Review of Microchip Analytical Methods Coupled with Aptamer-Based Signal Amplification Strategies for High-Sensitivity Bioanalytical Applications
Abstract
1. Introduction
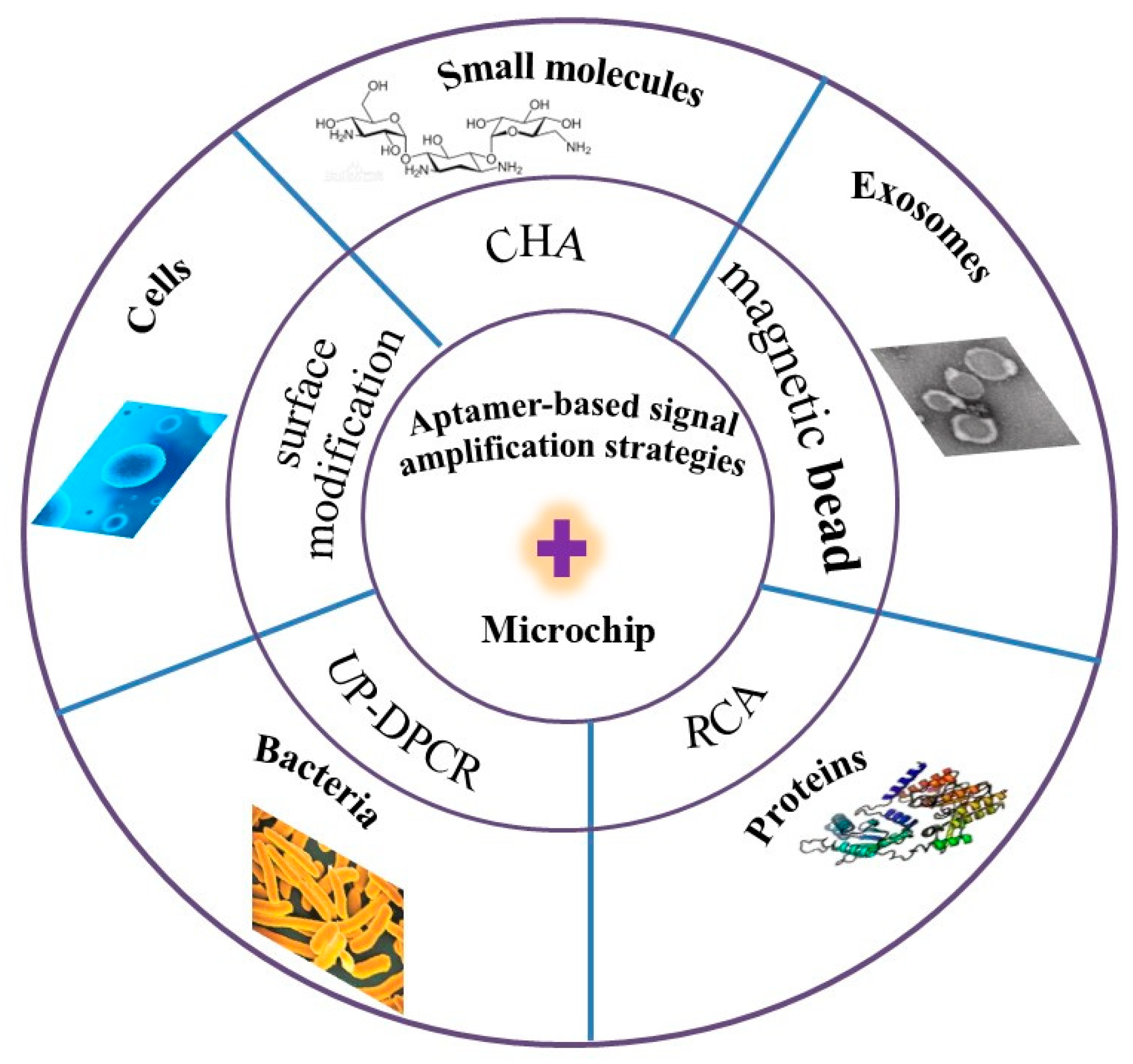
2. Signal Amplification Strategy Based on Aptamer-Integrated Microchips
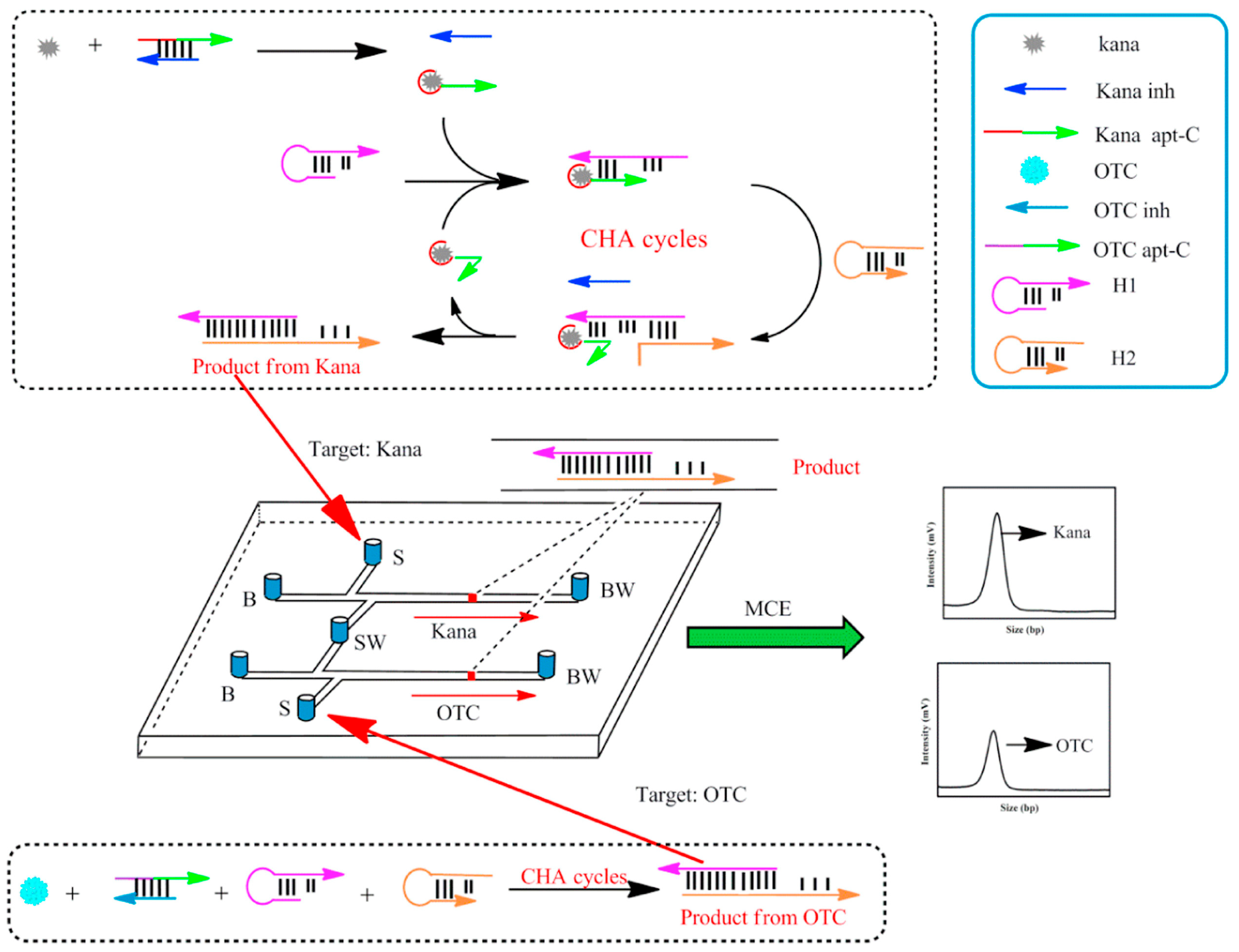
2.1. Detection of Small Molecules Using Amplification Strategies Based on Aptamer-Integrated Microchip
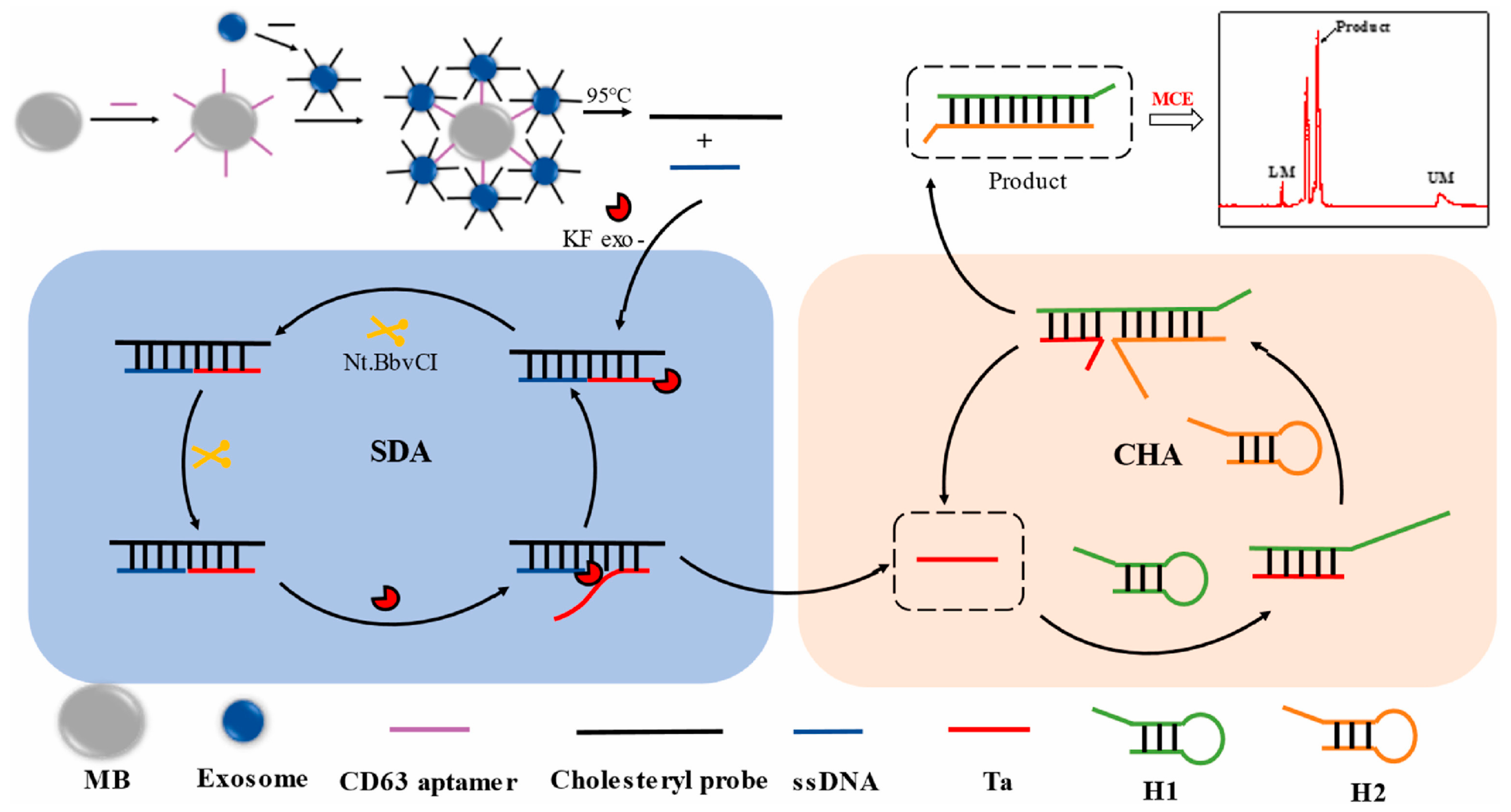
2.2. Detection of Exosomes Using Amplification Strategies Based on Aptamer-Integrated Microchip
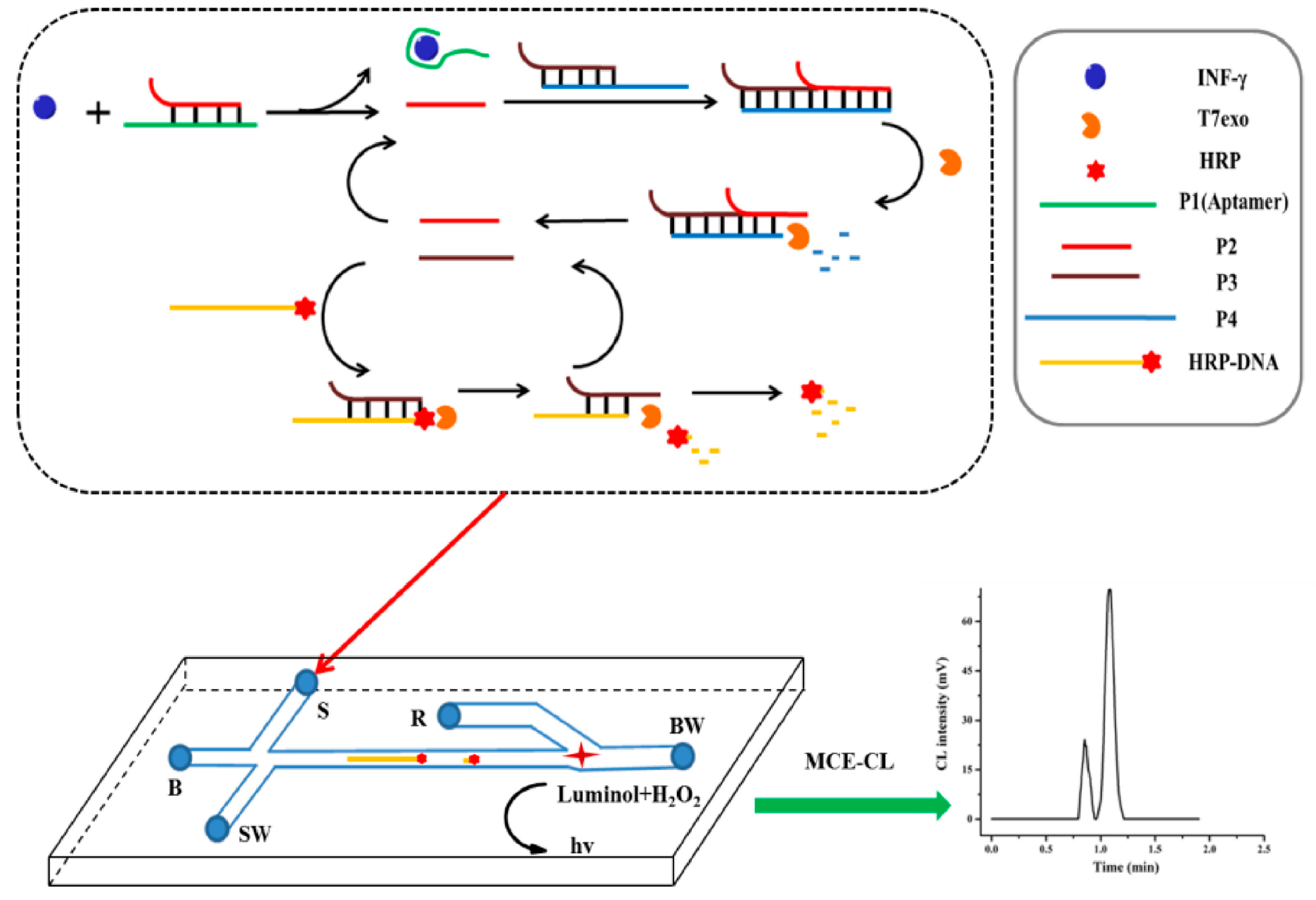
2.3. Detection of Proteins Using Amplification Strategies Based on Aptamer-Integrated Microchip
2.4. Detection of Bacteria Using Amplification Strategies Based on Aptamer-Integrated Microchip
2.5. Detection of Cells Using Amplification Strategies Based on Aptamer-Integrated Microchip
3. Conclusions, Future Prospects, and Challenges
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, X.; He, Y.; Ma, Y.; Bie, Z.; Liu, B.; Liu, Z. Hybrid Approach Combining Boronate Affinity Magnetic Nanoparticles and Capillary Electrophoresis for Efficient Selection of Glycoprotein-Binding Aptamers. Anal. Chem. 2016, 88, 9805–9812. [Google Scholar] [CrossRef]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gan, N.; Zhou, Y.; Li, T.; Hu, F.; Cao, Y.; Chen, Y. Novel label-free and high-throughput microchip electrophoresis platform for multiplex antibiotic residues detection based on aptamer probes and target catalyzed hairpin assembly for signal amplification. Biosens. Bioelectron. 2017, 97, 100–106. [Google Scholar] [CrossRef]
- Hu, Y.; Jiang, G.; Wen, Y.; Shao, Y.; Yang, G.; Qu, F. Selection of aptamers targeting small molecules by capillary electrophoresis: Advances, challenges, and prospects. Biotechnol. Adv. 2025, 78, 108491. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Zourob, M. In vitro selection of DNA aptamers targeting β-lactoglobulin and their integration in graphene-based biosensor for the detection of milk allergen. Biosens. Bioelectron. 2017, 92, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liu, T.; Wang, Y.; Miao, P. Electrochemical aptasensors for detection of small molecules, macromolecules, and cells. Rev. Anal. Chem. 2016, 35, 201–211. [Google Scholar] [CrossRef]
- Bhardwaj, T.; Dalal, P.; Rathore, A.S.; Jha, S.K. An aptamer based microfluidic chip for impedimetric detection of Ranibizumab in a bioreactor. Sens. Actuators B Chem. 2020, 312, 127941. [Google Scholar] [CrossRef]
- Tivon, Y.; Falcone, G.; Deiters, A. Protein Labeling and Crosslinking by Covalent Aptamers. Angew. Chem. Int. Ed. 2021, 60, 15899–15904. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, F.; Zhang, Y.; Zhu, L.; Li, Y.; Zhao, S.; He, P.; Wang, Q. A sensitive assay based on specific aptamer binding for the detection of Salmonella enterica serovar Typhimurium in milk samples by microchip capillary electrophoresis. J. Chromatogr. A 2018, 1534, 188–194. [Google Scholar] [CrossRef]
- Bouvier-Mueller, A.; Duconge, F. Application of aptamers for in vivo molecular imaging and theranostics. Adv. Drug Deliv. Rev. 2018, 134, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef]
- Yuhan, J.; Zhu, L.; Zhu, L.; Huang, K.; He, X.; Xu, W. Cell-specific aptamers as potential drugs in therapeutic applications: A review of current progress. J. Control. Release 2022, 346, 405–420. [Google Scholar] [CrossRef]
- Kadioglu, O.; Malczyk, A.H.; Greten, H.J.; Efferth, T. Aptamers as a novel tool for diagnostics and therapy. Investig. New Drugs 2015, 33, 513–520. [Google Scholar] [CrossRef]
- McKeague, M.; Derosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012, 748913. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Chang, D.; He, S.; Li, Y. Aptamers from random sequence space: Accomplishments, gaps and future considerations. Anal. Chim. Acta 2022, 1196, 339511. [Google Scholar] [CrossRef]
- Walton, B.M.; Jackson, G.W.; Deutz, N.; Cote, G. Surface-enhanced Raman spectroscopy competitive binding biosensor development utilizing surface modification of silver nanocubes and a citrulline aptamer. J. Biomed. Opt. 2017, 22, 075002. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-H.; Eom, Y.-S.; Kim, T.-H. Recent Advances in Aptamer-Based Sensors for Sensitive Detection of Neurotransmitters. Biosensors 2023, 13, 413. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, X.; Wang, Q. Review of microchip analytical methods for the determination of pathogenic Escherichia coli. Talanta 2021, 232, 122410. [Google Scholar] [CrossRef]
- Xu, R.; Cheng, Y.; Li, X.; Zhang, Z.; Zhu, M.; Qi, X.; Chen, L.; Han, L. Aptamer-based signal amplification strategies coupled with microchips for high-sensitivity bioanalytical applications: A review. Anal. Chim. Acta 2022, 1209, 339893. [Google Scholar] [CrossRef]
- Jia, X.; Dong, S.; Wang, E. Engineering the bioelectrochemical interface using functional nanomaterials and microchip technique toward sensitive and portable electrochemical biosensors. Biosens. Bioelectron. 2016, 76, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Hennig, S.; Shu, Z.; Gutzweiler, L.; Koltay, P.; von Stetten, F.; Zengerle, R.; Früh, S.M. Paper-based open microfluidic platform for protein electrophoresis and immunoprobing. Electrophoresis 2022, 43, 621–631. [Google Scholar] [CrossRef]
- He, H.; Wang, X.; Tan, H.; Xiang, S.; Xu, Y. The culture of A549 cells and its secreted cytokine IL-6 monitoring on the designed multifunctional microfluidic chip. Talanta 2025, 285, 127395. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, M.; Li, X.; Ye, J.; Feng, Z.; Sun, W.; Hu, W. Gold nanoparticle-decorated fluorine-doped tin oxide substrate for sensitive label-free OIRD microarray chips. Anal. Bioanal. Chem. 2024, 416, 3775–3783. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Hu, X.; Wang, Q.; Zhang, Y. Recent advances in microchip-based methods for the detection of pathogenic bacteria. Chin. Chem. Lett. 2022, 33, 2817–2831. [Google Scholar] [CrossRef]
- Dawod, M.; Arvin, N.E.; Kennedy, R.T. Recent advances in protein analysis by capillary and microchip electrophoresis. Analyst 2017, 142, 1847–1866. [Google Scholar] [CrossRef]
- Olivares, C.; Ruppe, E.; Ferreira, S.; Corbel, T.; Andremont, A.; de Gunzburg, J.; Guedj, J.; Burdet, C. A modelling framework to characterize the impact of antibiotics on the gut microbiota diversity. Gut Microbes 2025, 17, 2442523. [Google Scholar] [CrossRef]
- Zhang, C.; Ning, K.; Zhang, W.; Guo, Y.; Chen, J.; Liang, C. Determination and removal of antibiotics in secondary effluent using a horizontal subsurface flow constructed wetland. Environ. Sci. Process. Impacts 2013, 15, 709–714. [Google Scholar] [CrossRef]
- Wang, Q.; Xue, Q.; Chen, T.; Li, J.; Liu, Y.; Shan, X.; Liu, F.; Jia, J. Recent advances in electrochemical sensors for antibiotics and their applications. Chin. Chem. Lett. 2021, 32, 609–619. [Google Scholar] [CrossRef]
- Li, Y.; Liu, C.; Li, Q.; Mao, S. Fluorescence analysis of antibiotics and antibiotic-resistance genes in the environment: A mini review. Chin. Chem. Lett. 2024, 35, 109541. [Google Scholar] [CrossRef]
- Dolati, S.; Ramezani, M.; Nabavinia, M.S.; Soheili, V.; Abnous, K.; Taghdisi, S.M. Selection of specific aptamer against enrofloxacin and fabrication of graphene oxide based label-free fluorescent assay. Anal. Biochem. 2018, 549, 124–129. [Google Scholar] [CrossRef]
- Hu, M.; Yue, F.; Dong, J.; Tao, C.; Bai, M.; Liu, M.; Zhai, S.; Chen, S.; Liu, W.; Qi, G.; et al. Screening of broad-spectrum aptamer and development of electrochemical aptasensor for simultaneous detection of penicillin antibiotics in milk. Talanta 2024, 269, 125508. [Google Scholar] [CrossRef]
- Zhao, T.; Chen, Q.; Wen, Y.; Bian, X.; Tao, Q.; Liu, G.; Yan, J. A competitive colorimetric aptasensor for simple and sensitive detection of kanamycin based on terminal deoxynucleotidyl transferase-mediated signal amplification strategy. Food Chem. 2022, 377, 132072. [Google Scholar] [CrossRef]
- Zhou, L.; Gan, N.; Zhou, Y.; Li, T.; Cao, Y.; Chen, Y. A label-free and universal platform for antibiotics detection based on microchip electrophoresis using aptamer probes. Talanta 2017, 167, 544–549. [Google Scholar] [CrossRef]
- Zhou, L.; Gan, N.; Hu, F.; Li, T.; Cao, Y.; Wu, D. Microchip electrophoresis array-based aptasensor for multiplex antibiotic detection using functionalized magnetic beads and polymerase chain reaction amplification. Sens. Actuators B-Chem. 2018, 263, 568–574. [Google Scholar] [CrossRef]
- Zhang, K.; Gan, N.; Shen, Z.; Cao, J.; Hu, F.; Li, T. Microchip electrophoresis based aptasensor for multiplexed detection of antibiotics in foods via a stir-bar assisted multi-arm junctions recycling for signal amplification. Biosens. Bioelectron. 2019, 130, 139–146. [Google Scholar] [CrossRef]
- Zhang, K.; Gan, N.; Hu, F.; Chen, X.; Li, T.; Cao, J. Microfluidic electrophoretic non-enzymatic kanamycin assay making use of a stirring bar functionalized with gold-labeled aptamer, of a fluorescent DNA probe, and of signal amplification via hybridization chain reaction. Microchim. Acta 2018, 185, 181. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hong, F.; Cao, Y.; Hu, F.; Wu, Y.; Wu, D.; Li, T.; Lin, J.; Gan, N. A microchip electrophoresis-based assay for ratiometric detection of kanamycin by R-shape probe and exonuclease-assisted signal amplification. Talanta 2018, 189, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Gan, N.; Wu, Y.; Hu, F.; Lin, J.; Cao, Y.; Wu, D. Multiplex detection of quality indicator molecule targets in urine using programmable hairpin probes based on a simple double-T type microchip electrophoresis platform and isothermal polymerase-catalyzed target recycling. Analyst 2018, 143, 2696–2704. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, J.; Chen, S.; Huang, Y.; Zhaok, S. An ultrasensitive microchip electrophoresis chemiluminescence assay platform for detection of trace biomolecules. J. Chromatogr. A 2020, 1613, 460693. [Google Scholar] [CrossRef]
- Xie, L.; Li, T.; Hu, F.; Jiang, Q.; Wang, Q.; Gan, N. A novel microfluidic chip and antibody-aptamer based multianalysis method for simultaneous determination of several tumor markers with polymerization nicking reactions for homogenous signal amplification. Microchem. J. 2019, 147, 454–462. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.T.; Johnstone, R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983, 33, 967–978. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.; Wang, Z.; Irfan, M.; Qu, F. Recent advances of aptasensors for exosomes detection. Biosens. Bioelectron. 2020, 160, 112213. [Google Scholar] [CrossRef]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schafer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In Vivo Imaging Reveals Extracellular Vesicle-Mediated Phenocopying of Metastatic Behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, R.; Sarvnaz, H.; Arabpour, M.; Ramshe, S.M.; Asef-Kabiri, L.; Yousefi, H.; Akbari, M.E.; Eskandari, N. Cancer exosomes and natural killer cells dysfunction: Biological roles, clinical significance and implications for immunotherapy. Mol. Cancer 2022, 21, 15. [Google Scholar] [CrossRef]
- Gao, X.; Teng, X.; Dai, Y.; Li, J. Rolling Circle Amplification-Assisted Flow Cytometry Approach for Simultaneous Profiling of Exosomal Surface Proteins. ACS Sens. 2021, 6, 3611–3620. [Google Scholar] [CrossRef]
- Gandhi, J.; Sushma, M.V.; Rengan, A.K.; Naik, M.N.; Mishra, D.K.; Boyinpally, S.R.; Joseph, J. Proteomic profiling of exosomes in a mouse model of Candida albicans endophthalmitis. Exp. Cell Res. 2022, 417, 113222. [Google Scholar] [CrossRef]
- Iha, K.; Tsurusawa, N.; Tsai, H.-Y.; Lin, M.-W.; Sonoda, H.; Watabe, S.; Yoshimura, T.; Ito, E. Ultrasensitive ELISA detection of proteins in separated lumen and membrane fractions of cancer cell exosomes. Anal. Biochem. 2022, 654, 114831. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, K.; Huang, X.; Qiao, L.; Liu, B. Mass Spectrometry Imaging of Mass Tag Immunoassay Enables the Quantitative Profiling of Biomarkers from Dozens of Exosomes. Anal. Chem. 2021, 93, 709–714. [Google Scholar] [CrossRef]
- Sina, A.A.I.; Vaidyanathan, R.; Wuethrich, A.; Carrascosa, L.G.; Trau, M. Label-free detection of exosomes using a surface plasmon resonance biosensor. Anal. Bioanal. Chem. 2019, 411, 1311–1318. [Google Scholar] [CrossRef]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between Cancerous Exosomes and Protein Markers Based on Surface-Enhanced Raman Spectroscopy (SERS) and Principal Component Analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhu, Y.; Zhao, R.; Wei, X.; Xin, X. Visualization of exosomes from mesenchymal stem cells in vivo by magnetic resonance imaging. Magn. Reson. Imaging 2020, 68, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, J.; Xie, Q.; Chu, Z.; Zhang, F.; Wang, Q. Ultrasensitive detection of exosomes by microchip electrophoresis combining with triple amplification strategies. Talanta 2023, 265, 124930. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, H.; Fu, J.; Wu, X.; Liang, X.-Y.; Liu, X.-Y.; Wu, X.; Cao, L.-L.; Xu, Z.-Y.; Dong, M. Microfluidic-based exosome isolation and highly sensitive aptamer exosome membrane protein detection for lung cancer diagnosis. Biosens. Bioelectron. 2022, 214, 114487. [Google Scholar] [CrossRef]
- Ismail, B.P.; Senaratne-Lenagala, L.; Stube, A.; Brackenridge, A. Protein demand: Review of plant and animal proteins used in alternative protein product development and production. Anim. Front. 2020, 10, 53–63. [Google Scholar] [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int. J. Anal. Chem. 2016, 2016, 7436849. [Google Scholar] [CrossRef]
- Lin, X.; Sun, X.; Luo, S.; Liu, B.; Yang, C. Development of DNA-based signal amplification and microfluidic technology for protein assay: A review. Trac-Trends Anal. Chem. 2016, 80, 132–148. [Google Scholar] [CrossRef]
- Tarasova, I.A.; Masselon, C.D.; Gorshkov, A.V.; Gorshkov, M.V. Predictive chromatography of peptides and proteins as a complementary tool for proteomics. Analyst 2016, 141, 4816–4832. [Google Scholar] [CrossRef]
- Aydin, S. A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA. Peptides 2015, 72, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Yu, Q.; Lv, Z.; Du, X. Tandem Assays of Protein and Glucose with Functionalized Core/Shell Particles Based on Magnetic Separation and Surface-Enhanced Raman Scattering. Small 2013, 9, 3259–3264. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Mao, S.; Li, H.-F.; Lin, J.-M. Multi-DNAzymes-functionalized gold nanoparticles for ultrasensitive chemiluminescence detection of thrombin on microchip. Anal. Chim. Acta 2018, 1027, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Chen, Q.; Liu, W.; Yi, L.; Li, H.; Wang, Z.; Lin, J.-M. Assay of multiplex proteins from cell metabolism based on tunable aptamer and microchip electrophoresis. Biosens. Bioelectron. 2015, 63, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhao, J.; Huang, Y.; Zhao, S.; Liu, Y.-M. Aptamer-based microchip electrophoresis assays for amplification detection of carcinoembryonic antigen. Clin. Chim. Acta 2015, 450, 304–309. [Google Scholar] [CrossRef] [PubMed]
- Geng, X.; Chen, J.; Chu, Z.; Zhang, J.; Zhang, F.; Wang, Q. Highly sensitive detection of MUC1 by microchip electrophoresis combining with target recycling amplification and strand displacement amplification. J. Pharm. Biomed. Anal. 2022, 219, 114967. [Google Scholar] [CrossRef]
- Xu, Y.; Hassan, M.M.; Sharma, A.S.; Li, H.; Chen, Q. Recent advancement in nano-optical strategies for detection of pathogenic bacteria and their metabolites in food safety. Crit. Rev. Food Sci. Nutr. 2023, 63, 486–504. [Google Scholar] [CrossRef]
- Voitoux, E.; Lafarge, V.; Collette, C.; Lombard, B. Applicability of the draft standard method for the detection of Escherichia coli O157 in dairy products. Int. J. Food Microbiol. 2002, 77, 213–221. [Google Scholar] [CrossRef]
- Dimov, N.; McDonnell, M.B.; Munro, I.; McCluskey, D.K.; Johnston, I.D.; Tan, C.K.L.; Coudron, L. Electrowetting-based Digital Microfluidics Platform for Automated Enzyme-linked Immunosorbent Assay. Jove-J. Vis. Exp. 2020, 2020, e60489. [Google Scholar]
- Furutani, S.; Naruishi, N.; Hagihara, Y.; Nagai, H. Development of an on-site rapid real-time polymerase chain reaction system and the characterization of suitable DNA polymerases for TaqMan probe technology. Anal. Bioanal. Chem. 2016, 408, 5641–5649. [Google Scholar] [CrossRef]
- Kawasaki, S.; Yamashoji, S.; Asakawa, A.; Isshiki, K.; Kawamoto, S. Menadione-catalyzed luminol chemiluminescence assay for the rapid detection of viable bacteria in foods under aerobic conditions. J. Food Prot. 2004, 67, 2767–2771. [Google Scholar] [CrossRef]
- Andrei, C.-C.; Moraillon, A.; Lau, S.; Felidj, N.; Yamakawa, N.; Bouckaert, J.; Larquet, E.; Boukherroub, R.; Ozanam, F.; Szunerits, S.; et al. Rapid and sensitive identification of uropathogenic Escherichia coli using a surface-enhanced-Raman-scattering-based biochip. Talanta 2020, 219, 121174. [Google Scholar] [CrossRef]
- Xu, M.; Wang, R.; Li, Y. Electrochemical biosensors for rapid detection of Escherichia coli O157:H7. Talanta 2017, 162, 511–522. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, L.; He, P.; Zi, F.; Hu, X.; Wang, Q. Sensitive assay of Escherichia coli in food samples by microchip capillary electrophoresis based on specific aptamer binding strategy. Talanta 2019, 197, 284–290. [Google Scholar] [CrossRef]
- Luo, F.; Li, Z.; Dai, G.; Lu, Y.; He, P.; Wang, Q. Ultrasensitive biosensing pathogenic bacteria by combining aptamer-induced catalysed hairpin assembly circle amplification with microchip electrophoresis. Sens. Actuators B-Chem. 2020, 306, 127577. [Google Scholar] [CrossRef]
- Luo, F.; Li, Z.; Dai, G.; Lu, Y.; He, P.; Wang, Q. Simultaneous detection of different bacteria by microchip electrophoresis combined with universal primer-duplex polymerase chain reaction. J. Chromatogr. A 2020, 1615, 460734. [Google Scholar] [CrossRef]
- Jiang, Y.; Qiu, Z.; Le, T.; Zou, S.; Cao, X. Developing a dual-RCA microfluidic platform for sensitive E. coli O157:H7 whole-cell detections. Anal. Chim. Acta 2020, 1127, 79–88. [Google Scholar] [CrossRef]
- Li, S.; Jiang, Y.; Yang, X.; Lin, M.; Dan, H.; Zou, S.; Cao, X. In situ rolling circle amplification surface modifications to improve E. coli O157:H7 capturing performances for rapid and sensitive microfluidic detection applications. Anal. Chim. Acta 2021, 1150, 338229. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Zou, S.; Cao, X. A simple dendrimer-aptamer based microfluidic platform for E. coli O157:H7 detection and signal intensification by rolling circle amplification. Sens. Actuators B-Chem. 2017, 251, 976–984. [Google Scholar] [CrossRef]
- Li, T.; Ou, G.; Chen, X.; Li, Z.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Naked-eye based point-of-care detection of E. coli O157: H7 by a signal-amplified microfluidic aptasensor. Anal. Chim. Acta 2020, 1130, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Su, M.; Li, L.; Lan, F.; Yang, G.; Ge, S.; Yu, J.; Song, X. Aptamer-based fluorescent and visual biosensor for multiplexed monitoring of cancer cells in microfluidic paper-based analytical devices. Sens. Actuators B-Chem. 2016, 229, 347–354. [Google Scholar] [CrossRef]
- Sheng, W.; Chen, T.; Tan, W.; Fan, Z.H. Multivalent DNA Nanospheres for Enhanced Capture of Cancer Cells in Microfluidic Devices. ACS Nano 2013, 7, 7067–7076. [Google Scholar] [CrossRef]
- Zhang, W.; He, Z.; Yi, L.; Mao, S.; Li, H.; Lin, J.-M. A dual-functional microfluidic chip for on-line detection of interleukin-8 based on rolling circle amplification. Biosens. Bioelectron. 2018, 102, 652–660. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, L.; Nie, W.; Huang, L.; Zhang, J.; Li, F.; Xie, H.-Y. Biomimetic Microfluidic System for Fast and Specific Detection of Circulating Tumor Cells. Anal. Chem. 2019, 91, 15726–15731. [Google Scholar] [CrossRef]
- Ngoc-Viet, N.; Jen, C.-P. Selective Detection of Human Lung Adenocarcinoma Cells Based on the Aptamer-Conjugated Self-Assembled Monolayer of Gold Nanoparticles. Micromachines 2019, 10, 195. [Google Scholar]
- Ngoc-Viet, N.; Yang, C.-H.; Liu, C.-J.; Kuo, C.-H.; Wu, D.-C.; Jen, C.-P. An Aptamer-Based Capacitive Sensing Platform for Specific Detection of Lung Carcinoma Cells in the Microfluidic Chip. Biosensors 2018, 8, 98. [Google Scholar] [CrossRef]
- Khaksari, S.; Ameri, A.R.; Taghdisi, S.M.; Sabet, M.; Bami, S.M.J.G.; Abnous, K.; Shaegh, S.A.M. A microfluidic electrochemical aptasensor for highly sensitive and selective detection of A549 cells as integrin α604-containing cell model via IDA aptamers. Talanta 2023, 252, 123781. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tao, C.; Yin, J.; Wang, Y.; Li, Y. Enhancing the response rate of strand displacement-based electrochemical aptamer sensors using bivalent binding aptamer-cDNA probes. Biosens. Bioelectron. 2018, 103, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.; Baker, B.R.; Wachsmann-Hogiu, S.; Pagba, C.V.; Laurence, T.A.; Lane, S.M.; Lee, L.P.; Tok, J.B.H. Aptamer-Based SERRS Sensor for Thrombin Detection. Nano Lett. 2008, 8, 4386–4390. [Google Scholar] [CrossRef]
- Tang, T.; Liu, Y.; Jiang, Y. Recent Progress on Highly Selective and Sensitive Electrochemical Aptamer-based Sensors. Chem. Res. Chin. Univ. 2022, 38, 866–878. [Google Scholar] [CrossRef]
- Xie, C.; Chen, N.; Yang, Y.; Yuan, Q. Recent Progress of Aptamer Functionalized Two-dimensional Materials Field Effect Transistor Sensors. Chem. J. Chin. Univ.-Chin. 2021, 42, 3406–3420. [Google Scholar]
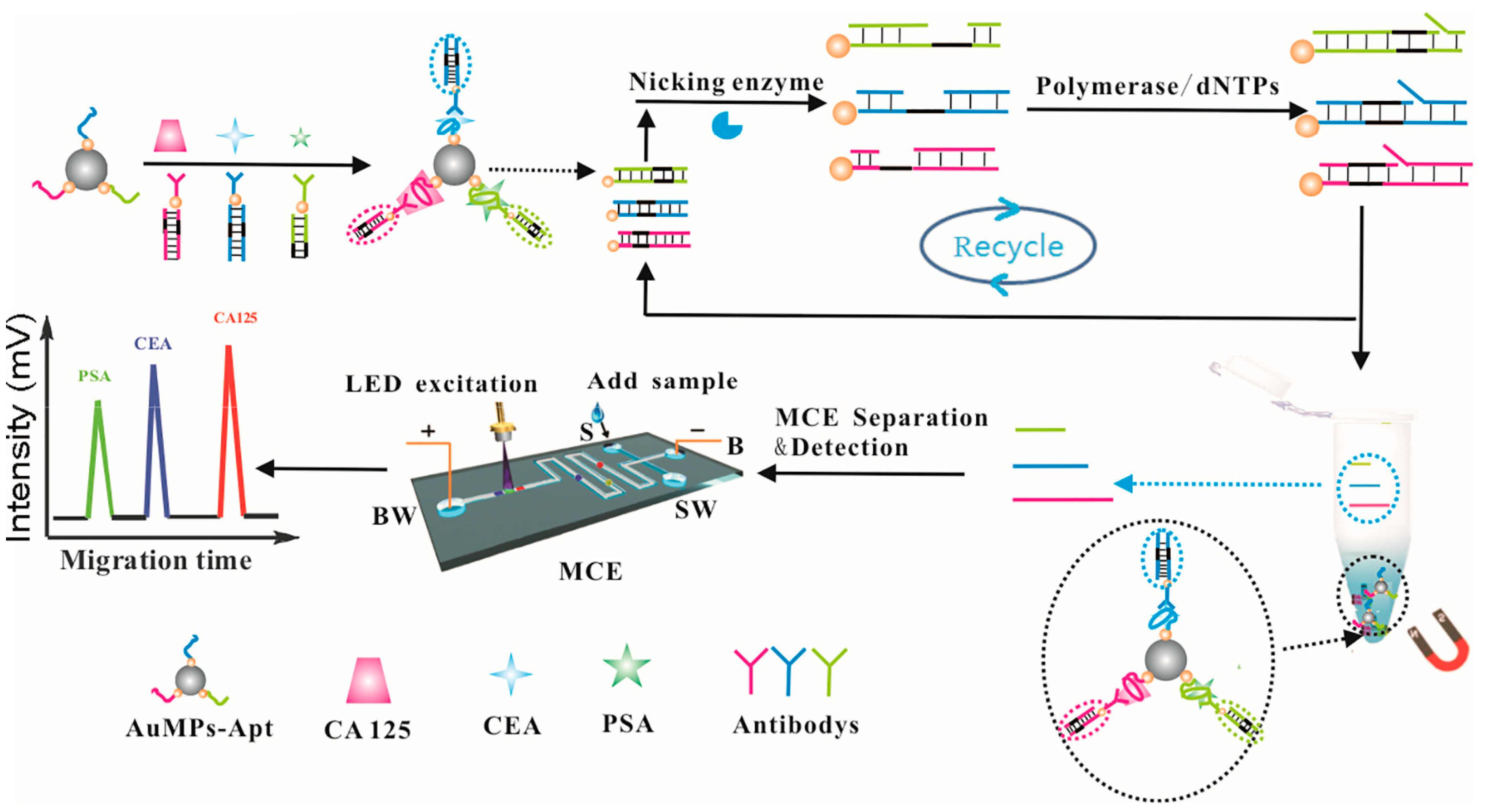

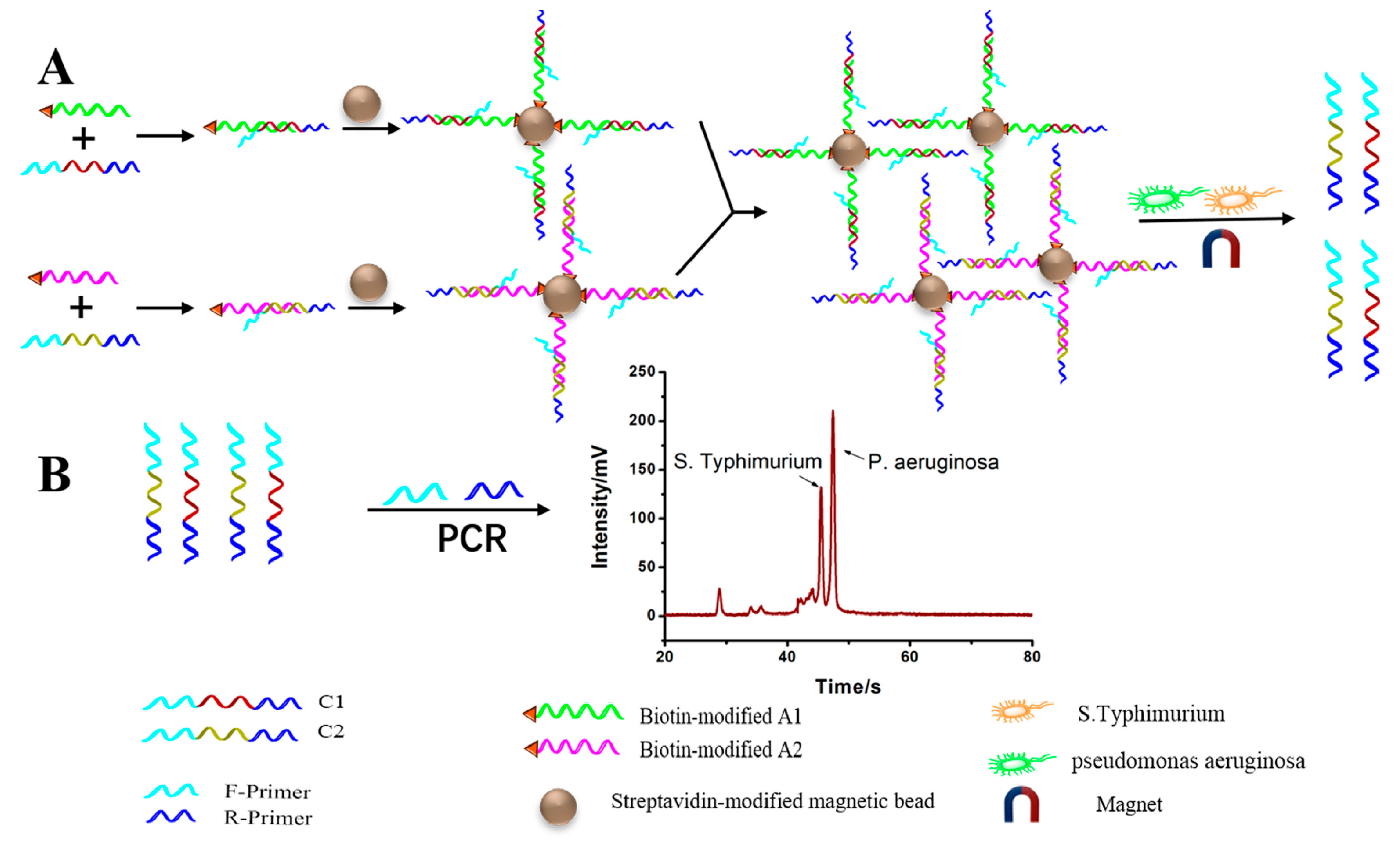
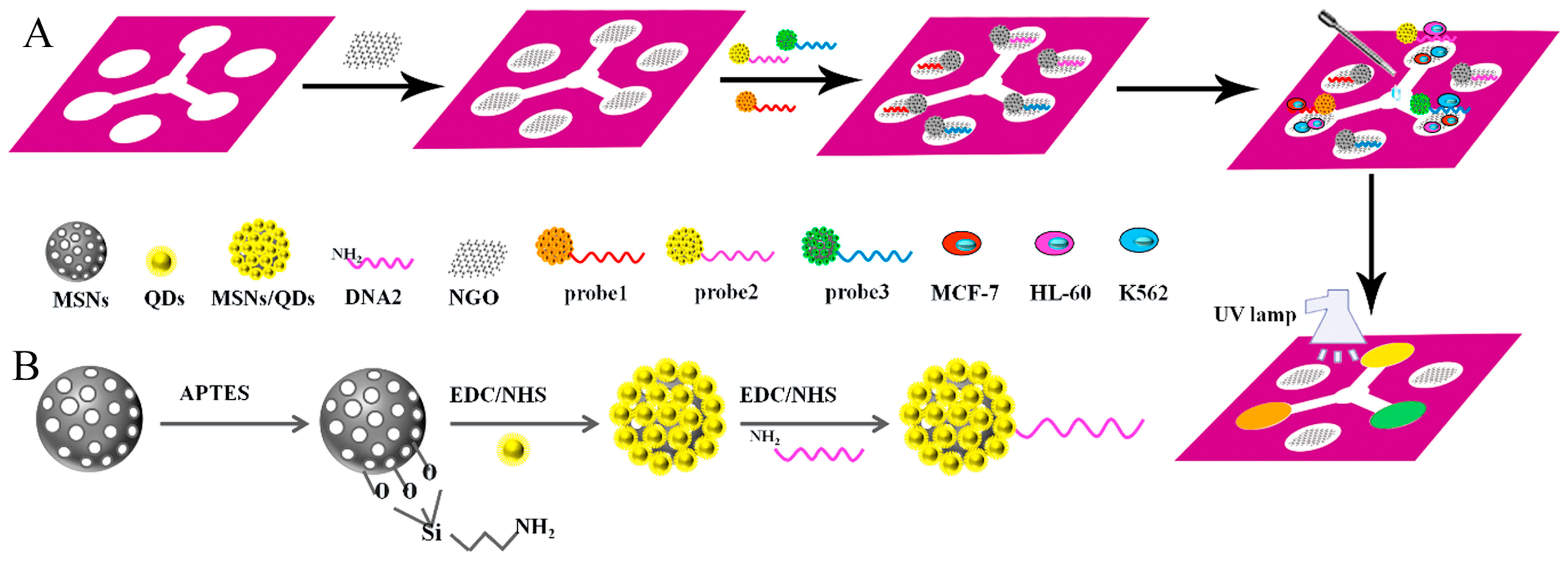
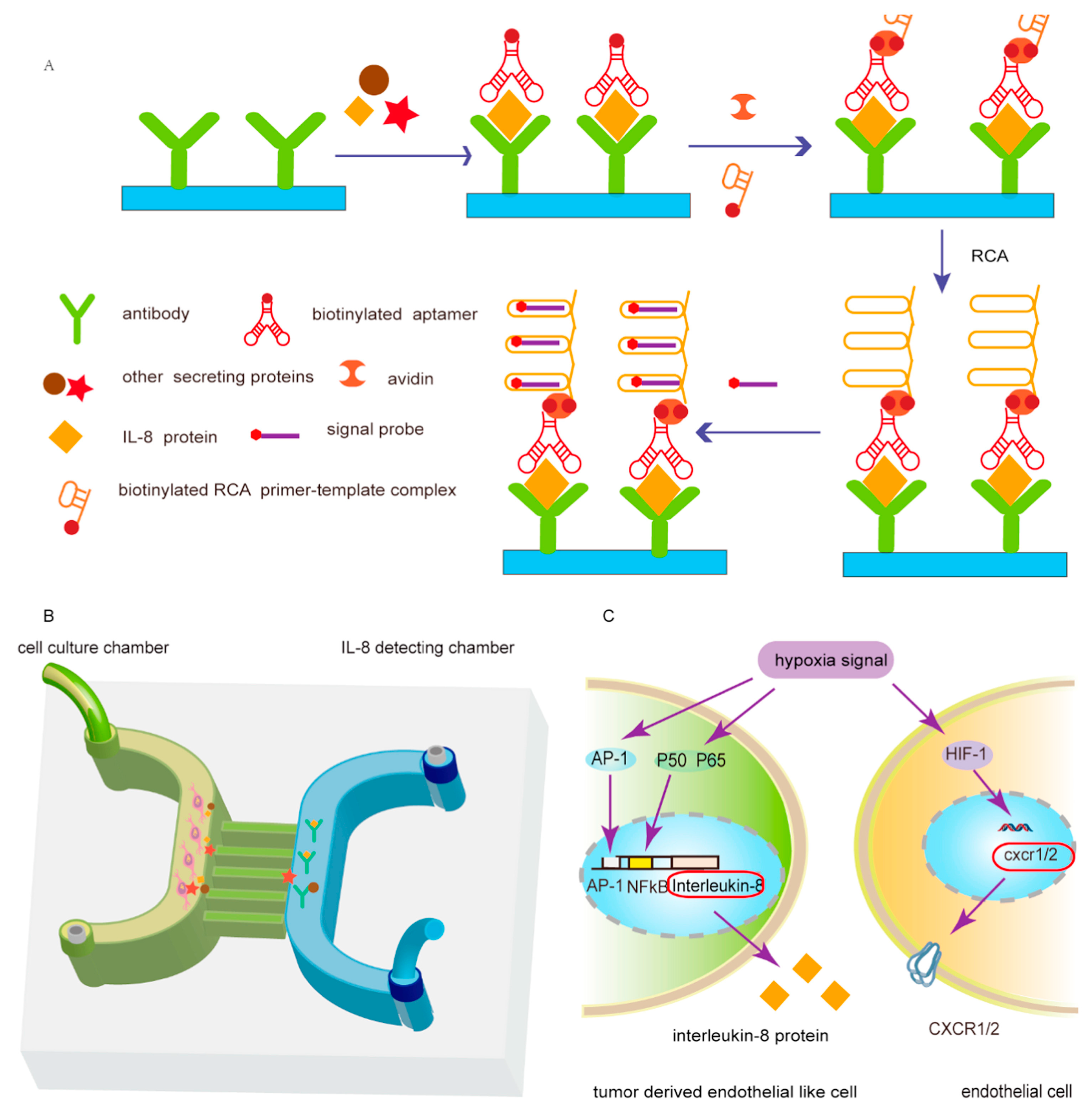
| Applications | Target | Signal Amplification | Linear Range | LOD | Reference |
|---|---|---|---|---|---|
| small molecules | chloramphenicol | combining aptamer and MCE | 0.008–1.0 ng/mL | 0.003 ng mL−1 | [34] |
| Kana and OTC | CHA | 0.001–10.0 ng/mL | 0.7 pg mL−1 and 0.9 pg mL−1 | [4] | |
| KANA and CAP | magnetic beads and PCR | 0.0025–10 nM and 0.006–10 nM | 1.2 pg mL−1 (0.0025 nM) and 1.9 pg mL−1 (0.006 nM) | [35] | |
| CAP and Kana | a stir-bar assisted multi-arm junctions recycling | 0.001 to 40 ng mL−1 | 0.52 pg mL−1 and 0.41 pg mL−1 | [36] | |
| Kana | HCR | 1.0 to 10.0 ng mL−1 | 0.29 pg mL−1 | [37] | |
| Kana | exonuclease assisted signal amplification | 0.5 to 10.0 ng mL−1 | 150 fg mL−1 | [38] | |
| AMPI, ATP, E2 | isothermal polymerase-catalyzed target recycling | 1–100 nM, 0.05–10 nM, and 0.1–50 nM | 0.017 pg mL−1 (0.05 nM), 5.07 pg mL−1 (1 nM), and 0.027 pg mL−1 (0.1 nM) | [39] | |
| PSA, CEA, and CA125 | polymerization nicking reactions | 1 pg mL−1–1.0 ng mL−1 | 0.1 pg mL−1, 0.15 pg mL−1 and 0.12 pg mL−1 | [41] | |
| exosomes | human breast cancer cell | SDA and CHA | 44–8.75 × 105 particle μL−1 | 26 particle μL−1 | [54] |
| PD-L1 | magnetic bead | --- | 5.1 × 109 particles mL−1 | [55] | |
| proteins | thrombin | RCA | 1–25 pM | 0.55 pM | [62] |
| PDGF-BB and VEGF165 | micellar enrichment | 5.15–2.03 nM, and 3.14–2.53 nM | 5.00–150.0 nM | [63] | |
| CEA | magnetic bead assisted target-induced strand circle | 0.13–8.0 ng mL−1 | 0.068 ng mL−1 | [64] | |
| MUC1 | TRA and SDA | 1.0–1.0 × 103 pg mL−1 | 0.23 pg mL−1 | [65] | |
| bacteria | S. Typhimurium | micellar enrichment | 1.2 × 103–7.5 × 105 CFU mL−1 | 3.37 × 102 CFU mL−1 | [10] |
| E. coli O157:H7 | CHA | 2 × 102–2 × 105 CFU mL−1 | 75 CFU mL−1 | [74] | |
| S. typhimurium and P. aeruginosa | UP-DPCR | 5 × 101–5 × 106 CFU mL−1 and 1 × 101–5 × 104 CFU mL−1 | 15 CFU mL−1 and 5 CFU mL−1 | [75] | |
| E. coli O157:H7 | RCA | 102 to 105 cells mL−1 | 120 cells mL−1 | [76] | |
| E. coli O157:H7 | cRCA | 102 to 105 cells mL−1 | 5 cells mL−1 | [77] | |
| E. coli O157:H7 | RCA | 102 to 105 cells mL−1 | 102 cells mL−1 | [78] | |
| E. coli O157:H7 | HCR | 500–5 × 107 CFU mL−1 | 250 CFU mL−1 | [79] | |
| cells | MCF-7 cells, K562 cells, and HL-60 cells | graphene oxide | 180–8 × 107, 210–7 × 107 and 200–7 × 107 cells mL−1 | 62 cells mL−1, 65 cells mL−1, and 70 cells mL−1 | [80] |
| CTCs | multivalent DNA nanospheres | 100–105 cells mL−1 | 100 cells mL−1 | [81] | |
| IL-8 | RCA | 7.5–120 pg mL−1 | 0.84 pmol L−1 | [82] | |
| circulating tumor cells | Fe3O4 magnetic nanoclusters | 5–103 cells mL−1 | 5 cells mL−1 | [83] | |
| A549 cells and HeLa cells | gold nanoparticles | 105–106 cells mL−1 | 5 × 105 cells mL−1 and 106 cells mL−1 | [84] | |
| A549 cells | a process of self-assembled monolayers | 1 × 105–5 × 105 cells mL−1 | 1.5 × 104 cells mL−1 | [85] | |
| A549 cells | surface modification | 50–5 × 105 cells mL−1 | 14 cells mL−1 | [86] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xue, X.; Hou, Y.; Hu, C.; Zhang, Y. Review of Microchip Analytical Methods Coupled with Aptamer-Based Signal Amplification Strategies for High-Sensitivity Bioanalytical Applications. Biosensors 2025, 15, 653. https://doi.org/10.3390/bios15100653
Xue X, Hou Y, Hu C, Zhang Y. Review of Microchip Analytical Methods Coupled with Aptamer-Based Signal Amplification Strategies for High-Sensitivity Bioanalytical Applications. Biosensors. 2025; 15(10):653. https://doi.org/10.3390/bios15100653
Chicago/Turabian StyleXue, Xudong, Yanli Hou, Caihua Hu, and Yan Zhang. 2025. "Review of Microchip Analytical Methods Coupled with Aptamer-Based Signal Amplification Strategies for High-Sensitivity Bioanalytical Applications" Biosensors 15, no. 10: 653. https://doi.org/10.3390/bios15100653
APA StyleXue, X., Hou, Y., Hu, C., & Zhang, Y. (2025). Review of Microchip Analytical Methods Coupled with Aptamer-Based Signal Amplification Strategies for High-Sensitivity Bioanalytical Applications. Biosensors, 15(10), 653. https://doi.org/10.3390/bios15100653





