-Modified ePADs for Simultaneous Detection of Homocysteine and C-Reactive Protein in Saliva toward Early Diagnosis of Cardiovascular Disease. ACS Sens. 2024, 9, 4265–4276. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, L.; Xuan, T.; Wang, J.; Wang, X. Label-free Electrochemical Impedance Spectroscopy Aptasensor for Ultrasensitive Detection of Lung Cancer Biomarker Carcinoembryonic Antigen. Front. Chem. 2021, 9, 721008. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Wang, X.; Li, M.; Qi, H.; Gao, Q.; Zhang, C. Ultrasensitive Electrochemiluminescence Aptasensor for Assessment of Protein Heterogeneity in Small Cell Population. ACS Appl. Bio Mater. 2019, 2, 3052–3058. [Google Scholar] [CrossRef]
- Erkal-Aytemur, A.; Mülazımoğlu, İ.E.; Üstündağ, Z.; Caglayan, M.O. A novel aptasensor platform for the detection of carcinoembryonic antigen using quartz crystal microbalance. Talanta 2024, 277, 126376. [Google Scholar] [CrossRef]
- Lu, T.; Wang, L.; Xia, Y.; Jin, Y.; Zhang, L.; Du, S. A multimer-based SERS aptasensor for highly sensitive and homogeneous assay of carcinoembryonic antigens. Analyst 2021, 146, 3016–3024. [Google Scholar] [CrossRef]
- Villalonga, A.; Sánchez, A.; Vilela, D.; Mayol, B.; Martínez-Ruíz, P.; Villalonga, R. Electrochemical aptasensor based on anisotropically modified (Janus-type) gold nanoparticles for determination of C-reactive protein. Microchim. Acta 2022, 189, 309. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.; Datta, D.; Chaudhry, S.; Dutta, M.; Stroscio, M.A. Rapid Detection of Tumor Necrosis Factor-Alpha Using Quantum Dot-Based Optical Aptasensor. IEEE Trans. Nanobiosci. 2018, 17, 417–423. [Google Scholar] [CrossRef]
- Xu, X.; Ji, J.; Chen, P.; Wu, J.; Jin, Y.; Zhang, L.; Du, S. Salt-induced gold nanoparticles aggregation lights up fluorescence of DNA-silver nanoclusters to monitor dual cancer markers carcinoembryonic antigen and carbohydrate antigen 125. Anal. Chim. Acta 2020, 1125, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Rana, M.S.; Meng, F.; Zhang, R.; Huang, L.; Qian, K. Integrating zirconia-gold hybrid into aptasensor capable of ultrasensitive and robust carcinoembryonic antigen determination. Electrochim. Acta 2024, 507, 145063. [Google Scholar] [CrossRef]
- Li, H.; Zhang, Y.; Chang, A.; Cai, S.; Zhou, N. Amperometric sandwich-type aptasensor based on peroxidase mimetic Au@Pt for the carcinoembryonic antigen. Electroanalysis 2023, 35, e202200443. [Google Scholar] [CrossRef]
- Jin, J.; Guo, J.; Guo, J.; Li, D. Carbon-Based Biosensor in Point of Care Setting. Adv. Sens. Res. 2024, 3, 2400037. [Google Scholar] [CrossRef]
- Mahani, M.; Faghihi-Fard, M.; Divsar, F.; Torkzadeh-Mahani, M.; Khakbaz, F. Ultrasensitive FRET-based aptasensor for interleukin-6 as a biomarker for COVID-19 progression using nitrogen-doped carbon quantum dots and gold nanoparticles. Microchim. Acta 2022, 189, 472. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, X.; Zhang, C.; Meng, H.; Liu, B.; Wei, X. A rapid fluorescent aptasensor for point-of-care detection of C-reactive protein. Talanta 2022, 249, 123661. [Google Scholar] [CrossRef]
- Ansari, M.A.; Mohd-Naim, N.F.; Ahmed, M.U. Electrochemical Nanoaptasensor Based on Graphitic Carbon Nitride/Zirconium Dioxide/Multiwalled Carbon Nanotubes for Matrix Metalloproteinase-9 in Human Serum and Saliva. ACS Appl. Bio Mater. 2024, 7, 1579–1587. [Google Scholar] [CrossRef]
- Li, Y.; Hua, X.; Wang, J.; Jin, B. cMWCNT/CoHCF/AuNPs nanocomposites aptasensor for electrochemical detection of interleukin-6. Talanta Open 2023, 7, 100188. [Google Scholar] [CrossRef]
- Hou, Y.; Wang, J.; Liu, S.; Sun, Y.; Dai, Y.; Luo, C.; Wang, X. A novel flower-shaped Ag@ZIF-67 chemiluminescence sensor for sensitive detection of CEA. Talanta 2023, 253, 123938. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, H.; Xu, J.; Zhao, X.; Song, S.; Zhang, H. Myocardial infarction biomarker C-reactive protein detection on nanocomposite aptasensor. Biotechnol. Appl. Biochem. 2022, 69, 166–171. [Google Scholar] [CrossRef]
- Hoseynidokht, F.; Mazloum-Ardakani, M.; Sahraei, N.; Hakimian, F.; Behnam Rad, M. Highly sensitive electrochemical detection of carcinoembryonic antigen by silver nanoparticles/carboxylated single-walled carbon nanotubes embedded in mesoporous carbon foam. J. Solid State Electrochem. 2024, 28, 377–387. [Google Scholar] [CrossRef]
- Mishyn, V.; Aslan, M.; Hugo, A.; Rodrigues, T.; Happy, H.; Sanyal, R.; Knoll, W.; Baudoux, F.; Bouchiat, V.; Bilyy, R.O.; et al. Catch and release strategy of matrix metalloprotease aptamers via thiol–disulfide exchange reaction on a graphene based electrochemical sensor. Sens. Diagn. 2022, 1, 739–749. [Google Scholar] [CrossRef]
- Shen, Z.; Ni, S.; Yang, W.; Sun, W.; Yang, G.; Liu, G. Redox probes tagged electrochemical aptasensing device for simultaneous detection of multiple cytokines in real time. Sens. Actuators B Chem. 2021, 336, 129747. [Google Scholar] [CrossRef]
- Ma, R.; Gopinath, S.C.B.; Lakshmipriya, T.; Chen, Y. Carbon Material Hybrid Construction on an Aptasensor for Monitoring Surgical Tumors. J. Anal. Methods Chem. 2022, 2022, 9740784. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.K.; Chao, C.H.; Yeh, Y.S. A Graphene-PEDOT:PSS Modified Paper-Based Aptasensor for Electrochemical Impedance Spectroscopy Detection of Tumor Marker. Sensors 2020, 20, 1372. [Google Scholar] [CrossRef]
- Mazloum-Ardakani, M.; Hosseinzadeh, L. A Sensitive Electrochemical Aptasensor for TNF-α Based on Bimetallic Ag@Pt Core-Shell Nanoparticle Functionalized Graphene Nanostructures as Labels for Signal Amplification. J. Electrochem. Soc. 2016, 163, B119. [Google Scholar] [CrossRef]
- Mahyari, M.; Hooshmand, S.E.; Sepahvand, H.; Gholami, S.; Rezayan, A.H.; Zarei, M.A. Gold nanoparticles anchored onto covalent poly deep eutectic solvent functionalized graphene: An electrochemical aptasensor for the detection of C-reactive protein. Mater. Chem. Phys. 2021, 269, 124730. [Google Scholar] [CrossRef]
- Villalonga, A.; Vegas, B.; Paniagua, G.; Eguílaz, M.; Mayol, B.; Parrado, C.; Rivas, G.; Díez, P.; Villalonga, R. Amperometric aptasensor for carcinoembryonic antigen based on a reduced graphene oxide/gold nanoparticles modified electrode. J. Electroanal. Chem. 2020, 877, 114511. [Google Scholar] [CrossRef]
- Yan, M.; Fu, L.L.; Feng, H.C.; Namadchian, M. Application of Ag nanoparticles decorated on graphene nanosheets for electrochemical sensing of CEA as an important cancer biomarker. Environ. Res. 2023, 239, 117363. [Google Scholar] [CrossRef]
- Agrahari, S.; Kumar Singh, A.; Tiwari, I.; Srikrishna, S. Label free and ultrasensitive electrochemical detection of carcinoembryonic antigen using a disposable aptasensor based on Pd and graphene. Microchem. J. 2024, 205, 111359. [Google Scholar] [CrossRef]
- Xu, L.; Zou, L.; Guo, J.; Cao, Y.; Feng, C.; Ye, B. Simple “Signal-Off” Electrochemical Aptasensor Based on Aptamer-Cu3(PO4)2 Hybrid Nanoflowers/Graphene Oxide for Carcinoembryonic Antigen Detection. ChemElectroChem 2020, 7, 1660–1665. [Google Scholar] [CrossRef]
- Lin, F.; Li, X.; Zhang, J.; Zhang, H.; Zhang, Z.; Hou, L.; Shen, B.; Jin, L. Ratiometric electrochemical aptasensor based on functionalized graphene nanocomposites for detection of CA19-9. Anal. Sci. 2025, 41, 825–838. [Google Scholar] [CrossRef]
- Li, J.; Zhao, L.; Wang, W.; Liu, Y.; Yang, H.; Kong, J.; Si, F. Polymer-functionalized carbon nanotubes prepared via ring-opening polymerization for electrochemical detection of carcinoembryonic antigen. Sens. Actuators B Chem. 2021, 328, 129031. [Google Scholar] [CrossRef]
- Tabrizi, M.A.; Acedo, P. Highly Sensitive RNA-Based Electrochemical Aptasensor for the Determination of C-Reactive Protein Using Carbon Nanofiber-Chitosan Modified Screen-Printed Electrode. Nanomaterials 2022, 12, 415. [Google Scholar] [CrossRef]
- Rahmati, Z.; Roushani, M.; Hosseini, H. Thionine functionalized hollow N-doped carbon nanoboxes: As a high-performance substrate for fabrication of label-free electrochemical aptasensor toward ultrasensitive detection of carcinoembryonic antigen. J. Electroanal. Chem. 2021, 903, 115858. [Google Scholar] [CrossRef]
- Mazloum-Ardakani, M.; Hosseinzadeh, L.; Taleat, Z. Synthesis and electrocatalytic effect of Ag@Pt core–shell nanoparticles supported on reduced graphene oxide for sensitive and simple label-free electrochemical aptasensor. Biosens. Bioelectron. 2015, 74, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Moharramnejad, M.; Karim, A.; Gharanli, S.; Malekshah, R.E.; Amini, A.H.; Sharifi, M.S.; Basmenj, Z.S.; Salariyeh, Z.; Mohammadkhani, M.; Shahi, M.; et al. A comprehensive review of MOFs based on electrochemical biosensors as smart platforms in cancer biomarkers detection. Microchem. J. 2025, 208, 112498. [Google Scholar] [CrossRef]
- Wang, Z.; Guo, L.; Tian, J.; Han, Y.; Zhai, D.; Cui, L.; Zhang, P.; Zhang, X.; Yang, S.; Zhang, L. Aversatile MOF as an electrochemical/fluorescence/colorimetric signal probe for the tri-modal detection of MMP-9 secretion in the extracellular matrix to identify the efficacy of chemotherapeutic drugs. Anal. Chim. Acta 2024, 1315, 342798. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Huang, S.; Yan, Y.; Pang, W.; Zhong, F.; Huang, Q.; Caddeo, F.; Zhang, M.; Jin, M.; Shui, L. Multiplex signal amplification for ultrasensitive CRP assay via integrated electrochemical biosensor array using MOF-derived carbon material and aptamers. Talanta 2024, 272, 125735. [Google Scholar] [CrossRef]
- Ali, G.K.; Omer, K.M. Ultrasensitive aptamer-functionalized Cu-MOF fluorescent nanozyme as an optical biosensor for detection of C-reactive protein. Anal. Biochem. 2022, 658, 114928. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Chen, K.; Li, S.; He, L.; Wang, M.; Zhou, N.; Du, M. Impedimetric aptasensor based on zirconium–cobalt metal–organic framework for detection of carcinoembryonic antigen. Microchim. Acta 2022, 189, 338. [Google Scholar] [CrossRef]
- Huang, S.; Liu, Z.; Yan, Y.; Chen, J.; Yang, R.; Huang, Q.; Jin, M.; Shui, L. Triple signal-enhancing electrochemical aptasensor based on rhomboid dodecahedra carbonized-ZIF67 for ultrasensitive CRP detection. Biosens. Bioelectron. 2022, 207, 114129. [Google Scholar] [CrossRef]
- Chandio, I.; Cheung, S.; Ai, Y.; Liang, Q. High-entropy alloy nanoparticles integrated with metal-organic framework for enhanced aptasensing of carcinoembryonic antigen. Microchem. J. 2025, 209, 112760. [Google Scholar] [CrossRef]
- He, P.; Zhang, Q.; Liu, Q. Impedimetric aptasensor based on MOF based composite for measuring of carcinoembryonic antigen as a tumor biomarker. Chemosphere 2023, 338, 139339. [Google Scholar] [CrossRef]
- Li, J.; Liu, L.; Ai, Y.; Liu, Y.; Sun, H.; Liang, Q. Self-Polymerized Dopamine-Decorated Au NPs and Coordinated with Fe-MOF as a Dual Binding Sites and Dual Signal-Amplifying Electrochemical Aptasensor for the Detection of CEA. ACS Appl. Mater. Interfaces 2020, 12, 5500–5510. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.H.; Qiao, X.; Fan, J.; Hao, Y.Q.; Zhang, Y.T.; Zhou, Y.L.; Xu, M.T. A label-free ECL aptasensor for sensitive detection of carcinoembryonic antigen based on CdS QDs@MOF and TEOA@Au as bi-coreactants of Ru(bpy)32+. Microchem. J. 2022, 173, 106910. [Google Scholar] [CrossRef]
- Chen, S.; Yan, H.; Tang, Q.; Gu, Y.; Zhang, J.; Yang, Y.; Wang, H.; Qian, Z.; Li, L.; Guo, L.; et al. A novel FAM-based fluorescent aptasensor with covalent organic framework as quencher for sensitive determination of carcinoembryonic antigen. Microchem. J. 2025, 215, 114197. [Google Scholar] [CrossRef]
- Zhang, X.; Chi, K.N.; Li, D.L.; Deng, Y.; Ma, Y.C.; Xu, Q.Q.; Hu, R.; Yang, Y.H. 2D-porphrinic covalent organic framework-based aptasensor with enhanced photoelectrochemical response for the detection of C-reactive protein. Biosens. Bioelectron. 2019, 129, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Han, Q.; Xi, F. The Fabrication of a Probe-Integrated Electrochemiluminescence Aptasensor Based on Double-Layered Nanochannel Array with Opposite Charges for the Sensitive Determination of C-Reactive Protein. Molecules 2023, 28, 7867. [Google Scholar] [CrossRef]
- Ma, N.; Xu, S.; Wu, W.; Liu, J. Electrochemiluminescence Aptasensor with Dual Signal Amplification by Silica Nanochannel-Based Confinement Effect on Nanocatalyst and Efficient Emitter Enrichment for Highly Sensitive Detection of C-Reactive Protein. Molecules 2023, 28, 7664. [Google Scholar] [CrossRef]
- Zhao, J.; Gu, X.; Liu, J. Boosted electrochemiluminescence of luminol/oxygen by nanochannel-confined nanocatalyst for sensitive aptasensing of tumor biomarker. Microchem. J. 2025, 216, 114671. [Google Scholar] [CrossRef]
- An, J.; Zhang, C.; Yan, F.; Ma, P. Nanochannel-confined platinum nanostructure for enhancement of luminol-dissolved oxygen electrochemiluminescence coupled with gated aptasensor for sensitive detection of carcinoembryonic antigen. Microchem. J. 2024, 206, 111413. [Google Scholar] [CrossRef]
- Shi, Z.; Li, K.; Wang, Y.; Li, Z.; Zhu, Z.; Wang, L. Development of a dual-channel electrochemical aptasensor for simultaneous detection of CRP and IL-6 in sepsis diagnosis. Electrochim. Acta 2025, 539, 147110. [Google Scholar] [CrossRef]
- Shi, Z.; Li, K.; Wang, Y.; Hu, Y.; Li, Z.; Zhu, Z. An innovative label-free electrochemical aptamer sensor: Utilizing Ti3C2Tx/MoS2/Au NPs for accurate interleukin-6 detection. Talanta 2024, 276, 126281. [Google Scholar] [CrossRef]
- Li, K.; Shi, Z.; Wang, Y.; Yan, F.; Li, Z.; Wang, Z.; Zhu, Z. A label-free electrochemical aptasensor based on Ti3C2Tx-Ag/Au nanoparticles as a signal amplification strategy for CRP detection. Microchem. J. 2023, 195, 109479. [Google Scholar] [CrossRef]
- Shi, S.S.; Li, X.J.; Ma, R.N.; Shang, L.; Zhang, W.; Zhao, H.Q.; Jia, L.P.; Wang, H.S. A novel dual-signal output strategy for POCT of CEA based on a smartphone electrochemical aptasensing platform. Microchim. Acta 2024, 191, 407. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Fan, B.; Chang, S.; Guo, D.; Wang, F.; Pan, Q. An ultrasensitive photoelectrochemical assay for tumor necrosis factor-alpha based on hollow CdS cubes as a signal generator and NiCo2O4-Au as a signal extinguisher. Analyst 2023, 148, 4746–4752. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Sun, X.; Li, Y.; Zeng, Y.; Gong, J.; Wang, Z.; An, Y.; Li, H. Biomineralized Mn3(PO4)2/aptamer nanosheets for enhanced electrochemical determination of C-reactive protein. Sens. Actuators B Chem. 2021, 333, 129510. [Google Scholar] [CrossRef]
- Oranzie, M.; January, J.L.; Sanga, N.A.; Leve, Z.D.; Mini, S.; Cupido, C.; Douman, S.F.; Iwuoha, E.I. Aptamer-Driven Biosensor Technology for the Quantitative Analysis of C-Reactive Protein. ChemElectroChem 2025, 12, e202400667. [Google Scholar] [CrossRef]
- Chen, Q.; Liu, Z.; Luo, D.; Ren, F.; Ran, F.; Chen, W.; Zhang, B.; Wang, C.; Chen, H.; Wei, J. Ultrasensitive fluorescent aptasensor for CRP detection based on the RNase H assisted DNA recycling signal amplification strategy. RSC Adv. 2019, 9, 11960–11967. [Google Scholar] [CrossRef]
- Liu, M.; Liu, L.; Du, M.; Li, Q.; Wu, S.; Su, S.; Jian, N.; Wu, Y.; Wang, Y. A fluorescent platform integrated with a “one-pot” nicking endonuclease signal amplification and magnetic separation for simultaneous detection of tumor markers. Talanta 2025, 282, 127011. [Google Scholar] [CrossRef]
- Zhang, S.; Li, Z.; Wang, Y.; Guo, C.; Guo, R.R.; Guo, Y.; Zhang, Z. Development of an electrochemiluminescence aptasensor combining covalent-triazine framework emitter with exonuclease III-driven DNA walker for sensitive CEA detection. Microchem. J. 2025, 215, 114388. [Google Scholar] [CrossRef]
- Wang, P.; Xie, Y.; Ma, H.; Liu, J.; Liu, C.; Feng, W.; Xi, S. A novel ratiometric electrochemical aptasensor for highly sensitive detection of carcinoembryonic antigen. Anal. Biochem. 2022, 659, 114957. [Google Scholar] [CrossRef] [PubMed]
- Niu, C.; Lin, X.; Jiang, X.; Guo, F.; Liu, J.; Liu, X.; Huang, H.; Huang, Y. An electrochemical aptasensor for highly sensitive detection of CEA based on exonuclease III and hybrid chain reaction dual signal amplification. Bioelectrochemistry 2022, 143, 107986. [Google Scholar] [CrossRef]
- Ji, Y.; Guo, J.; Ye, B.; Peng, G.; Zhang, C.; Zou, L. An ultrasensitive carcinoembryonic antigen electrochemical aptasensor based on 3D DNA nanoprobe and Exo III. Biosens. Bioelectron. 2022, 196, 113741. [Google Scholar] [CrossRef]
- Shang, L.; Shi, B.J.; Zhang, W.; Jia, L.P.; Ma, R.N.; Xue, Q.W.; Wang, H.S.; Yan, W. Electrochemical stripping chemiluminescence coupled with recycling amplification strategy for sensitive detection of carcinoembryonic antigen. Sens. Actuators B Chem. 2022, 368, 132191. [Google Scholar] [CrossRef]
- Ma, H.; Wang, P.; Xie, Y.; Liu, J.; Feng, W.; Li, S. Ratiometric electrochemical aptasensor for the sensitive detection of carcinoembryonic antigen based on a hairpin DNA probe and exonuclease I-assisted target recycling. Anal. Biochem. 2022, 649, 114694. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ma, H.; Zhu, Y.; Feng, W.; Su, M.; Li, S.; Mao, H. A new ratiometric electrochemical aptasensor for the sensitive detection of carcinoembryonic antigen based on exonuclease I-assisted target recycling and hybridization chain reaction amplification strategy. J. Electroanal. Chem. 2022, 910, 116167. [Google Scholar] [CrossRef]
- Huang, S.; Wang, Y.; Liu, S.; Li, H.; Yang, M.; Fang, Y.; Xiao, Q. Triblock polyadenine-based electrochemical aptasensor for ultra-sensitive detection of carcinoembryonic antigen via exonuclease III-assisted target recycling and hybridization chain reaction. Bioelectrochemistry 2024, 159, 108749. [Google Scholar] [CrossRef]
- Díaz-Fernández, A.; Ferapontova, E.E. Covalent Hemin/G4 complex-linked sandwich bioassay on magnetic beads for femtomolar HER-2/neu detection in human serum via direct electrocatalytic reduction of oxygen. Anal. Chim. Acta 2022, 1219, 340049. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lu, W.; Che, S.; Zhang, X.; Yuan, X. Self-powered dual-photoelectrode photoelectrochemical aptasensor amplified by hemin/G-quadruplex-based DNAzyme. Microchim. Acta 2025, 192, 74. [Google Scholar] [CrossRef]
- Zhai, X.J.; Wang, Q.L.; Cui, H.F.; Song, X.; Lv, Q.Y.; Guo, Y. A DNAzyme-catalyzed label-free aptasensor based on multifunctional dendrimer-like DNA assembly for sensitive detection of carcinoembryonic antigen. Biosens. Bioelectron. 2021, 194, 113618. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, H.; Guo, Z.; Wang, X.; Zhou, N. Immobilization-free electrochemical homogeneous aptasensor for highly sensitive detection of carcinoembryonic antigen by dual amplification strategy. Anal. Chim. Acta 2023, 1274, 341586. [Google Scholar] [CrossRef]
- Gao, Z.; Shang, L.; Zhang, W.; Jia, L.P.; Ma, R.N.; Li, X.J.; Wang, H.S. Simplified ratiometric electrochemiluminescence aptasensor for carcinoembryonic antigen detection based on unmodified aptamer using single luminophore and coreactant. Microchim. Acta 2025, 192, 566. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Weng, C.; Wang, J.; Yang, W.; Lu, Q.; Yan, X.; Sakran, M.A.; Hong, J.; Zhu, W.; Zhou, X. A label-free electrochemical magnetic aptasensor based on exonuclease III–assisted signal amplification for determination of carcinoembryonic antigen. Microchim. Acta 2020, 187, 492. [Google Scholar] [CrossRef]
- Bai, Y.; Zhang, H.; Zhao, L.; Wang, Y.; Chen, X.; Zhai, H.; Tian, M.; Zhao, R.; Wang, T.; Xu, H.; et al. A novel aptasensor based on HCR and G-quadruplex DNAzyme for fluorescence detection of Carcinoembryonic Antigen. Talanta 2021, 221, 121451. [Google Scholar] [CrossRef]
- Díaz-Fernández, A.; Lorenzo-Gómez, R.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Electrochemical aptasensors for cancer diagnosis in biological fluids—A review. Anal. Chim. Acta 2020, 1124, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.M.; Lv, S.; Zhang, W.; Cui, Y. Microfluidic Point-of-Care (POC) Devices in Early Diagnosis: A Review of Opportunities and Challenges. Sensors 2022, 22, 1620. [Google Scholar] [CrossRef]
- Zhao, P.; Zheng, J.; Liang, Y.; Tian, F.; Peng, L.; Huo, D.; Hou, C. Functionalized Carbon Nanotube-Decorated MXene Nanosheet-Enabled Microfluidic Electrochemical Aptasensor for Carcinoembryonic Antigen Determination. ACS Sustain. Chem. Eng. 2021, 9, 15386–15393. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, Y.; Matharu, Z.; Rahimian, A.; Revzin, A. Detecting multiple cell-secreted cytokines from the same aptamer-functionalized electrode. Biosens. Bioelectron. 2015, 64, 43–50. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, M.; Jiang, F.; Lu, C.; Zhu, Q.; Yang, Y.; Fu, L.; Li, L.; Liu, J.; Wang, Z.; et al. Microfluidic-SERS sensing system based on dual signal amplification and aptamer for gastric cancer detection. Microchim. Acta 2024, 191, 668. [Google Scholar] [CrossRef]
- Malekmohamadi, M.; Mirzaei, S.; Rezayan, A.H.; Abbasi, V.; Abouei Mehrizi, A. µPAD-based colorimetric nanogold aptasensor for CRP and IL-6 detection as sepsis biomarkers. Microchem. J. 2024, 197, 109744. [Google Scholar] [CrossRef]
- Conejo-Cuevas, G.; Pellitero, M.A.; Ruiz-Rubio, L.; Javier, F.; Campo, D. Microneedles for Continuous, Minimally Invasive Monitoring: A Technology Overview. Adv. Sens. Res. 2025, 4, 2500057. [Google Scholar] [CrossRef]
- Yuan, R.; Cai, J.; Li, J.; Xu, Y.; Ma, J.; Wang, L.; Su, S. Integrated Microneedle Aptasensing Platform toward Point-of-Care Monitoring of Bacterial Infections and Treatment. ACS Sens. 2025, 10, 5684–5693. [Google Scholar] [CrossRef] [PubMed]
- Overview of IVD Regulation|FDA. Available online: https://www.fda.gov/medical-devices/ivd-regulatory-assistance/overview-ivd-regulation (accessed on 25 September 2025).
- 510(k) Third Party Review Program|FDA. Available online: https://www.fda.gov/medical-devices/premarket-submissions-selecting-and-preparing-correct-submission/510k-third-party-review-program (accessed on 25 September 2025).
- Regulation—2017/746—EN—Medical Device Regulation—EUR-Lex. Available online: https://eur-lex.europa.eu/eli/reg/2017/746/oj/eng (accessed on 25 September 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).