Structure-Based Understanding of Cu2+ Coordination in Fluorescent Proteins for Metal Biosensor Applications—A Review
Abstract
1. Introduction
2. Fluorescent Proteins
3. Fluorescence Quenching and Reversibility of FP Fluorescence by Cu2+
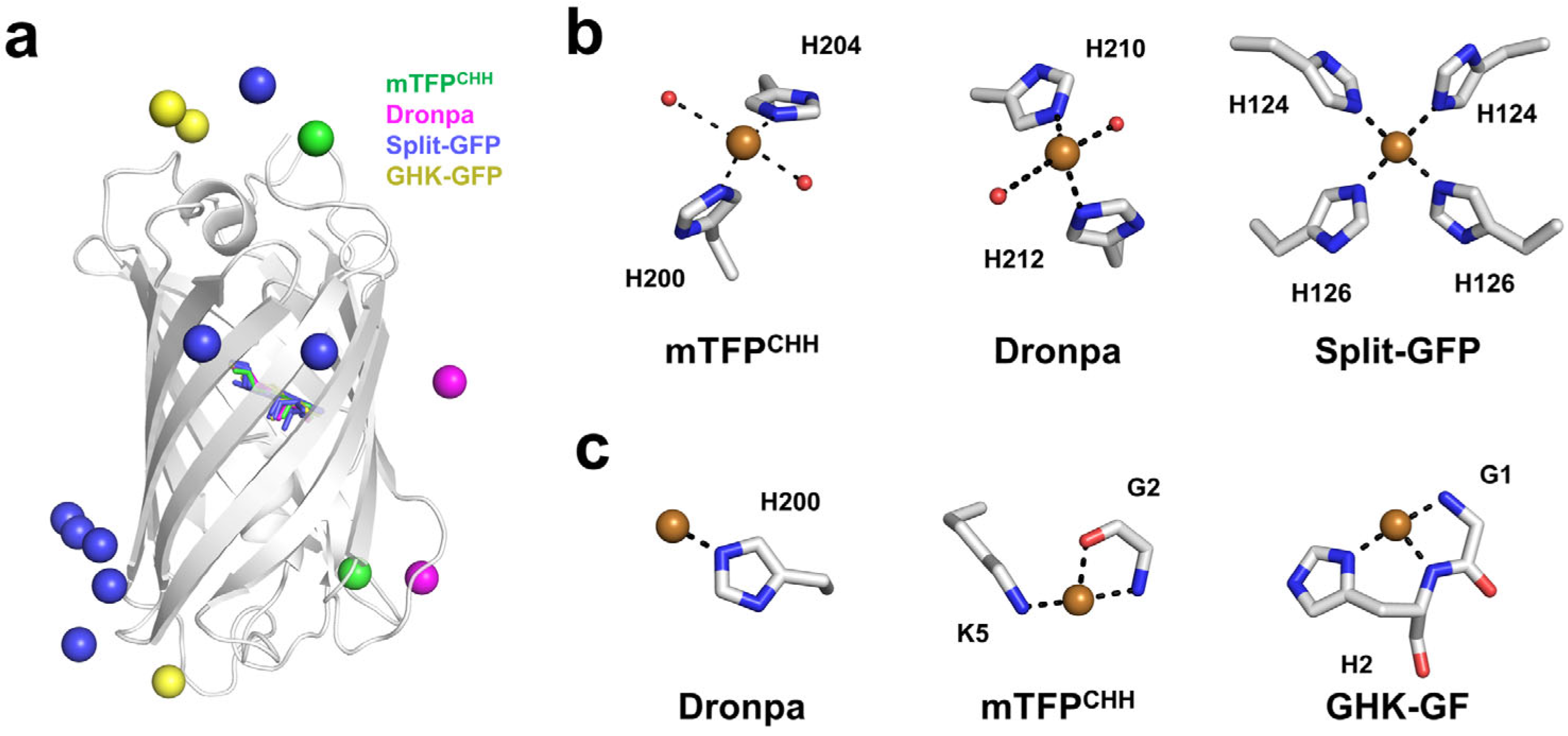
4. Structural Analysis of Cu2+ Binding to the FP
4.1. Quenchable Cu2+-Bound mTFPCHH
4.2. Quenchable Cu2+-Bound Dronpa
4.3. Cu2+ Bound to the Oligomeric Interface of Split-GFP
4.4. Cu2+-Binding GHK Tripeptide Fused to GFP
5. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapoor, S.; Brazete, D.; Pereira, I.C.; Bhatia, G.; Kaur, M.; Santos, L.F.; Banerjee, D.; Goel, A.; Ferreira, J.M.F. Impact of transition metal ions on the structure and bioactivity of alkali-free bioactive glasses. J. Non-Cryst. Solids 2019, 506, 98–108. [Google Scholar] [CrossRef]
- Kostenkova, K.; Scalese, G.; Gambino, D.; Crans, D.C. Highlighting the roles of transition metals and speciation in chemical biology. Curr. Opin. Chem. Biol. 2022, 69, 102155. [Google Scholar] [CrossRef]
- Thiele, D.J.; Gitlin, J.D. Assembling the pieces. Nat. Chem. Biol. 2008, 4, 145–147. [Google Scholar] [CrossRef]
- Hüttemann, M.; Helling, S.; Sanderson, T.H.; Sinkler, C.; Samavati, L.; Mahapatra, G.; Varughese, A.; Lu, G.; Liu, J.; Ramzan, R.; et al. Regulation of mitochondrial respiration and apoptosis through cell signaling: Cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim. Biophys. Acta-Bioenerg. 2012, 1817, 598–609. [Google Scholar] [CrossRef]
- Wiggins, J.E.; Goyal, M.; Wharram, B.L.; Wiggins, R.C. Antioxidant Ceruloplasmin Is Expressed by Glomerular Parietal Epithelial Cells and Secreted into Urine in Association with Glomerular Aging and High-Calorie Diet. J. Am. Soc. Nephrol. 2006, 17, 1382–1387. [Google Scholar] [CrossRef]
- Harris, E.D. Copper as a Cofactor and Regulator of Copper, Zinc Superoxide Dismutase. J. Nutr. 1992, 122, 636–640. [Google Scholar] [CrossRef]
- Postma, G.C.; Nicastro, C.N.; Valdez, L.B.; Rukavina Mikusic, I.A.; Grecco, A.; Minatel, L. Decrease lysyl oxidase activity in hearts of copper-deficient bovines. J. Trace Elem. Med. Biol. 2021, 65, 126715. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N. Copper Deficiency Myelopathy (Human Swayback). Mayo Clin. Proc. 2006, 81, 1371–1384. [Google Scholar] [CrossRef]
- Menkes, J.H. Menkes disease and Wilson disease: Two sides of the same copper coin Part II: Wilson disease. Eur. J. Paediatr. Neurol. 1999, 3, 245–253. [Google Scholar] [CrossRef] [PubMed]
- de Bie, P.; Muller, P.; Wijmenga, C.; Klomp, L.W.J. Molecular pathogenesis of Wilson and Menkes disease: Correlation of mutations with molecular defects and disease phenotypes. J. Med. Genet. 2007, 44, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Scolari Grotto, F.; Glaser, V. Are high copper levels related to Alzheimer’s and Parkinson’s diseases? A systematic review and meta-analysis of articles published between 2011 and 2022. BioMetals 2023, 37, 3–22. [Google Scholar] [CrossRef]
- Barnham, K.J.; Bush, A.I. Metals in Alzheimer’s and Parkinson’s Diseases. Curr. Opin. Chem. Biol. 2008, 12, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Izydorczyk, G.; Mikula, K.; Skrzypczak, D.; Moustakas, K.; Witek-Krowiak, A.; Chojnacka, K. Potential environmental pollution from copper metallurgy and methods of management. Environ. Res. 2021, 197, 111050. [Google Scholar] [CrossRef] [PubMed]
- Rehman, M.; Liu, L.; Wang, Q.; Saleem, M.H.; Bashir, S.; Ullah, S.; Peng, D. Copper environmental toxicology, recent advances, and future outlook: A review. Environ. Sci. Pollut. Res. 2019, 26, 18003–18016. [Google Scholar] [CrossRef]
- Mahamoud Ahmed, A.; Tardy, V.; Bonnineau, C.; Billard, P.; Pesce, S.; Lyautey, E. Changes in sediment microbial diversity following chronic copper-exposure induce community copper-tolerance without increasing sensitivity to arsenic. J. Hazard. Mater. 2020, 391, 122197. [Google Scholar] [CrossRef]
- Shrivas, K. Monitoring of copper level in water and soil samples by using liquid–liquid extraction. Environ. Monit. Assess. 2009, 168, 315–319. [Google Scholar] [CrossRef]
- Umbría-Salinas, K.; Valero, A.; Martins, S.E.; Wallner-Kersanach, M. Copper ecological risk assessment using DGT technique and PNEC: A case study in the Brazilian coast. J. Hazard. Mater. 2021, 403, 123918. [Google Scholar] [CrossRef]
- Narin, I. Determination of trace metal ions by AAS in natural water samples after preconcentration of pyrocatechol violet complexes on an activated carbon column. Talanta 2000, 52, 1041–1046. [Google Scholar] [CrossRef]
- Bulska, E.; Wagner, B. Quantitative aspects of inductively coupled plasma mass spectrometry. Philos. Trans. R. Soc. A 2016, 374, 20150369. [Google Scholar] [CrossRef]
- Chen, D.; Darabedian, N.; Li, Z.; Kai, T.; Jiang, D.; Zhou, F. An improved Bathocuproine assay for accurate valence identification and quantification of copper bound by biomolecules. Anal. Biochem. 2016, 497, 27–35. [Google Scholar] [CrossRef]
- Sulthana, S.F.; Iqbal, U.M.; Suseela, S.B.; Anbazhagan, R.; Chinthaginjala, R.; Chitathuru, D.; Ahmad, I.; Kim, T.-h. Electrochemical Sensors for Heavy Metal Ion Detection in Aqueous Medium: A Systematic Review. ACS Omega 2024, 9, 25493–25512. [Google Scholar] [CrossRef]
- Wainwright, P.; Wadey, D.; Cook, P. An inductively coupled plasma mass spectrometry method for relative free copper determination and generation of a paediatric reference interval. Ann. Clin. Biochem. 2017, 55, 485–490. [Google Scholar] [CrossRef] [PubMed]
- Ratnasekhar, C.; Laur, N.; Kinscherf, R.; Pomytkin, K.; Kaiser, L.; Knes, O.; Deigner, H.-P. ICP-MS trace element analysis in serum and whole blood. PLoS ONE 2020, 15, e0233357. [Google Scholar] [CrossRef]
- Cui, L.; Wu, J.; Li, J.; Ge, Y.; Ju, H. Electrochemical detection of Cu2+ through Ag nanoparticle assembly regulated by copper-catalyzed oxidation of cysteamine. Biosens. Bioelectron. 2014, 55, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Dai, J.; Guo, S.; Sun, R.; Zhang, C.; Zhao, X.; Zhou, L.; Zhang, F.; Li, N.; Wang, M.; et al. Efficient photoelectrochemical sensor of Cu2+ based on ZnO-graphene nanocomposite sensitized with hexagonal CdS by calcination method. J. Electroanal. Chem. 2021, 893, 115330. [Google Scholar] [CrossRef]
- Dinu, A.; Bounegru, A.V.; Iticescu, C.; Georgescu, L.P.; Apetrei, C. Electrochemical Detection of Cd2+, Pb2+, Cu2+ and Hg2+ with Sensors Based on Carbonaceous Nanomaterials and Fe3O4 Nanoparticles. Nanomaterials 2024, 14, 702. [Google Scholar] [CrossRef]
- Zheng, X.; Cheng, W.; Ji, C.; Zhang, J.; Yin, M. Detection of metal ions in biological systems: A review. Rev. Anal. Chem. 2020, 39, 231–246. [Google Scholar] [CrossRef]
- Forzani, E.S.; Zhang, H.; Chen, W.; Tao, N. Detection of Heavy Metal Ions in Drinking Water Using a High-Resolution Differential Surface Plasmon Resonance Sensor. Environ. Sci. Technol. 2004, 39, 1257–1262. [Google Scholar] [CrossRef]
- Malitesta, C.; Guascito, M.R. Heavy metal determination by biosensors based on enzyme immobilised by electropolymerisation. Biosens. Bioelectron. 2005, 20, 1643–1647. [Google Scholar] [CrossRef]
- Zhu, X.; Xu, L.; Lou, Y.; Yu, H.; Li, X.; Blake, D.A.; Liu, F. Preparation of Specific Monoclonal Antibodies (MAbs) against Heavy Metals: MAbs That Recognize Chelated Cadmium Ions. J. Agric. Food Chem. 2007, 55, 7648–7653. [Google Scholar] [CrossRef]
- Lee, J.-S.; Han, M.S.; Mirkin, C.A. Colorimetric Detection of Mercuric Ion (Hg2+) in Aqueous Media using DNA-Functionalized Gold Nanoparticles. Angew. Chem. Int. Ed. 2007, 46, 4093–4096. [Google Scholar] [CrossRef]
- Li, T.; Shi, L.; Wang, E.; Dong, S. Silver-Ion-Mediated DNAzyme Switch for the Ultrasensitive and Selective Colorimetric Detection of Aqueous Ag+ and Cysteine. Chem. Eur. J. 2009, 15, 3347–3350. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Riedel, K.; Adler, K.; Kunze, G. Amperometric measurement of copper ions with a deputy substrate using a novel Saccharomyces cerevisiae sensor. Biosens. Bioelectron. 2000, 15, 211–219. [Google Scholar] [CrossRef]
- Yan, L.; Bao, K.; Xu, X.; Li, L.; Wu, X. Recent progress in fluorescent probes for Cu2+ based on small organic molecules. J. Mol. Struct. 2024, 1316, 139100. [Google Scholar] [CrossRef]
- Hsiao, W.W.-W.; Pham, U.K.; Le, T.-N.; Lam, X.M.; Chiang, W.-H. Advances in aggregation-induced emission luminogens for biomedicine: From luminescence mechanisms to diagnostic applications. Biosens. Bioelectron. 2025, 270, 116942. [Google Scholar] [CrossRef]
- Saedi, Z.; Roushani, M.; Khaleghian-Moghadam, R.; Darabi, A. Selective and sensitive Detection of Cu2+ in aqueous solution based on cation exchange by Metal−Organic framework TMU-16 as a fluorescent sensor. J. Lumin. 2022, 251, 119165. [Google Scholar] [CrossRef]
- Fonseca, R.R.F.d.; Santos, G.F.S.d.; Rodrigues, J.G.A.; Ferreira, R.d.Q.; Luz, P.P. A functional FeIII-based NH2-MOF as an electrochemical sensor for Cu2+ detection in ethanol fuel. Ionics 2024, 30, 2397–2407. [Google Scholar] [CrossRef]
- Sargazi, S.; Fatima, I.; Hassan Kiani, M.; Mohammadzadeh, V.; Arshad, R.; Bilal, M.; Rahdar, A.; Díez-Pascual, A.M.; Behzadmehr, R. Fluorescent-based nanosensors for selective detection of a wide range of biological macromolecules: A comprehensive review. Int. J. Biol. Macromol. 2022, 206, 115–147. [Google Scholar] [CrossRef]
- Fang, X.; Zheng, Y.; Duan, Y.; Liu, Y.; Zhong, W. Recent Advances in Design of Fluorescence-Based Assays for High-Throughput Screening. Anal. Chem. 2018, 91, 482–504. [Google Scholar] [CrossRef]
- Jensen, G.C.; Janis, M.K.; Nguyen, H.N.; David, O.W.; Zastrow, M.L. Fluorescent Protein-Based Sensors for Detecting Essential Metal Ions across the Tree of Life. ACS Sens. 2024, 9, 1622–1643. [Google Scholar] [CrossRef]
- Wang, M.; Da, Y.; Tian, Y. Fluorescent proteins and genetically encoded biosensors. Chem. Soc. Rev. 2023, 52, 1189–1214. [Google Scholar] [CrossRef]
- Stepanenko, O.V.; Verkhusha, V.V.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Fluorescent proteins as biomarkers and biosensors: Throwing color lights on molecular and cellular processes. Curr. Protein Pept. Sci. 2008, 9, 338–369. [Google Scholar] [CrossRef] [PubMed]
- Frei, M.S.; Mehta, S.; Zhang, J. Next-Generation Genetically Encoded Fluorescent Biosensors Illuminate Cell Signaling and Metabolism. Annu. Rev. Biophys. 2024, 53, 275–297. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H.; Xu, Y. Structural Analysis of the Large Stokes Shift Red Fluorescent Protein tKeima. Molecules 2024, 29, 2579. [Google Scholar] [CrossRef] [PubMed]
- Tian, F.; Xu, G.; Zhou, S.; Chen, S.; He, D. Principles and applications of green fluorescent protein-based biosensors: A mini-review. Analyst 2023, 148, 2882–2891. [Google Scholar] [CrossRef]
- Li, S.-A.; Meng, X.-Y.; Zhang, Y.-J.; Chen, C.-L.; Jiao, Y.-X.; Zhu, Y.-Q.; Liu, P.-P.; Sun, W. Progress in pH-Sensitive sensors: Essential tools for organelle pH detection, spotlighting mitochondrion and diverse applications. Front. Pharmacol. 2024, 14, 1339518. [Google Scholar] [CrossRef]
- Chin, M.Y.; Patwardhan, A.R.; Ang, K.-H.; Wang, A.L.; Alquezar, C.; Welch, M.; Nguyen, P.T.; Grabe, M.; Molofsky, A.V.; Arkin, M.R.; et al. Genetically Encoded, pH-Sensitive mTFP1 Biosensor for Probing Lysosomal pH. ACS Sens. 2021, 6, 2168–2180. [Google Scholar] [CrossRef]
- Tutol, J.N.; Ong, W.S.Y.; Phelps, S.M.; Peng, W.; Goenawan, H.; Dodani, S.C. Engineering the ChlorON Series: Turn-On Fluorescent Protein Sensors for Imaging Labile Chloride in Living Cells. ACS Cent. Sci. 2023, 10, 77–86. [Google Scholar] [CrossRef]
- Yu, T.-G.; Lee, J.; Yoon, J.; Choi, J.M.; Kim, D.-G.; Heo, W.D.; Song, J.-J.; Kim, H.-S. Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State. JACS Au 2023, 3, 3055–3065. [Google Scholar] [CrossRef]
- Torres-Ocampo, A.P.; Palmer, A.E. Genetically encoded fluorescent sensors for metals in biology. Curr. Opin. Chem. Biol. 2023, 74, 102284. [Google Scholar] [CrossRef]
- Rahimi, Y.; Goulding, A.; Shrestha, S.; Mirpuri, S.; Deo, S.K. Mechanism of copper induced fluorescence quenching of red fluorescent protein, DsRed. Biochem. Biophys. Res. Commun. 2008, 370, 57–61. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Metal-Induced Fluorescence Quenching of Photoconvertible Fluorescent Protein DendFP. Molecules 2022, 27, 2922. [Google Scholar] [CrossRef]
- Choi, Y.-A.; Keem, J.O.; Kim, C.Y.; Yoon, H.R.; Heo, W.D.; Chung, B.H.; Jung, Y. A novel copper-chelating strategy for fluorescent proteins to image dynamic copper fluctuations on live cell surfaces. Chem. Sci. 2015, 6, 1301–1307. [Google Scholar] [CrossRef] [PubMed]
- Taki, M.; Iyoshi, S.; Ojida, A.; Hamachi, I.; Yamamoto, Y. Development of Highly Sensitive Fluorescent Probes for Detection of Intracellular Copper(I) in Living Systems. J. Am. Chem. Soc. 2010, 132, 5938–5939. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, O.; Johnson, F.H.; Saiga, Y. Extraction, Purification and Properties of Aequorin, a Bioluminescent Protein from the Luminous Hydromedusan, Aequorea. J. Cell. Comp. Physiol. 1962, 59, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M. Green fluorescent protein (GFP): Applications, structure, and related photophysical behavior. Chem. Rev. 2002, 102, 759–781. [Google Scholar] [CrossRef]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Piatkevich, K.D.; Verkhusha, V.V. Advances in engineering of fluorescent proteins and photoactivatable proteins with red emission. Curr. Opin. Chem. Biol. 2010, 14, 23–29. [Google Scholar] [CrossRef]
- Seo, P.-W.; Kim, G.-J.; Kim, J.-S. A short guide on blue fluorescent proteins: Limits and perspectives. Appl. Microbiol. Biotechnol. 2024, 108, 208. [Google Scholar] [CrossRef]
- Day, R.N.; Davidson, M.W. The fluorescent protein palette: Tools for cellular imaging. Chem. Soc. Rev. 2009, 38, 2887–2921. [Google Scholar] [CrossRef]
- Cheng, L.; Fu, J.; Tsukamoto, A.; Hawley, R.G. Use of green fluorescent protein variants to monitor gene transfer and expression in mammalian cells. Nat. Biotechnol. 1996, 14, 606–609. [Google Scholar] [CrossRef]
- Ogawa, H.; Inouye, S.; Tsuji, F.I.; Yasuda, K.; Umesono, K. Localization, trafficking, and temperature-dependence of the Aequorea green fluorescent protein in cultured vertebrate cells. Proc. Natl. Acad. Sci. USA 1995, 92, 11899–11903. [Google Scholar] [CrossRef] [PubMed]
- Mohr, M.A.; Argast, P.; Pantazis, P. Labeling cellular structures in vivo using confined primed conversion of photoconvertible fluorescent proteins. Nat. Protoc. 2016, 11, 2419–2431. [Google Scholar] [CrossRef] [PubMed]
- Piston, D.W.; Kremers, G.-J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem. Sci. 2007, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Kenworthy, A.K. Imaging Protein-Protein Interactions Using Fluorescence Resonance Energy Transfer Microscopy. Methods 2001, 24, 289–296. [Google Scholar] [CrossRef]
- dos Santos, N.V.; Saponi, C.F.; Ryan, T.M.; Primo, F.L.; Greaves, T.L.; Pereira, J.F.B. Reversible and irreversible fluorescence activity of the Enhanced Green Fluorescent Protein in pH: Insights for the development of pH-biosensors. Int. J. Biol. Macromol. 2020, 164, 3474–3484. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, M.-S.; Xing, X.-H. Temperature influence on fluorescence intensity and enzyme activity of the fusion protein of GFP and hyperthermophilic xylanase. Appl. Microbiol. Biotechnol. 2009, 84, 511–517. [Google Scholar] [CrossRef]
- Nam, K.H. Fluorescent Protein-Based Metal Biosensors. Chemosensors 2023, 11, 216. [Google Scholar] [CrossRef]
- Sun, N.; Malide, D.; Liu, J.; Rovira, I.I.; Combs, C.A.; Finkel, T. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat. Protoc. 2017, 12, 1576–1587. [Google Scholar] [CrossRef]
- Xu, Y.; Seo, Y.G.; Kim, I.J.; Nam, K.H. pH-Induced Conformational Change of the Chromophore of the Large Stokes Shift Fluorescent Protein tKeima. Molecules 2025, 30, 1623. [Google Scholar] [CrossRef]
- Bae, J.-E.; Kim, I.J.; Nam, K.H. Disruption of the hydrogen bonding network determines the pH-induced non-fluorescent state of the fluorescent protein ZsYellow by protonation of Glu221. Biochem. Biophys. Res. Commun. 2017, 493, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Richmond, T.A.; Takahashi, T.T.; Shimkhada, R.; Bernsdorf, J. Engineered Metal Binding Sites on Green Fluorescence Protein. Biochem. Biophys. Res. Commun. 2000, 268, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Eli, P.; Chakrabartty, A. Variants of DsRed fluorescent protein: Development of a copper sensor. Protein Sci. 2006, 15, 2442–2447. [Google Scholar] [CrossRef]
- Sumner, J.P.; Westerberg, N.M.; Stoddard, A.K.; Hurst, T.K.; Cramer, M.; Thompson, R.B.; Fierke, C.A.; Kopelman, R. DsRed as a highly sensitive, selective, and reversible fluorescence-based biosensor for both Cu+ and Cu2+ ions. Biosens. Bioelectron. 2006, 21, 1302–1308. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.J.; Kim, S.; Park, J.; Eom, I.; Kim, S.; Kim, J.H.; Ha, S.C.; Kim, Y.G.; Hwang, K.Y.; Nam, K.H. Crystal structures of Dronpa complexed with quenchable metal ions provide insight into metal biosensor development. FEBS Lett. 2016, 590, 2982–2990. [Google Scholar] [CrossRef]
- Bae, J.E.; Kim, I.J.; Nam, K.H. Spectroscopic Analysis of the Cu2+-Induced Fluorescence Quenching of Fluorescent Proteins AmCyan and mOrange2. Mol. Biotechnol. 2018, 60, 485–491. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic Analysis of Fe Ion-Induced Fluorescence Quenching of the Green Fluorescent Protein ZsGreen. J. Fluoresc. 2021, 31, 307–314. [Google Scholar] [CrossRef]
- Kim, I.J.; Xu, Y.; Nam, K.H. Spectroscopic and Structural Analysis of Cu2+-Induced Fluorescence Quenching of ZsYellow. Biosensors 2020, 10, 29. [Google Scholar] [CrossRef]
- Barondeau, D.P.; Kassmann, C.J.; Tainer, J.A.; Getzoff, E.D. Structural chemistry of a green fluorescent protein Zn biosensor. J. Am. Chem. Soc. 2002, 124, 3522–3524. [Google Scholar] [CrossRef]
- Yu, X.; Strub, M.P.; Barnard, T.J.; Noinaj, N.; Piszczek, G.; Buchanan, S.K.; Taraska, J.W. An engineered palette of metal ion quenchable fluorescent proteins. PLoS ONE 2014, 9, e95808. [Google Scholar] [CrossRef]
- Isarankura-Na-Ayudhya, C.; Tantimongcolwat, T.; Galla, H.J.; Prachayasittikul, V. Fluorescent protein-based optical biosensor for copper ion quantitation. Biol. Trace Elem. Res. 2010, 134, 352–363. [Google Scholar] [CrossRef]
- Ababou, A.; Bombarda, E. On the involvement of electron transfer reactions in the fluorescence decay kinetics heterogeneity of proteins. Protein Sci. 2008, 10, 2102–2113. [Google Scholar] [CrossRef]
- Mizuno, T.; Murao, K.; Tanabe, Y.; Oda, M.; Tanaka, T. Metal-ion-dependent GFP emission in vivo by combining a circularly permutated green fluorescent protein with an engineered metal-ion-binding coiled-coil. J. Am. Chem. Soc. 2007, 129, 11378–11383. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, Y.; Shrestha, S.; Deo, S.K. Metal Affinity-Based Purification of a Red Fluorescent Protein. Chromatographia 2007, 65, 429–433. [Google Scholar] [CrossRef]
- Fischer, J.; Renn, D.; Quitterer, F.; Radhakrishnan, A.; Liu, M.; Makki, A.; Ghorpade, S.; Rueping, M.; Arold, S.T.; Groll, M.; et al. Robust and Versatile Host Protein for the Design and Evaluation of Artificial Metal Centers. ACS Catal. 2019, 9, 11371–11380. [Google Scholar] [CrossRef]
- Jeschek, M.; Panke, S.; Ward, T.R. Artificial Metalloenzymes on the Verge of New-to-Nature Metabolism. Trends Biotechnol. 2018, 36, 60–72. [Google Scholar] [CrossRef]
- Davis, H.J.; Ward, T.R. Artificial Metalloenzymes: Challenges and Opportunities. ACS Cent. Sci. 2019, 5, 1120–1136. [Google Scholar] [CrossRef]
- Yu, X.; Wu, X.; Bermejo, G.A.; Brooks, B.R.; Taraska, J.W. Accurate High-Throughput Structure Mapping and Prediction with Transition Metal Ion FRET. Structure 2013, 21, 9–19. [Google Scholar] [CrossRef]
- Ando, R.; Mizuno, H.; Miyawaki, A. Regulated Fast Nucleocytoplasmic Shuttling Observed by Reversible Protein Highlighting. Science 2004, 306, 1370–1373. [Google Scholar] [CrossRef]
- Habuchi, S.; Ando, R.; Dedecker, P.; Verheijen, W.; Mizuno, H.; Miyawaki, A.; Hofkens, J. Reversible single-molecule photoswitching in the GFP-like fluorescent protein Dronpa. Proc. Natl. Acad. Sci. USA 2005, 102, 9511–9516. [Google Scholar] [CrossRef]
- Lee, H.; DeLoache, W.C.; Dueber, J.E. Spatial organization of enzymes for metabolic engineering. Metab. Eng. 2012, 14, 242–251. [Google Scholar] [CrossRef]
- Good, M.C.; Zalatan, J.G.; Lim, W.A. Scaffold Proteins: Hubs for Controlling the Flow of Cellular Information. Science 2011, 332, 680–686. [Google Scholar] [CrossRef]
- Lai, Y.-T.; King, N.P.; Yeates, T.O. Principles for designing ordered protein assemblies. Trends Cell Biol. 2012, 22, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Laganowsky, A.; Zhao, M.; Soriaga, A.B.; Sawaya, M.R.; Cascio, D.; Yeates, T.O. An approach to crystallizing proteins by metal-mediated synthetic symmetrization. Protein Sci. 2011, 20, 1876–1890. [Google Scholar] [CrossRef] [PubMed]
- Leibly, D.J.; Arbing, M.A.; Pashkov, I.; DeVore, N.; Waldo, G.S.; Terwilliger, T.C.; Yeates, T.O. A Suite of Engineered GFP Molecules for Oligomeric Scaffolding. Structure 2015, 23, 1754–1768. [Google Scholar] [CrossRef]
- Taylor, G.L. Introduction to phasing. Acta Crystallogr. D Struct. Biol. 2010, 66, 325–338. [Google Scholar] [CrossRef] [PubMed]
- Mehr, A.; Henneberg, F.; Chari, A.; Görlich, D.; Huyton, T. The copper(II)-binding tripeptide GHK, a valuable crystallization and phasing tag for macromolecular crystallography. Acta Crystallogr. D Struct. Biol. 2020, 76, 1222–1232. [Google Scholar] [CrossRef]
- Rubino, J.T.; Franz, K.J. Coordination chemistry of copper proteins: How nature handles a toxic cargo for essential function. J. Inorg. Biochem. 2012, 107, 129–143. [Google Scholar] [CrossRef]
- Ashraf, J.; Riaz, M.A. Biological potential of copper complexes: A review. Turk. J. Chem. 2022, 46, 595–623. [Google Scholar] [CrossRef]
- Nam, K.H. Evaluation of AlphaFold3 prediction for post-translational modification, oligomeric assembly, and quenchable metal binding of fluorescent proteins. J. Mol. Graph. Model. 2025, 142, 109169. [Google Scholar] [CrossRef]

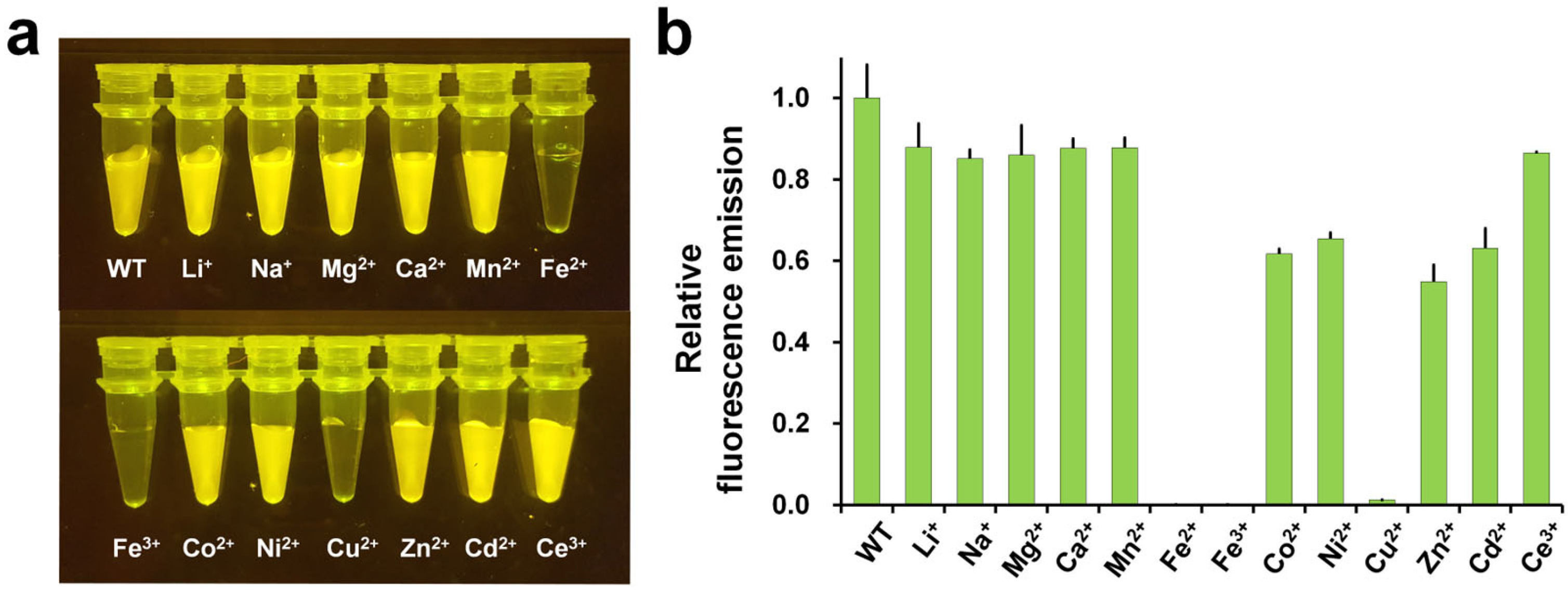
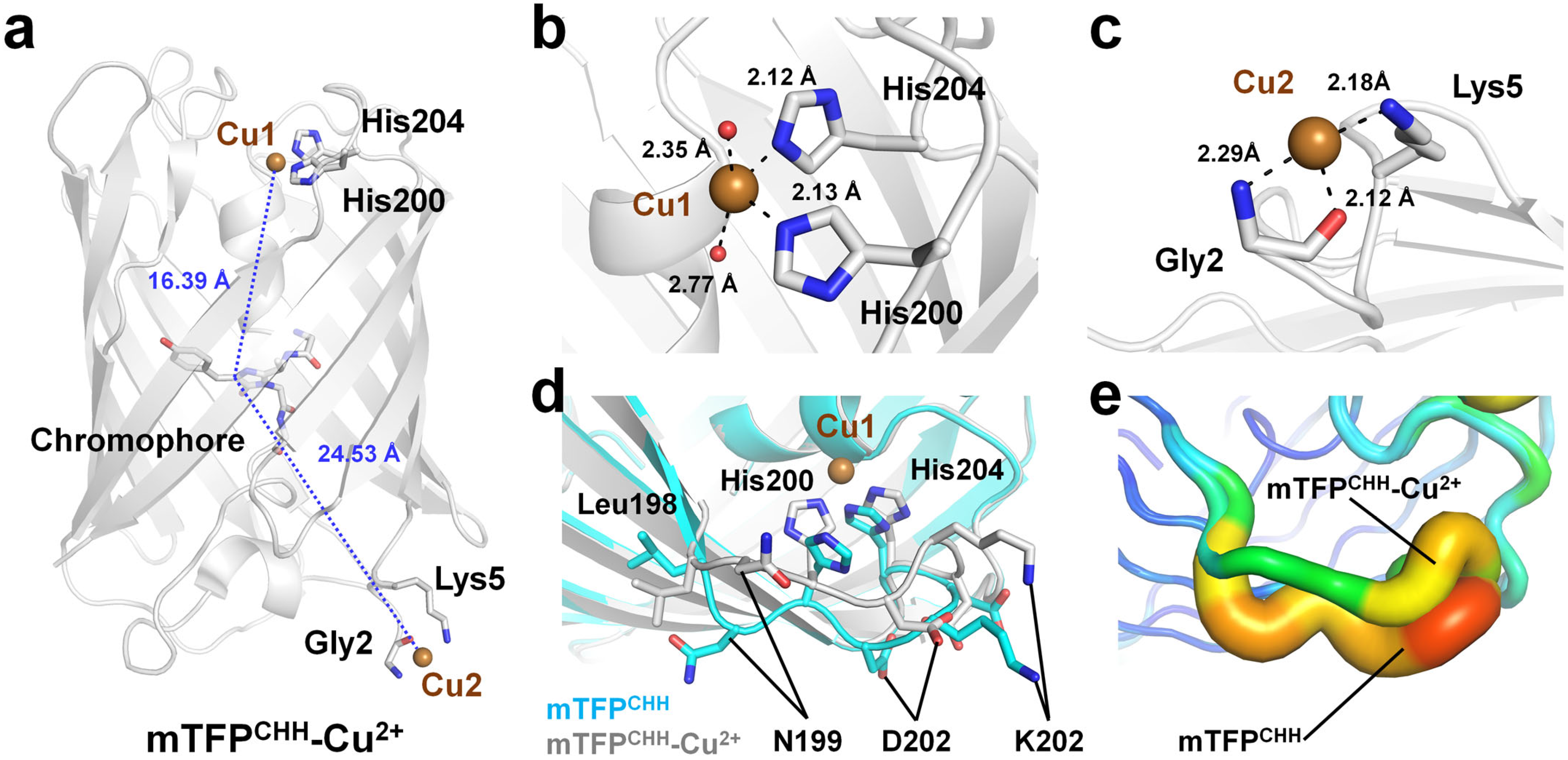
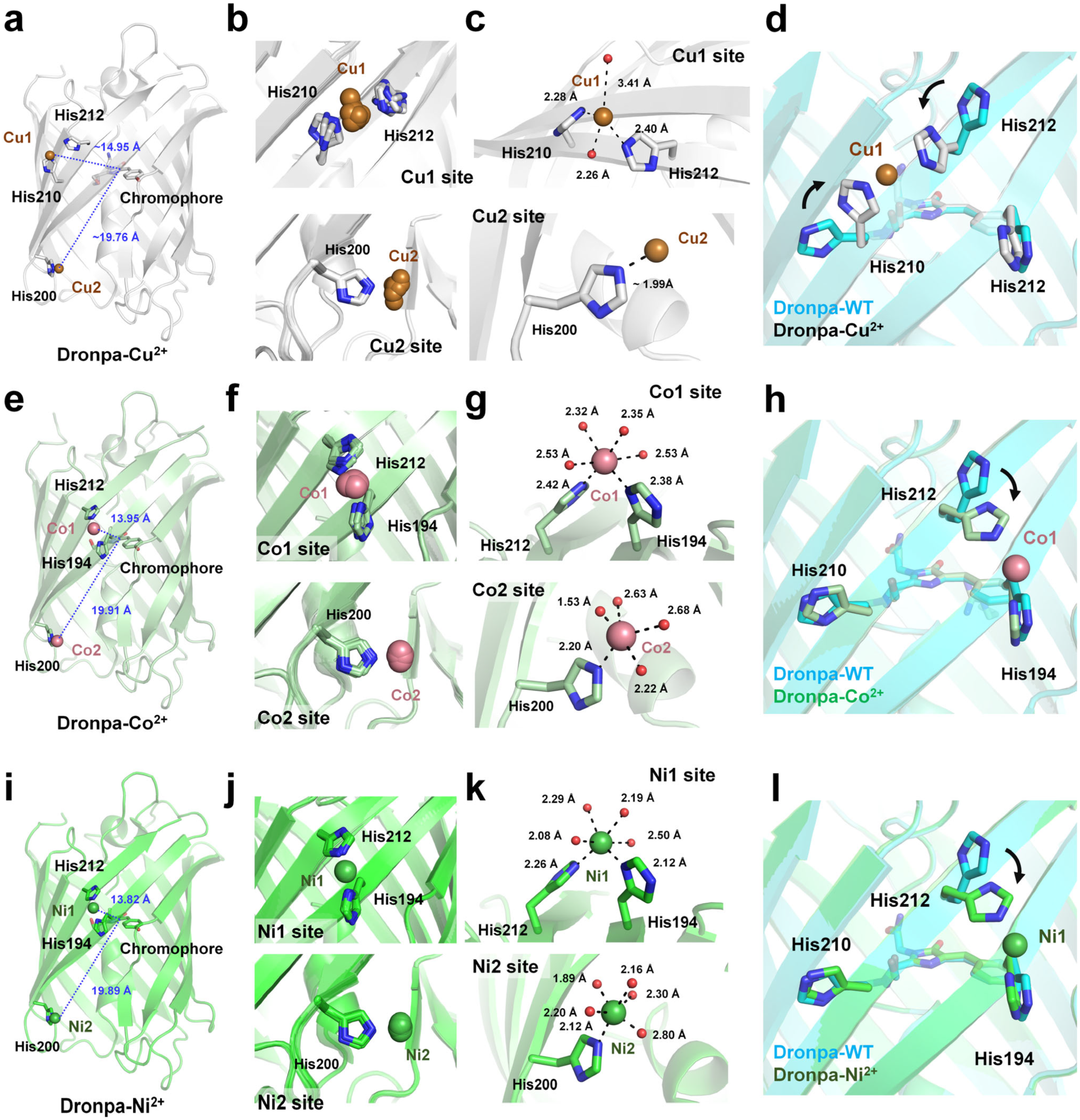

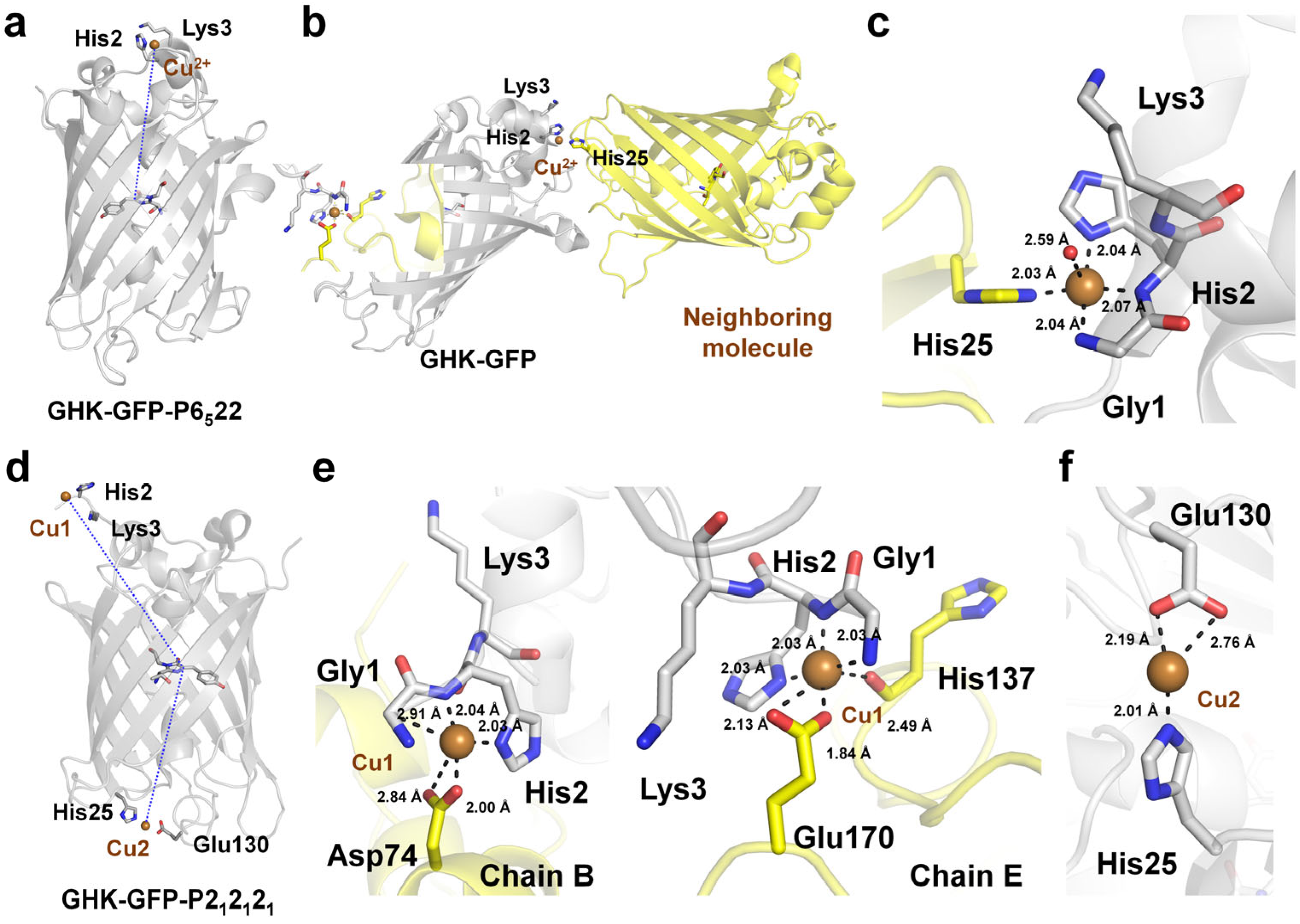
| FP | Quenching Efficiency (%) | Kd (μM) | Recovery (EDTA) | Limit of Detection (LOD) | Reference |
|---|---|---|---|---|---|
| DsRed | 90.0 ± 10 | 0.54 | ND | 45 ± 2 nM | [74] |
| drFP583 | 78.0 | 14.80 ± 1.68 | >90% (1 mM) | ND | [73] |
| Rmu13 | 66.0 | 10.90 ± 1.74 | ND | ND | [73] |
| Dronpa | 86.0 | ND | ND | ND | [75] |
| AmCyan | 80.0 | 56.10 | 89.4% (5 mM) | ND | [76] |
| mOrange2 | 89.0 | 21.46 | >100% (5 mM) | ND | [76] |
| ZsYellow | 81.4 | ND | ND | ND | [78] |
| ZsGreen | 77.2 | 68.2 | >90% (50 mM) | ND | [77] |
| DendFP | 98.8 | 137.18 | 44.2% (50 mM) | 3.2 μM | [52] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. Structure-Based Understanding of Cu2+ Coordination in Fluorescent Proteins for Metal Biosensor Applications—A Review. Biosensors 2025, 15, 675. https://doi.org/10.3390/bios15100675
Nam KH. Structure-Based Understanding of Cu2+ Coordination in Fluorescent Proteins for Metal Biosensor Applications—A Review. Biosensors. 2025; 15(10):675. https://doi.org/10.3390/bios15100675
Chicago/Turabian StyleNam, Ki Hyun. 2025. "Structure-Based Understanding of Cu2+ Coordination in Fluorescent Proteins for Metal Biosensor Applications—A Review" Biosensors 15, no. 10: 675. https://doi.org/10.3390/bios15100675
APA StyleNam, K. H. (2025). Structure-Based Understanding of Cu2+ Coordination in Fluorescent Proteins for Metal Biosensor Applications—A Review. Biosensors, 15(10), 675. https://doi.org/10.3390/bios15100675





