CRISPR-Cas12a and DNA Tetrahedron Assemblies Amplified Fluorescence Anisotropy for the Sensitive Detection of Hepatitis B Virus DNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Apparatus
2.2. FA Analysis
2.3. Polyacrylamide Gel Electrophoresis (PAGE)
2.4. Synthesis of DNA Cross Scaffolds
2.5. Steps for Testing HBV-DNA
2.6. Serum Sample Analysis
3. Results
3.1. Principles of HBV-DNA Detection
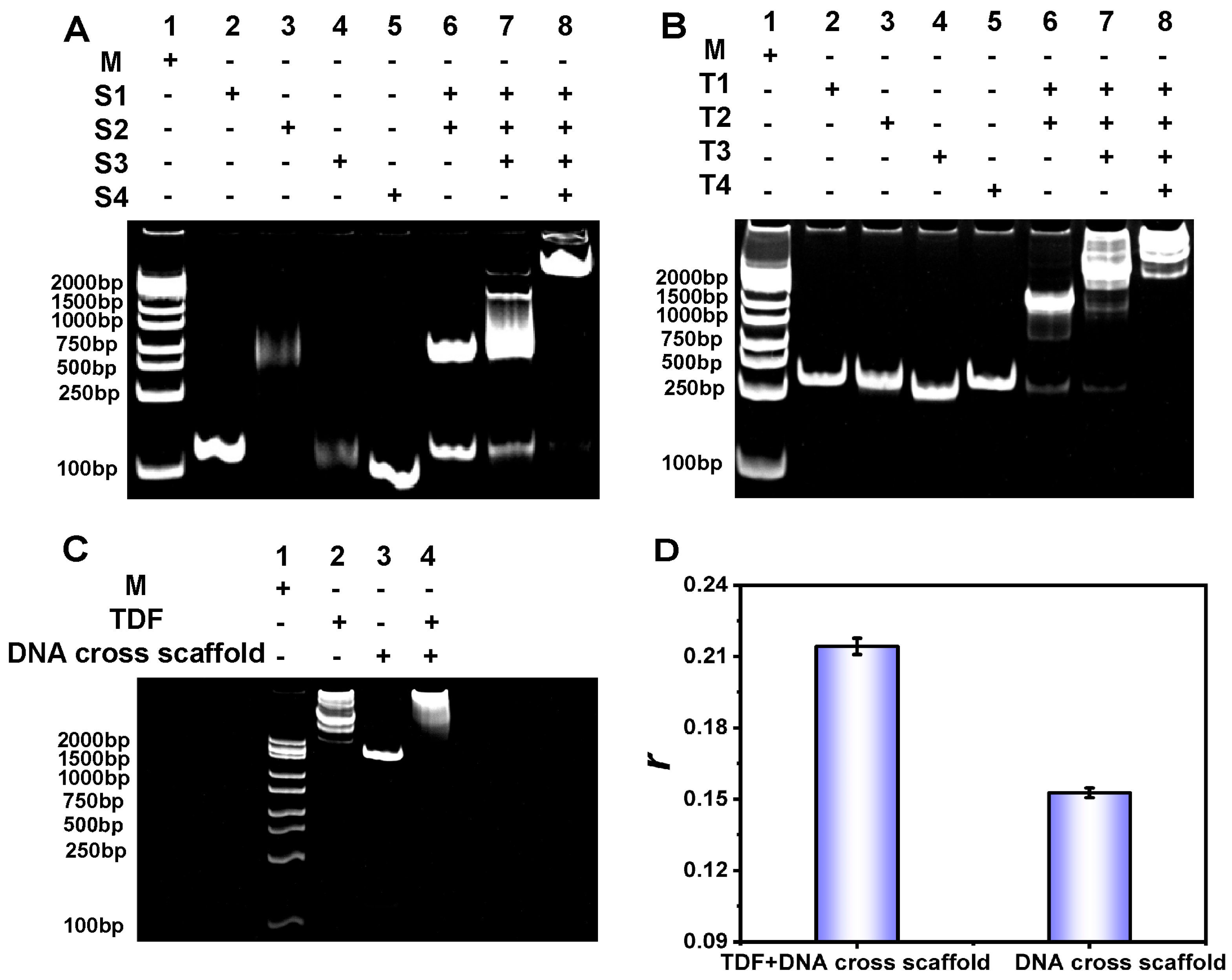
3.2. Characterization of DNA Nanostructures
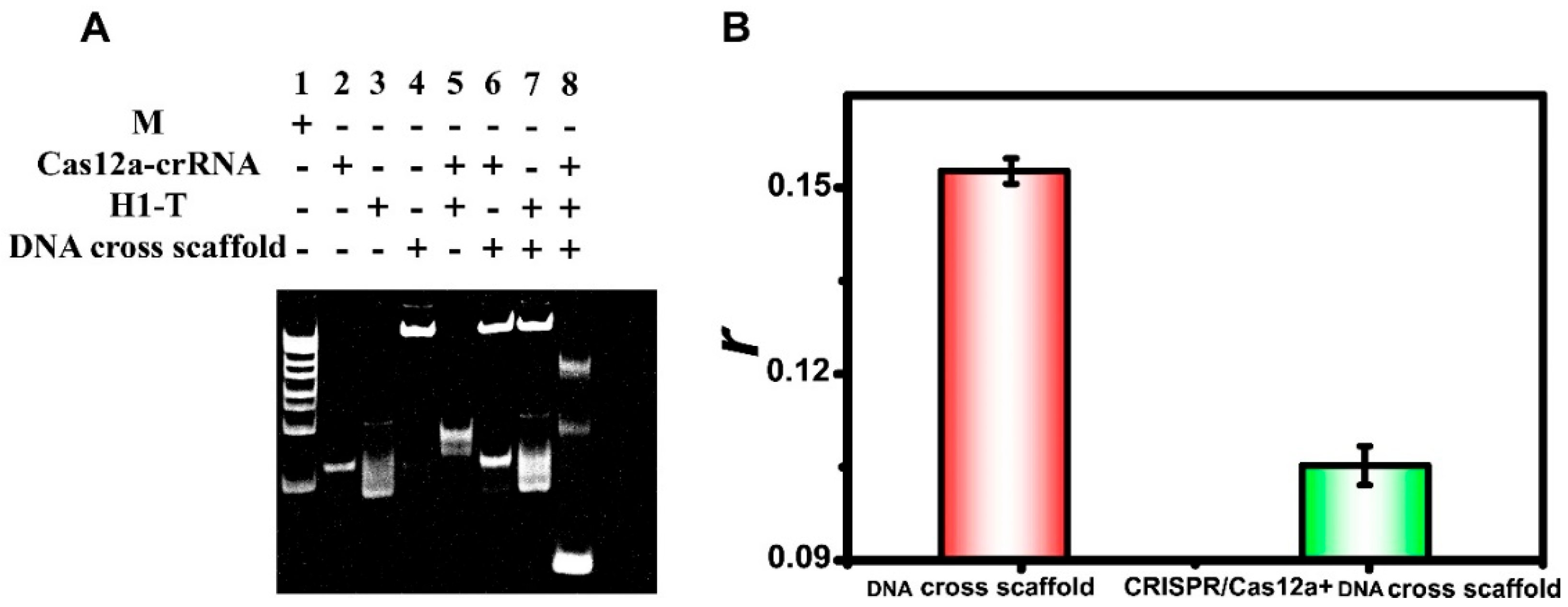
3.3. CRISPR/Cas12a Response
3.4. Feasibility Analysis
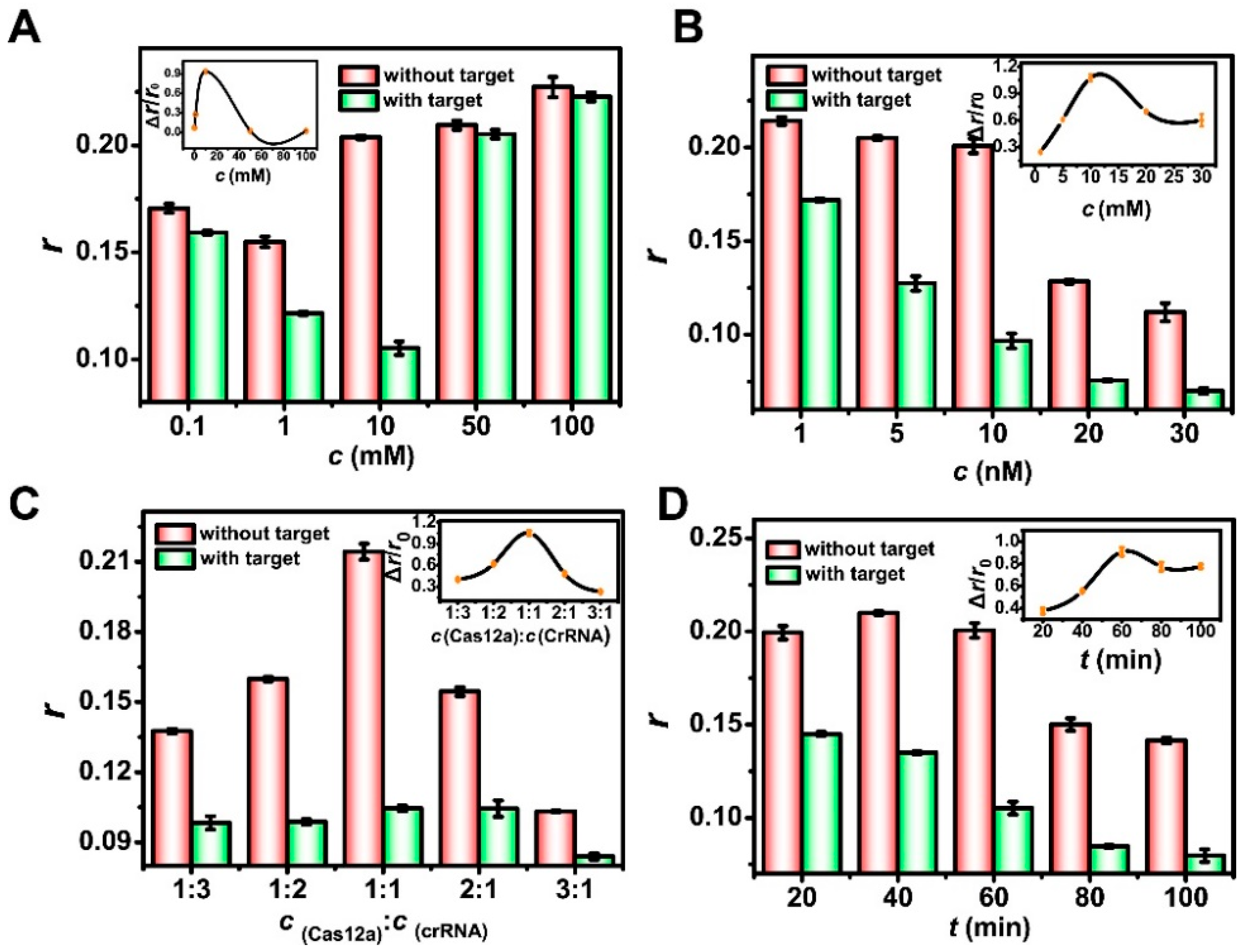
3.5. Optimization of the Experimental Conditions
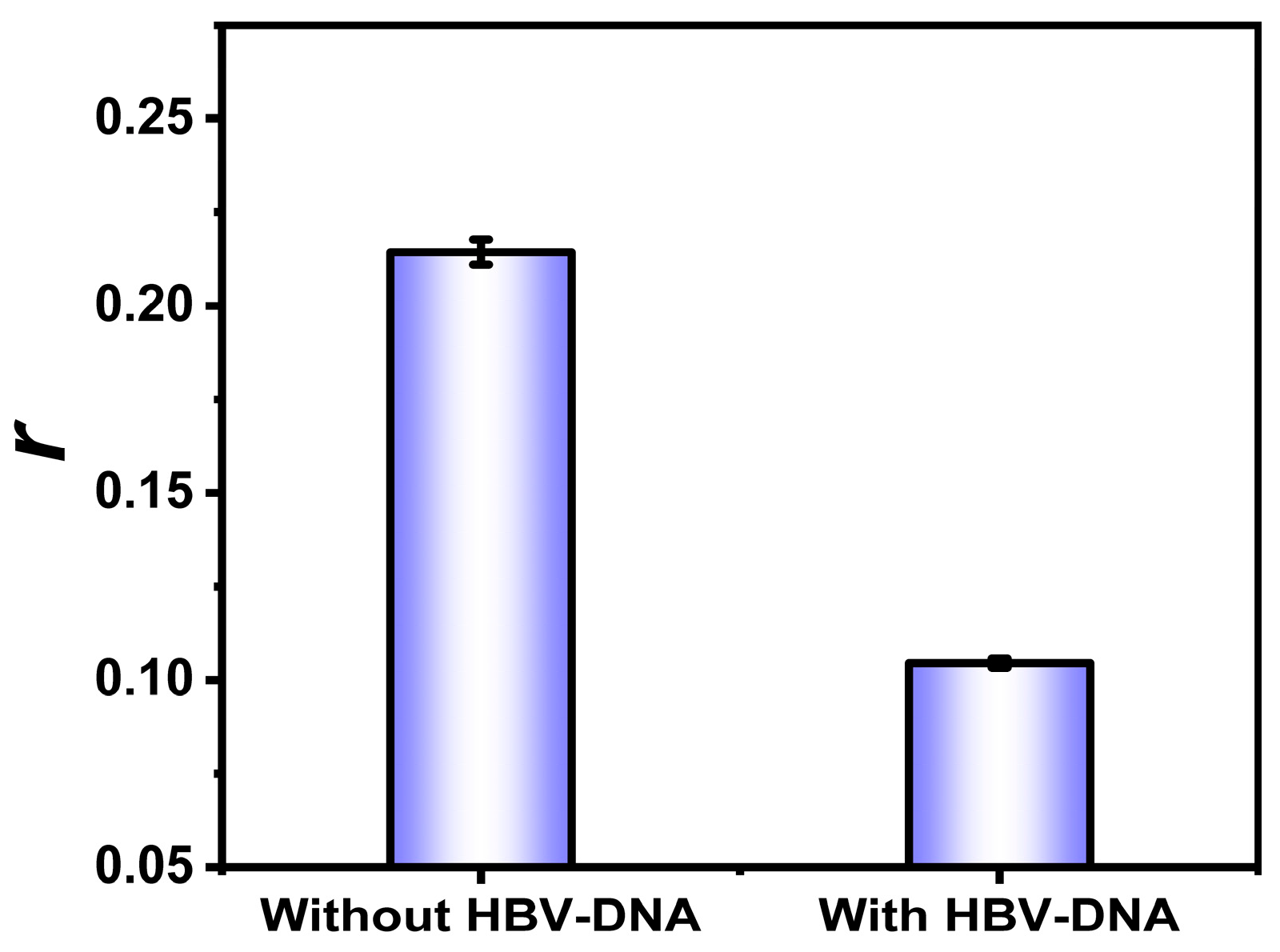
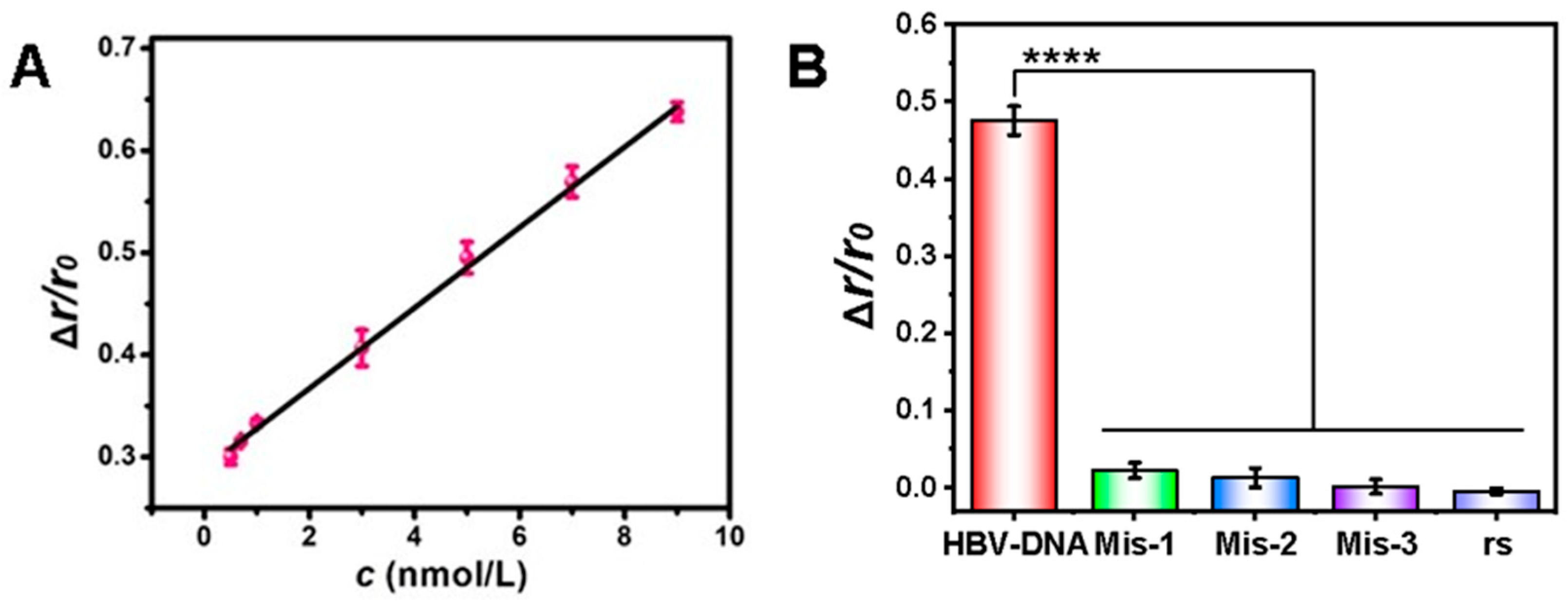
3.6. Sensitivity and Selectivity of HBV-DNA Detection
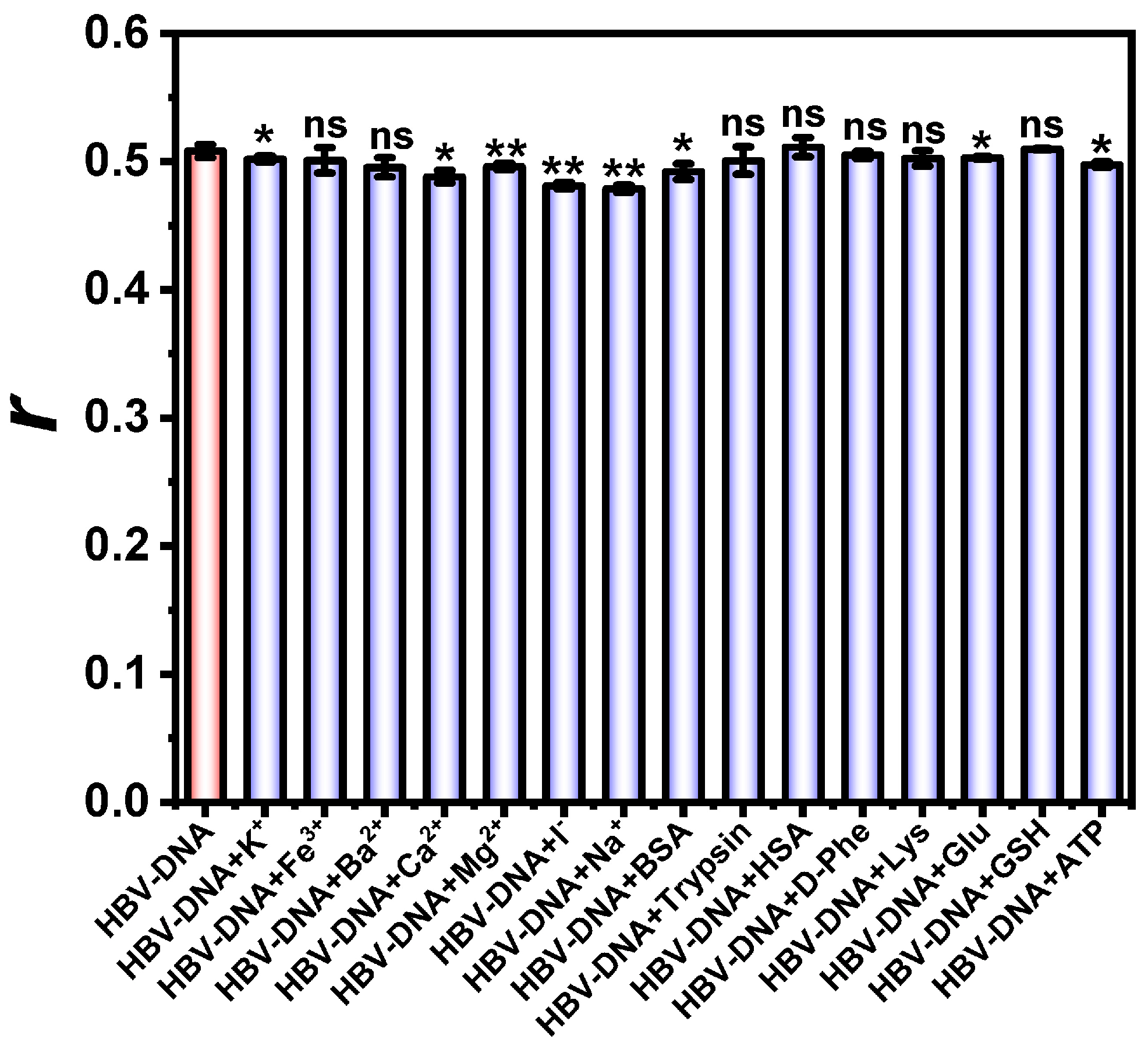
3.7. Interference During HBV-DNA Detection
3.8. Detection of HBV-DNA in Human Serum
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FA | fluorescence anisotropy |
| FP | fluorescence polarization |
| ssDNA | single-stranded deoxyribonucleic acid |
| RNA | ribonucleic acid |
| CRISPR | clusters of regularly interspaced short palindromic repeats |
| Cas | correlated protein |
| SBR | signal-to-background ratio |
| TDF | tetrahedral DNA frameworks |
| HBV-DNA | hepatitis B virus DNA |
| LOD | limit of detection |
| DEPC | diethyl pyrocarbonate |
| PBS | phosphate-buffered saline |
| TBE | tris-borate ethylene diamine tetraacetic acid |
| RSD | relative standard deviation |
| D-Phe | D-phenylalanine |
| Glu | glutamic acid |
| GSH | glutathione |
| Lys | lysine |
| ATP | adenosinetriphosphate |
| BSA | bovine serum albumin |
| HSA | human serum albumin |
| PAGE | polyacrylamide gel electrophoresis |
References
- Huang, Q.; Chen, B.; Shen, J.; Liu, L.; Li, J.; Shi, J.; Li, Q.; Zuo, X.; Wang, L.; Fan, C.; et al. Encoding fluorescence anisotropic barcodes with DNA frameworks. J. Am. Chem. Soc. 2021, 143, 10735–10742. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Masters, B.R. Principles of fluorescence spectroscopy, Third Edition. J. Biomed. Opt. 2008, 13, 029901. [Google Scholar] [CrossRef]
- Chen, J.; Liu, J.; Chen, X.; Qiu, H. Recent progress in nanomaterial-enhanced fluorescence polarization/anisotropy sensors. Chin. Chem. Lett. 2019, 30, 1575–1580. [Google Scholar] [CrossRef]
- Fan, Y.L.; Liu, Z.Y.; Zeng, Y.M.; Huang, L.Y.; Li, Z.; Zhang, Z.L.; Pang, D.W.; Tian, Z.Q. A near-infrared-II fluorescence anisotropy strategy for separation-free detection of adenosine triphosphate in complex media. Talanta 2021, 223, 121721–121728. [Google Scholar] [CrossRef]
- Zhang, D.; Fu, R.; Zhao, Q.; Rong, H.; Wang, H. nanoparticles-free fluorescence anisotropy amplification assay for detection of RNA nucleotide-cleaving DNAzyme activity. Anal. Chem. 2015, 87, 4903–4909. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, J.; Xing, X.; Yao, L.; Shang, H.; Chen, W. Framework nucleic acid-wrapped protein-inorganic hybrid nanoflowers with three-stage amplified fluorescence polarization for terminal deoxynucleotidyl transferase activity biosensing. Biosens. Bioelectron. 2021, 193, 113564–113569. [Google Scholar] [CrossRef]
- Fan, Y.Y.; Mou, Z.L.; Wang, M.; Li, J.; Zhang, J.; Dang, F.Q.; Zhang, Z.Q. Chimeric aptamers-based and MoS2 nanosheet-enhanced label-free fluorescence polarization strategy for adenosine triphosphate detection. Anal. Chem. 2018, 90, 13708–13713. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Zhang, X.B.; Kang, L.P.; Shen, G.L.; Yu, R.Q.; Tan, W. Target-triggered cyclic assembly of DNA–protein hybrid nanowires for dual-amplified fluorescence anisotropy assay of small molecules. Anal. Chem. 2013, 85, 11518–11523. [Google Scholar] [CrossRef]
- Ma, P.; Guo, H.; Duan, N.; Ma, X.; Yue, L.; Gu, Q.; Wang, Z. Label free structure-switching fluorescence polarization detection of chloramphenicol with truncated aptamer. Talanta 2021, 230, 122349–122357. [Google Scholar] [CrossRef]
- Zhu, Q.; Li, H.; Xu, D. Sensitive and enzyme-free fluorescence polarization detection for miRNA-21 based on decahedral sliver nanoparticles and strand displacement reaction. RSC Adv. 2020, 10, 17037–17044. [Google Scholar] [CrossRef]
- Chullipalliyalil, K.; Elkassas, K.; McAuliffe, M.A.P.; Vucen, S.; Crean, A. In-vial detection of protein denaturation using intrinsic fluorescence anisotropy. Anal. Chem. 2023, 95, 2774–2782. [Google Scholar] [CrossRef]
- Yakovchuk, P. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006, 34, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Pabo, C. Protein-DNA recognition. Ann. Rev. Biochem. 1984, 53, 293–321. [Google Scholar]
- Schweizer, K.; Léon, J.C.; Ravoo, B.J.; Müller, J. Thermodynamics of the formation of Ag(I)-mediated azole base pairs in DNA duplexes. J. Inorg. Biochem. 2016, 160, 256–263. [Google Scholar] [PubMed]
- Ma, X.; Zhang, Y.; Qiao, X.; Yuan, Y.; Sheng, Q.; Yue, T. Target-induced AIE Effect coupled with CRISPR/Cas12a system dual-signal biosensing for the ultrasensitive detection of gliotoxin. Anal. Chem. 2023, 95, 11723–11731. [Google Scholar]
- Liang, M.; Li, Z.; Wang, W.; Liu, J.; Liu, L.; Zhu, G.; Karthik, L.; Wang, M.; Wang, K.F.; Wang, Z.; et al. A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat. Commun. 2019, 10, 3672–3680. [Google Scholar] [CrossRef]
- Lau, C.H.; Liang, Q.L.; Chen, X.; Li, J.; Shi, Y.; Huang, Y.; Xia, Q.; Zhu, H.; Chen, G. CRISPR/Dx-Based Colorimetric and Electrochemical Detection Systems for POCT Applications. Biosens. Bioelectron. 2025, 288, 117778. [Google Scholar]
- Zhang, Z.X.; Rong, X.X.; Xie, T.J.; Li, Z.H.; Song, H.Z.; Zhen, S.J.; Wang, H.F.; Wu, J.H.; Jaffrey, S.R.; Li, X. Fluorogenic CRISPR for Genomic DNA Imaging. Nat. Commun. 2024, 15, 934. [Google Scholar] [CrossRef]
- Zhou, Y.; Che, S.; Wang, Z.; Zhang, X.; Yuan, X. Primer Exchange Reaction Assisted CRISPR/Cas9 Cleavage for Detection of Dual microRNAs with Electrochemistry Method. Microchim. Acta 2024, 191, 502. [Google Scholar] [CrossRef]
- Wei, L.; Luo, S.; Zhou, W.; Ren, B.; Li, M.; Liang, L.; Li, X.; Wei, G. Rapid Detection of Pseudomonas Aeruginosa by Glycerol One-Pot RAA/CRISPR-Cas12a Method. Front. Chem. 2025, 13, 1654270. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Zhang, X.; Liu, S.; Wang, S.; Li, X.; Zhang, X.; Li, B.; Huang, H. Signal Amplified Fluorometric Detection of Anti-CRISPR Proteins Based on Rolling Circle Amplification. Sens. Actuators B 2025, 425, 137007. [Google Scholar] [CrossRef]
- Nie, Y.; Li, X.; Yang, W.; Fei, S.; Wang, Y.; Li, Y.; Zhang, K.; Kang, J.; Cheng, Y.; Wang, H.; et al. Concanavalin-A-Assisted Extraction-Free One-Pot RPA–CRISPR/Cas12a Assay for Rapid Detection of HPV16. Microchim. Acta 2025, 192, 354. [Google Scholar] [CrossRef]
- Liu, G. Advancing CRISPR/Cas Biosensing with Integrated Devices. ACS Sens. 2025, 10, 575–576. [Google Scholar] [CrossRef]
- Li, Z.; Su, X.; Lin, Y.; Zhang, Y.; Zhang, A.; Wu, X.; Jiyu, X.; Li, Q.; Wei, Z. Expanding the Cell Quantity of CRISPR/Cas9 Gene Editing by Continuous Microfluidic Electroporation Chip. Bioelectrochemistry 2025, 161, 108840. [Google Scholar] [CrossRef]
- Zhuang, S.; Dong, Y.; Huang, X.; Liu, T.; Gu, D.; Liu, Y. A Universal Cas12a Activity Regulation Strategy via Photocleavable crRNA 3′-Terminal Additives Enables One-Pot CRISPR Detection of Arboviruses. Chem. Eng. J. 2025, 521, 166699–166708. [Google Scholar] [CrossRef]
- Dong, J.; Qi, L.; Wu, X.; Sun, C.; Hu, Q.; Su, Y.; Shao, G.; Zhang, Y.; Meng, F.; Du, Y.; et al. An Electrochemical, Fluorescent, and Colorimetric Triple-Mode Homogeneous Biosensor for the Simultaneous Detection of Influenza A, Influenza B, and SARS-CoV-2. Sens. Actuators B 2024, 411, 135723–135731. [Google Scholar] [CrossRef]
- Zheng, X.; Yao, S.; Yin, C.; Zhao, H.; Wang, J.; Su, T.; Li, H.; Wang, J.; Zhao, C. CRISPR-Integrated Nanoconfined Interparticle Catalytic Hairpin Assembly for Enhanced Dual-Mode SARS-CoV-2 Detection in Wastewater. Biosens. Bioelectron. 2025, 290, 118008–118015. [Google Scholar] [CrossRef]
- De, J.; Mukama, O.; Amissah, O.B.; Sun, Y.; Karangwa, E.; Liu, Y.; Mugisha, S.; Cheng, N.; Wang, L.; Chen, J.; et al. A rationally designed CRISPR/Cas12a assay using a multimodal reporter for various readouts. Anal. Chem. 2023, 95, 11741–11750. [Google Scholar]
- Yi, Z.; De, J.; Mukama, O.; Li, Z.; Odiwuor, N.; Jing, H.; Nie, C.; Hu, M.; Lin, Z.; Wei, H.; et al. Rational programming of Cas12a for early-stage detection of COVID-19 by lateral flow assay and portable real-time fluorescence readout facilities. Biosensors 2021, 12, 11. [Google Scholar] [CrossRef] [PubMed]
- Habimana, J.D.D.; Huang, R.; Muhoza, B.; Kalisa, Y.N.; Han, X.; Deng, W.; Li, Z. Mechanistic insights of CRISPR/Cas nucleases for programmable targeting and early-stage diagnosis: A review. Biosens. Bioelectron. 2022, 203, 114033–114064. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Chen, Z.; Li, C.; Hao, Y.; Tang, Y.; Yuan, Y.; Chai, L.; Fan, T.; Yu, J.; Ma, X.; et al. CRISPR-Cas12a-empowered electrochemical biosensor for rapid and ultrasensitive detection of SARS-CoV-2 delta variant. Nano Micro Lett. 2022, 14, 159–170. [Google Scholar] [CrossRef]
- Kellner, M.J.; Koob, J.G.; Gootenberg, J.S.; Abudayyeh, O.O.; Zhang, F. Sherlock: Nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019, 14, 2986–3012. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Chen, Z.; Xie, R.; Li, J.; Liu, C.; He, Y.; Ma, X.; Yang, H.; Xie, Z. CRISPR/Cas12a-assisted isothermal amplification for rapid and specific diagnosis of respiratory virus on an microfluidic platform. Biosens. Bioelectron. 2023, 237, 115523–115532. [Google Scholar] [PubMed]
- Duan, M.; Li, B.; He, Y.; Zhao, Y.; Liu, Y.; Zou, B.; Liu, Y.; Chen, J.; Dai, R.; Li, X.; et al. A CG@MXene nanocomposite-driven e-CRISPR biosensor for the rapid and sensitive detection of salmonella typhimurium in food. Talanta 2024, 266, 125011–125017. [Google Scholar]
- Xie, T.J.; Xie, J.L.; Luo, Y.J.; Mao, K.; Huang, C.Z.; Li, Y.F.; Zhen, S.J. CRISPR-Cas12a coupled with DNA nanosheet-amplified fluorescence anisotropy for sensitive detection of biomolecules. Anal. Chem. 2023, 95, 7237–7243. [Google Scholar] [CrossRef]
- Xie, J.L.; Xie, T.J.; Luo, Y.J.; Mao, K.; Huang, C.Z.; Li, Y.F.; Zhen, S.J. Octopus-like DNA nanostructure coupled with graphene oxide enhanced fluorescence anisotropy for hepatitis B virus DNA detection. Chin. Chem. Lett. 2024, 35, 109137–109143. [Google Scholar]
- Xiang, Q.; Huang, J.; Huang, H.; Mao, W.; Ye, Z. A label-free electrochemical platform for the highly sensitive detection of hepatitis B virus DNA using graphene quantum dots. RSC Adv. 2018, 8, 1820–1825. [Google Scholar] [CrossRef]
- Zhu, H.T.; Wang, J.X.; Xu, G.Y. Fast synthesis of Cu2O hollow microspheres and their application in DNA biosensor of hepatitis B virus. Cryst. Growth Des. 2009, 9, 633–638. [Google Scholar]
- Liu, Z.C.; Zhang, L.; Zhang, Y.M.; Liang, R.P.; Qiu, J.D. Exonuclease III-assisted recycling amplification detection of hepatitis B virus DNA by DNA-scaffolded silver nanoclusters probe. Sens. Actuators B 2014, 205, 219–226. [Google Scholar] [CrossRef]
- Wang, X.; Lou, Y.; Wang, Q.; Guo, Z.; Fang, X.; Zhong, H.; Mao, Q.; Jin, L.; Wu, H.; Zhao, Q. Ds-DNA nanosensor for the detection of hepatitis B virus DNA and the single-base mutants. Biosens. Bioelectron. 2010, 25, 1934–1940. [Google Scholar] [PubMed]

| Method | Linear Range | LOD | References |
|---|---|---|---|
| Electrochemistry | 10 nmol/L–500 nmol/L | 1 nmol/L | [38] |
| Electrochemistry | 0.1 nmol/L–1 μmol/L | 0.1 nmol/L | [39] |
| Fluorescence | 4 nmol/L–625 nmol/L | 0.97 nmol/L | [40] |
| Fluorescence | 0 nmol/L–500 nmol/L | 4 nmol/L | [41] |
| FA | 0.5 nmol/L–9 nmol/L | 48 pmol/L | This work |
| Sample | Add (nmol/L) | Found (nmol/L) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Human serum | 2 | 2.03 ± 0.08 | 97.6–104.1 | 4.8 |
| 5 | 5.05 ± 0.11 | 99.6–103.3 | 6.1 | |
| 8 | 8.10 ± 0.15 | 98.9–103.2 | 9.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, Y.; Xie, J.; Zhen, S. CRISPR-Cas12a and DNA Tetrahedron Assemblies Amplified Fluorescence Anisotropy for the Sensitive Detection of Hepatitis B Virus DNA. Biosensors 2025, 15, 700. https://doi.org/10.3390/bios15100700
Qin Y, Xie J, Zhen S. CRISPR-Cas12a and DNA Tetrahedron Assemblies Amplified Fluorescence Anisotropy for the Sensitive Detection of Hepatitis B Virus DNA. Biosensors. 2025; 15(10):700. https://doi.org/10.3390/bios15100700
Chicago/Turabian StyleQin, Yu, Jiali Xie, and Shujun Zhen. 2025. "CRISPR-Cas12a and DNA Tetrahedron Assemblies Amplified Fluorescence Anisotropy for the Sensitive Detection of Hepatitis B Virus DNA" Biosensors 15, no. 10: 700. https://doi.org/10.3390/bios15100700
APA StyleQin, Y., Xie, J., & Zhen, S. (2025). CRISPR-Cas12a and DNA Tetrahedron Assemblies Amplified Fluorescence Anisotropy for the Sensitive Detection of Hepatitis B Virus DNA. Biosensors, 15(10), 700. https://doi.org/10.3390/bios15100700





