Recent Advances in and Application of Fluorescent Microspheres for Multiple Nucleic Acid Detection
Abstract
1. Introduction
2. Fluorescent Microsphere Synthesis and Modification
2.1. Methods for the Preparation of Fluorescent Microspheres
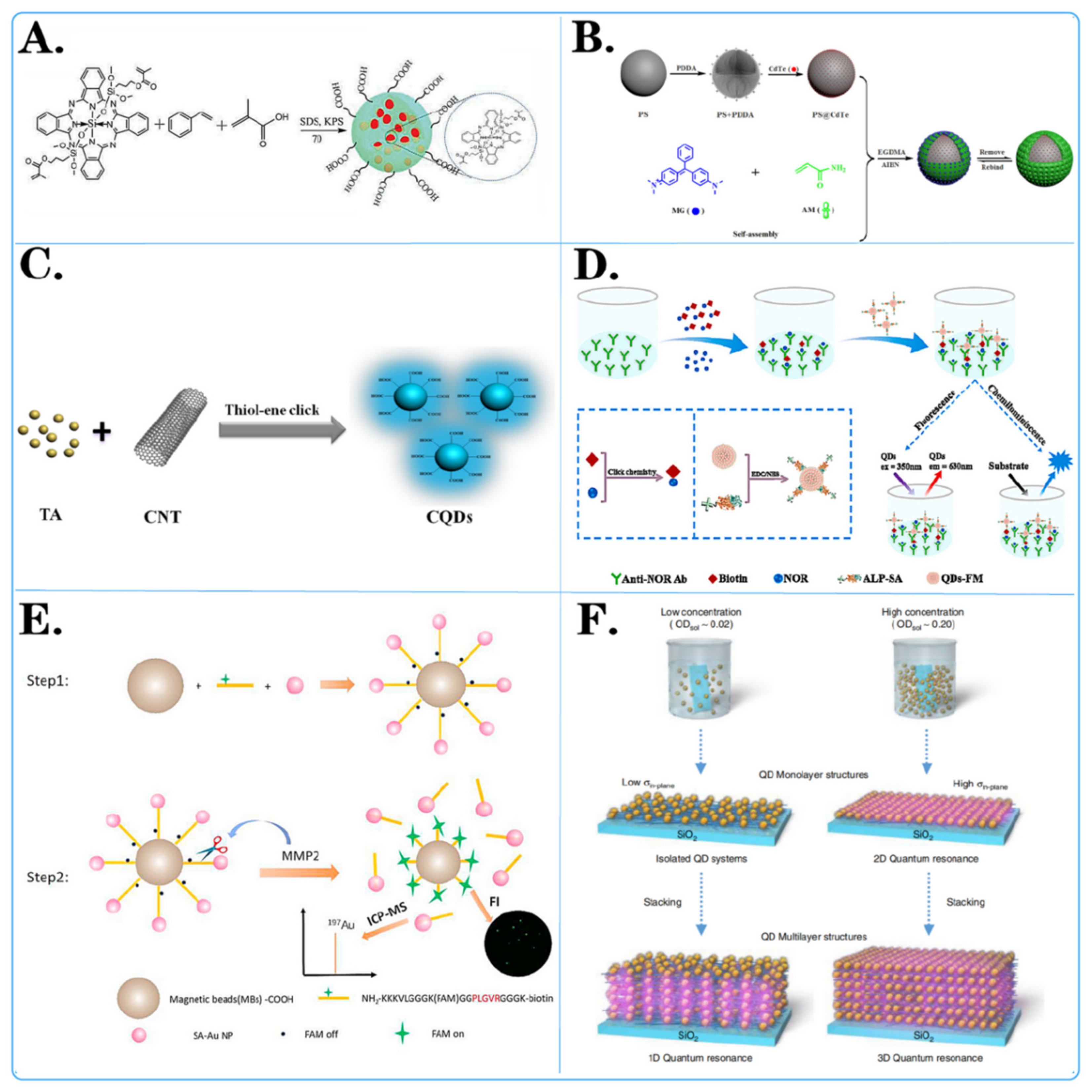
2.1.1. Adsorption Method
2.1.2. Embedding Method
2.1.3. Self-Assembly Method
2.1.4. Chemical Bonding Method
2.1.5. Copolymerization Method
2.2. Surface Modification Method for Fluorescent Microspheres
2.2.1. Chemical Modification Method
2.2.2. Biological Modification Method
2.2.3. Physical Modification Method
3. Applications of Fluorescent Microspheres in Multiplex Nucleic Acid Detection
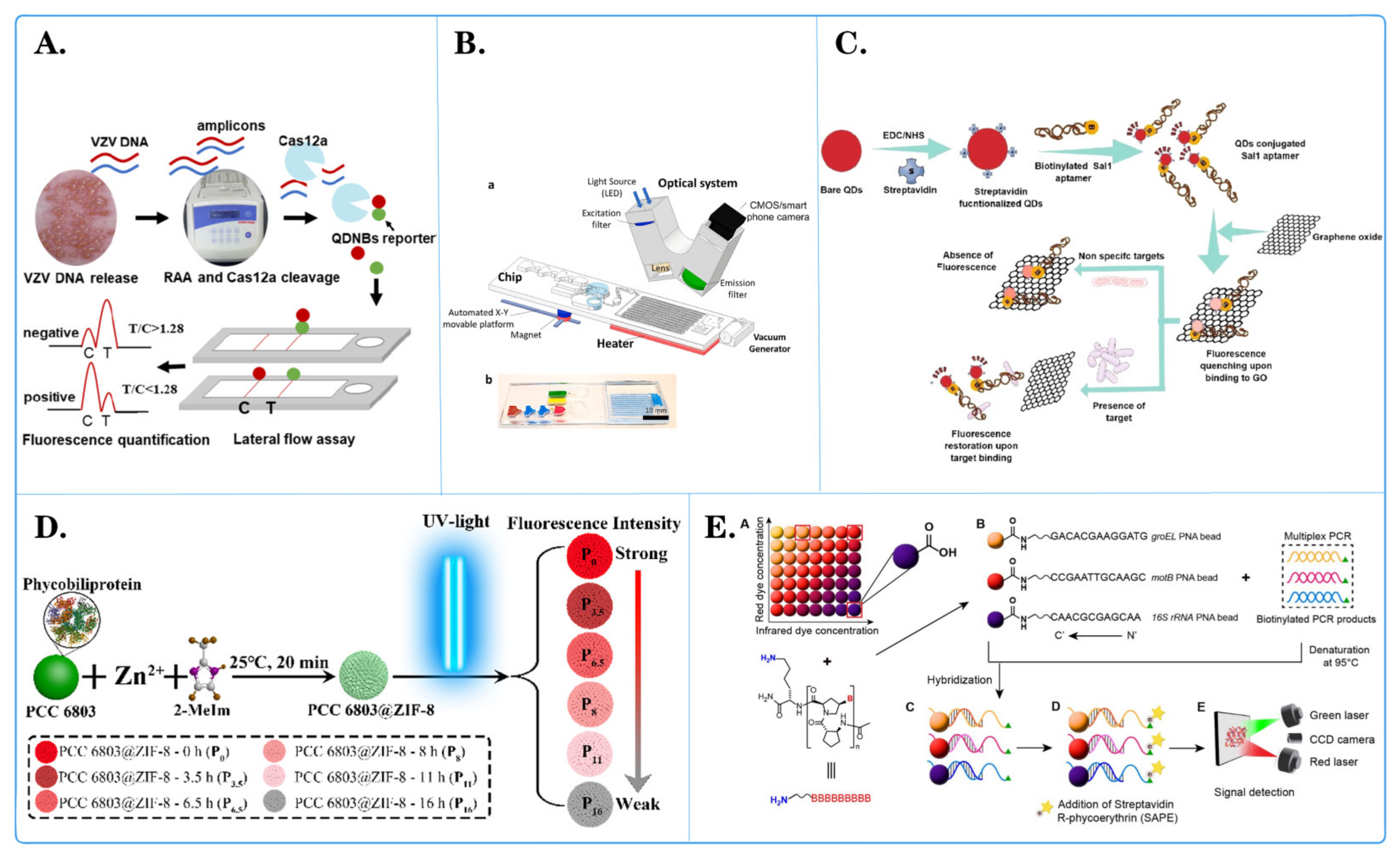
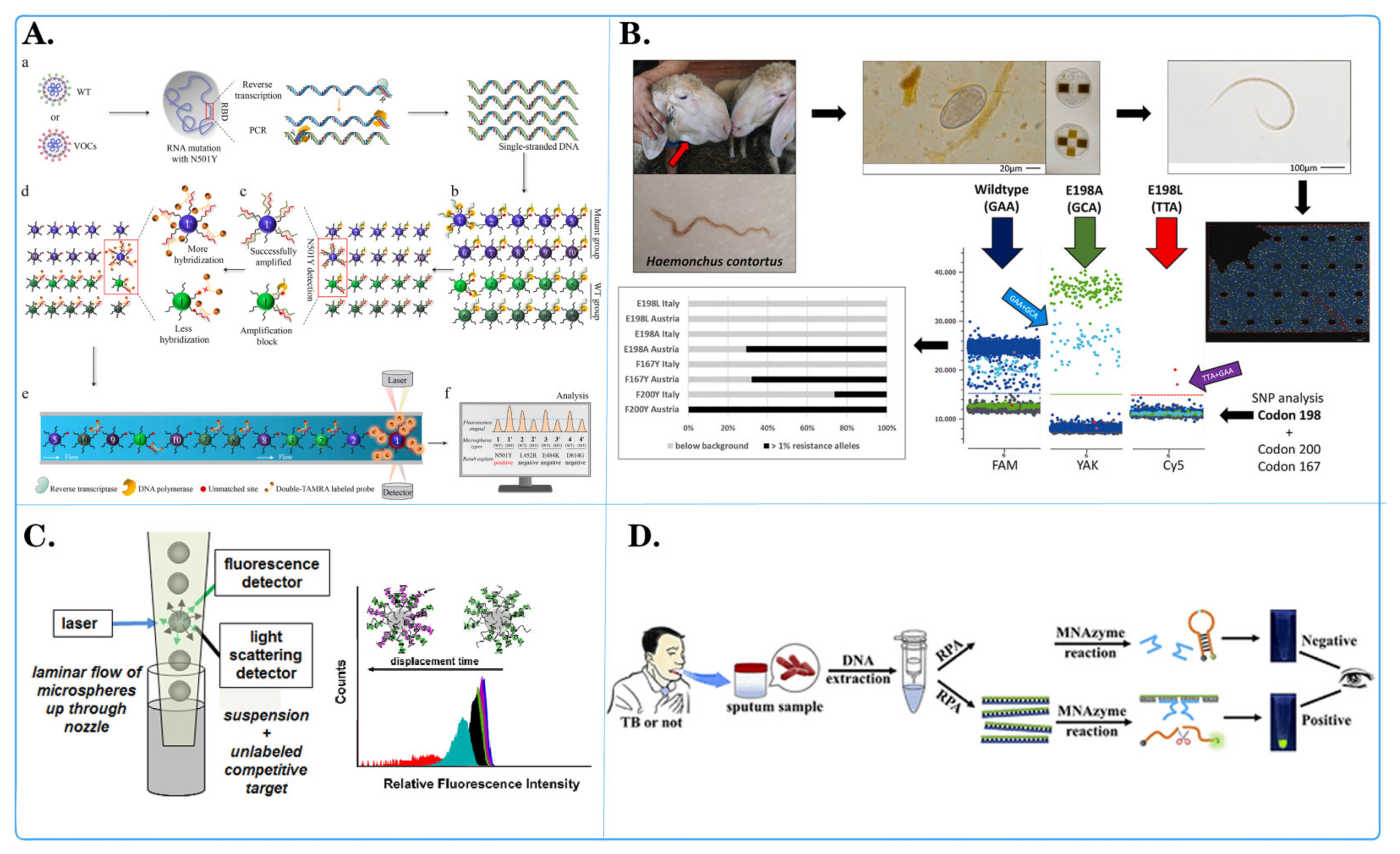
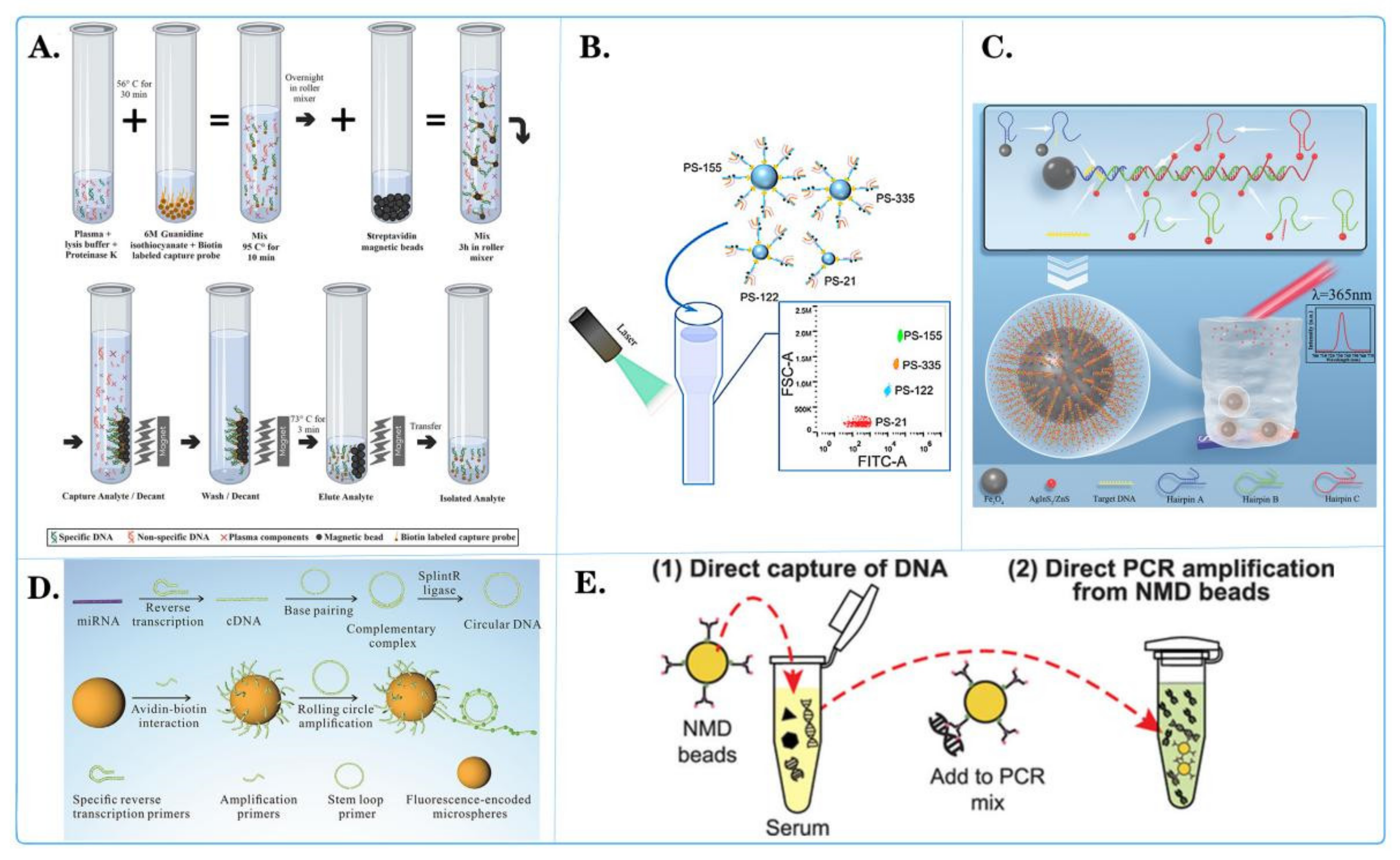
3.1. Application of Fluorescent Microspheres in Pathogen Detection
| Microsphere Type | Detection System | Detection Object | Advantage | Document Source |
|---|---|---|---|---|
| Magnetic microspheres with anti-TAG oligonucleotide coupling | Luminex xTAG | PKoV, PAstV, PEDV, PSaV, PSV, PTV, PDCoV, TGEV, BVDV, PoRV, and PToV virus DNA |
| [85] |
| Quantum dot nanobeads (QDNB) | Side-flow analysis (CQ-LFA) | VZV virus DNA |
| [86] |
| Quantum dots (QD) | A simple and versatile aptamer sensor based on fluorescence resonance energy transfer (FRET) | ssDNA of Salmonella paratyphi A |
| [87] |
| Fluorescent PCC 6803@ZIF-8 | A bead-based assay platform | DNA insertion sequence of Mycobacterium tuberculosis |
| [88] |
| RNA-bound magnetic beads | Digital reverse transcription recombinase polymerase amplification (DRT-RPA) | RNA of SARS-CoV-2 |
| [89] |
| #MC10012, #MC10015, #MC10021, Luminex | Microbead array method | DNA of Bacillus cereus |
| [90] |
| Polystyrene microsphere | Fluorescence encoding method | DNA sequences of COVID-19 (with related mutations) |
| [91] |
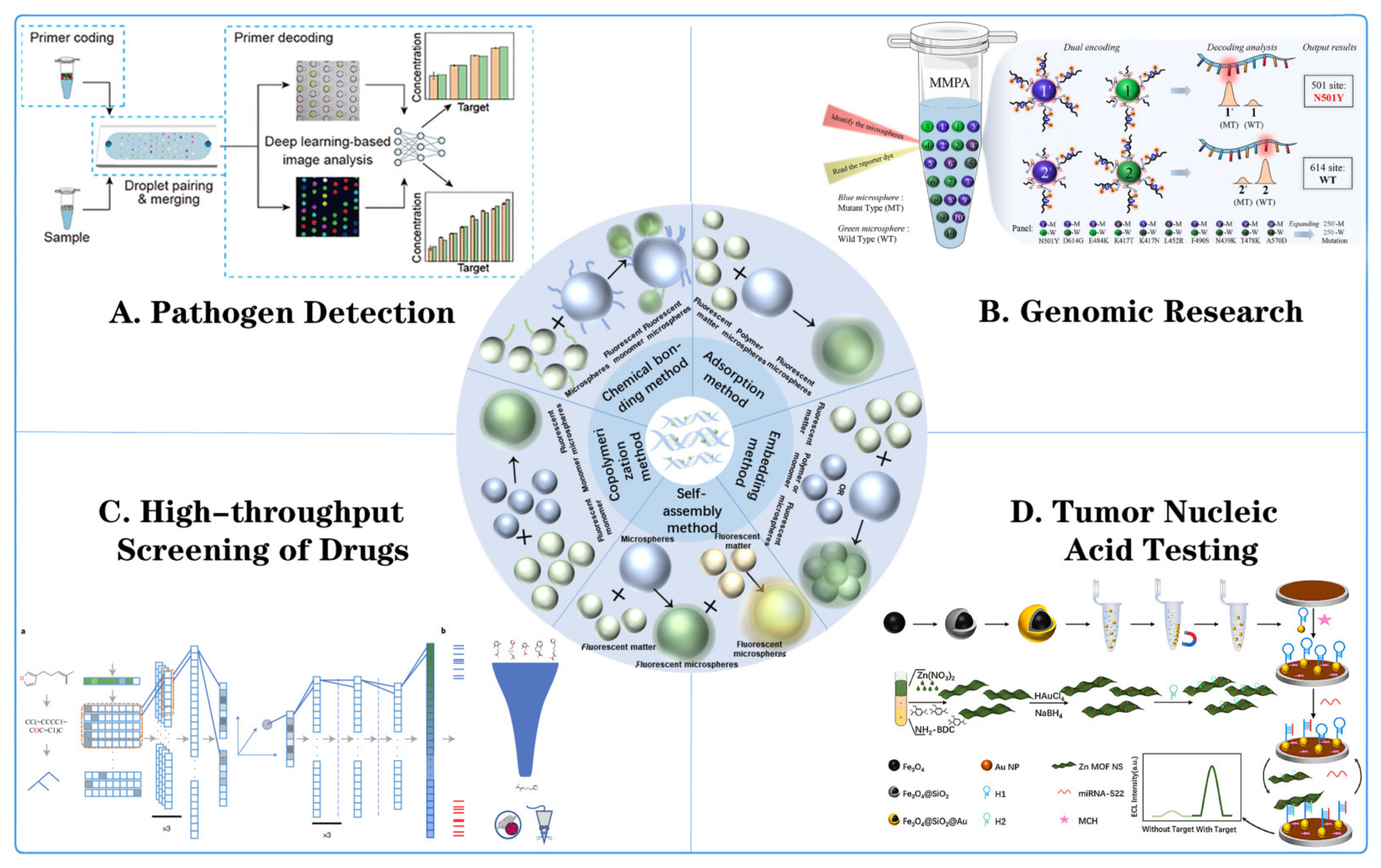
3.2. Application of Fluorescent Microspheres in Genomic Research
3.3. Application of Fluorescent Microspheres in High-Throughput Drug Screening
3.4. Application of Fluorescent Microspheres in Tumor Nucleic Acid Detection
4. Conclusions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Wang, H.; Liu, R.; Dong, K.; Zhang, L.; Zhang, J.; Zhang, X.; Zhang, J.; Xiao, X.; Zhang, W.; Wang, X. A universal and sensitive gene mutation detection method based on CRISPR-Cas12a. Anal. Chim. Acta 2023, 1246, 340886. [Google Scholar] [CrossRef] [PubMed]
- Ishige, T.; Itoga, S.; Matsushita, K. Locked Nucleic Acid Technology for Highly Sensitive Detection of Somatic Mutations in Cancer. Adv. Clin. Chem. 2018, 83, 53–72. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Hsiao, J.; Wahlestedt, C. Nucleic Acid-Based Therapeutics in Orphan Neurological Disorders: Recent Developments. Front. Mol. Biosci. 2021, 8, 643681. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; He, X.; Zhang, T.; Zhao, K.; Xiao, C.; Tong, Z.; Jin, L.; He, N.; Deng, Y.; Li, S.; et al. Highly sensitive smartphone-based detection of Listeria monocytogenes using SYTO9. Chin. Chem. Lett. 2022, 33, 1933–1935. [Google Scholar] [CrossRef]
- Song, W.; Feng, Y.; Zhang, J.; Kong, D.; Fan, J.; Zhao, M.; Hua, L.; Xiang, J.; Tang, X.; Xiao, S.; et al. Development of a multiplex reverse transcription-quantitative PCR (qPCR) method for detecting common causative agents of swine viral diarrhea in China. Porc. Health Manag. 2024, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, T.; Yang, G.; Deng, Y.; Mou, X.; He, N. A simple AuNPs-based colorimetric aptasensor for chlorpyrifos detection. Chin. Chem. Lett. 2022, 33, 1913–1916. [Google Scholar] [CrossRef]
- Wang, W.; He, S.; Dong, G.; Sheng, C. Nucleic-Acid-Based Targeted Degradation in Drug Discovery. J. Med. Chem. 2022, 65, 10217–10232. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Jia, F.; Wang, P.; Zhang, K. Nucleic acid-based drug delivery strategies. J. Control. Release 2020, 323, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.G.; Shen, J.; Yang, J.; Wang, J.W.; Zhao, R.C.; Zhang, T.L.; Guo, J.; Zhang, X. Nucleic acid drug vectors for diagnosis and treatment of brain diseases. Signal Transduct. Target. Ther. 2023, 8, 39. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.M.; Kim, I.H.; Kim, S. Nucleic Acid Testing of SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 6150. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ma, X.; Zhang, X.; Hu, G.; Deng, Y.; Li, S.; Chen, Z.; He, N.; Wu, Y.; Jiang, Z. Novel aerosol detection platform for SARS-CoV-2: Based on specific magnetic nanoparticles adsorption sampling and digital droplet PCR detection. Chin. Chem. Lett. 2023, 34, 107701. [Google Scholar] [CrossRef] [PubMed]
- Rai, P.; Kumar, B.K.; Deekshit, V.K.; Karunasagar, I.; Karunasagar, I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021, 105, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Safiabadi Tali, S.H.; LeBlanc, J.J.; Sadiq, Z.; Oyewunmi, O.D.; Camargo, C.; Nikpour, B.; Armanfard, N.; Sagan, S.M.; Jahanshahi-Anbuhi, S. Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection. Clin. Microbiol. Rev. 2021, 34, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.P.; Zar, H.J. Advances in the diagnosis of pulmonary tuberculosis in children. Paediatr. Respir. Rev. 2020, 36, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Krebs, M.G.; Malapelle, U.; Andre, F.; Paz-Ares, L.; Schuler, M.; Thomas, D.M.; Vainer, G.; Yoshino, T.; Rolfo, C. Practical Considerations for the Use of Circulating Tumor DNA in the Treatment of Patients with Cancer: A Narrative Review. JAMA Oncol. 2022, 8, 1830–1839. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xiao, B.; Xuan, J.; Sun, C.; Li, L. Current and future clinical applications of multiple nucleic acid detection technology. Chin. J. Lab. Med. 2022, 45, 428–432. [Google Scholar] [CrossRef]
- Liu, W.; Tong, M.; Lin, F.Y.; Gao, X.D.; Liu, J.Y. Progress in clinical application of molecular diagnostic techniques. Biotechnol. Lett. 2020, 31, 240–250. [Google Scholar]
- Zhu, H.; Zhang, H.; Xu, Y.; Lassakova, S.; Korabecna, M.; Neuzil, P. PCR past, present and future. Biotechniques 2020, 69, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2018, 144, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Jensen, E. Technical review: In situ hybridization. Anat. Rec. 2014, 297, 1349–1353. [Google Scholar] [CrossRef] [PubMed]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: A review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and Emerging Technologies. Semin. Thromb. Hemost. 2019, 45, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Prasad, D. Isothermal nucleic acid amplification and its uses in modern diagnostic technologies. 3 Biotech 2023, 13, 200. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Mahanama, A.; Wilson-Davies, E. Insight into PCR testing for surgeons. Surgery 2021, 39, 759–768. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Parvin, R.; Fan, Q.; Ye, F. Emerging digital PCR technology in precision medicine. Biosens. Bioelectron. 2022, 211, 114344. [Google Scholar] [CrossRef] [PubMed]
- Gall, J.G. The origin of in situ hybridization—A personal history. Methods 2016, 98, 4–9. [Google Scholar] [CrossRef] [PubMed]
- Zito Marino, F.; Brunelli, M.; Rossi, G.; Calabrese, G.; Calio, A.; Nardiello, P.; Martignoni, G.; Squire, J.A.; Cheng, L.; Massi, D.; et al. Multitarget fluorescence in situ hybridization diagnostic applications in solid and hematological tumors. Expert. Rev. Mol. Diagn. 2021, 21, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kang, Y.; Luo, J.; Pang, K.; Xu, X.; Wu, J.; Li, X.; Jin, S. Next-Generation Sequencing Reveals the Progression of COVID-19. Front. Cell. Infect. Microbiol. 2021, 11, 632490. [Google Scholar] [CrossRef] [PubMed]
- Wensel, C.R.; Pluznick, J.L.; Salzberg, S.L.; Sears, C.L. Next-generation sequencing: Insights to advance clinical investigations of the microbiome. J. Clin. Investig. 2022, 132. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Xu, F.; Wu, J.; Schubert, J.; Li, M.M. Application of Next Generation Sequencing in Laboratory Medicine. Ann. Lab. Med. 2021, 41, 25–43. [Google Scholar] [CrossRef] [PubMed]
- Obande, G.A.; Banga Singh, K.K. Current and Future Perspectives on Isothermal Nucleic Acid Amplification Technologies for Diagnosing Infections. Infect. Drug Resist. 2020, 13, 455–483. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Liao, C.; Liang, L.; Yi, X.; Zhou, Z.; Wei, G. Recent advances in recombinase polymerase amplification: Principle, advantages, disadvantages and applications. Front. Cell. Infect. Microbiol. 2022, 12, 1019071. [Google Scholar] [CrossRef] [PubMed]
- Largy, E.; Konig, A.; Ghosh, A.; Ghosh, D.; Benabou, S.; Rosu, F.; Gabelica, V. Mass Spectrometry of Nucleic Acid Noncovalent Complexes. Chem. Rev. 2022, 122, 7720–7839. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Chen, X.; Wang, Y. Mass Spectrometry for Assessing Protein-Nucleic Acid Interactions. Anal. Chem. 2023, 95, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Koblan, L.W.; Liu, D.R. Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 2020, 38, 824–844. [Google Scholar] [CrossRef] [PubMed]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed]
- Collias, D.; Beisel, C.L. CRISPR technologies and the search for the PAM-free nuclease. Nat. Commun. 2021, 12, 555. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas based virus detection: Recent advances and perspectives. Biosens. Bioelectron. 2021, 193, 113541. [Google Scholar] [CrossRef] [PubMed]
- Gabelica, V. Native Mass Spectrometry and Nucleic Acid G-Quadruplex Biophysics: Advancing Hand in Hand. Acc. Chem. Res. 2021, 54, 3691–3699. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, J.L. Recent developments in DNA adduct analysis using liquid chromatography coupled with mass spectrometry. J. Sep. Sci. 2020, 43, 31–55. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, Y.; Zhu, L.; Liu, H.; Su, X.; Liu, Y.; Chen, Z.; Chen, H.; He, N. A novel cartridge for nucleic acid extraction, amplification and detection of infectious disease pathogens with the help of magnetic nanoparticles. Chin. Chem. Lett. 2023, 34, 108092. [Google Scholar] [CrossRef]
- He, Y.; Hu, C.; Li, Z.; Wu, C.; Zeng, Y.; Peng, C. Multifunctional carbon nanomaterials for diagnostic applications in infectious diseases and tumors. Mater. Today Bio 2022, 14, 100231. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Gu, J.; Jiang, Z.; Cao, Y.; Mao, F.; Xue, Y.; Wang, J.; Dai, K.; Qin, L.; Liu, K.; et al. Application of nanotechnology in the early diagnosis and comprehensive treatment of gastrointestinal cancer. J. Nanobiotechnol. 2022, 20, 415. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Wang, P.; Ke, Y. DNA Nanotechnology-Based Biosensors and Therapeutics. Adv. Healthc. Mater. 2021, 10, e2002205. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Kong, N.; Zhang, X.; Cao, Y.; Langer, R.; Tao, W. The landscape of mRNA nanomedicine. Nat. Med. 2022, 28, 2273–2287. [Google Scholar] [CrossRef] [PubMed]
- Graham, H.; Chandler, D.J.; Dunbar, S.A. The genesis and evolution of bead-based multiplexing. Methods 2019, 158, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Houser, B. Bio-Rad’s Bio-Plex(R) suspension array system, xMAP technology overview. Arch. Physiol. Biochem. 2012, 118, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Parsa, S.F.; Vafajoo, A.; Rostami, A.; Salarian, R.; Rabiee, M.; Rabiee, N.; Rabiee, G.; Tahriri, M.; Yadegari, A.; Vashaee, D.; et al. Early diagnosis of disease using microbead array technology: A review. Anal. Chim. Acta 2018, 1032, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.B.; Xu, Y.L.; Liu, Y.; Wang, Y.; Sui, Y.; Liu, J.G.; Wang, X. Inherently fluorescent polystyrene microspheres for coating, sensing and cellular imaging. Colloids Surf. B Biointerfaces 2017, 152, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Shikha, S.; Mei, Q.; Liu, J.; Zhang, Y. Fluorescent microbeads for point-of-care testing: A review. Mikrochim. Acta 2019, 186, 361. [Google Scholar] [CrossRef] [PubMed]
- Leng, Y.; Sun, K.; Chen, X.; Li, W. Suspension arrays based on nanoparticle-encoded microspheres for high-throughput multiplexed detection. Chem. Soc. Rev. 2015, 44, 5552–5595. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Fang, Q.; Zhao, Z.; Li, Z. CoID-LAMP: Color-Encoded, Intelligent Digital LAMP for Multiplex Nucleic Acid Quantification. Anal. Chem. 2023, 95, 5069–5078. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Wang, J.; He, S.; Su, X.; Huang, W.; Chen, M.; Zhuo, Z.; Zhu, X.; Fang, M.; Li, T.; et al. An encodable multiplex microsphere-phase amplification sensing platform detects SARS-CoV-2 mutations. Biosens. Bioelectron. 2022, 203, 114032. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, J.; Wang, X.; Gao, M.; Guo, B.; Gao, M.; Liu, J.; Yu, Y.; Wang, L.; Kong, W.; et al. Prediction of drug efficacy from transcriptional profiles with deep learning. Nat. Biotechnol. 2021, 39, 1444–1452. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Zhang, Y.; Zhao, H.; Liang, Z.; Shi, J.; Ma, Q. High electrochemical active Au-NP/2D zinc-metal organic frameworks heterostructure-based ECL sensor for the miRNA-522 detection in triple negative breast cancer. Talanta 2023, 265, 124875. [Google Scholar] [CrossRef] [PubMed]
- Vladisavljevic, G.T. Structured microparticles with tailored properties produced by membrane emulsification. Adv. Colloid Interface Sci. 2015, 225, 53–87. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Sun, L.; Zhan, H.; Fan, L.J. Preparation of Fluorescence-Encoded Microspheres Based on Hydrophobic Conjugated Polymer-Dye Combination and the Immunoassay. ACS Appl. Bio Mater. 2019, 2, 3009–3018. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, N.; Liu, P.; Liu, Z.; Gao, L.; Jiao, T. Preparation of Fluorescently Labeled Chitosan-Quercetin Drug-Loaded Nanoparticles with Excellent Antibacterial Properties. J. Funct. Biomater. 2022, 13, 141. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumar, V.; Alsawalha, M.; Alomayri, T.; Allehyani, S.; Hu, Y.B.; Fu, M.L.; Yuan, B. MWCNT supported V2O5 quantum dot nanoparticles decorated Bi2O3 nanosheets hybrid system: Efficient visible light driven photocatalyst for degradation of ciprofloxacin. Chemosphere 2022, 306, 135505. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Szymanski, C.; McNeill, J. Preparation and encapsulation of highly fluorescent conjugated polymer nanoparticles. Langmuir 2006, 22, 2956–2960. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.S.; Zhang, B.; Xu, L.D.; Bao, N.; Zhang, Q.; Ding, S.N. CdSe/ZnS quantum dot-encoded maleic anhydride-grafted PLA microspheres prepared through membrane emulsification for multiplexed immunoassays of tumor markers. Analyst 2022, 147, 1873–1880. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.; Zhang, X.; Zhang, L.; Li, J.; Lv, Y.; Li, N.; Wang, L.; Wu, R.; Li, L.S. Fabrication of CuInZnS/ZnS Quantum Dot Microbeads by a Two-Step Approach of Emulsification-Solvent Evaporation and Surfactant Substitution and Its Application for Quantitative Detection. Inorg. Chem. 2023, 62, 3474–3484. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Chen, Y.; Liu, W.; Fang, H.; Li, X.; Hou, L.; Liu, Y.; Lai, W.; Huang, X.; Xiong, Y. Development of a rapid and sensitive quantum dot nanobead-based double-antigen sandwich lateral flow immunoassay and its clinical performance for the detection of SARS-CoV-2 total antibodies. Sens. Actuators B Chem. 2021, 343, 130139. [Google Scholar] [CrossRef] [PubMed]
- Qiu, T.; Song, J.; Fan, L.J. Preparation of fluorescent microspheres via layer-by-layer self-assembly. J. Control. Release 2015, 213, e103–e104. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shen, W.; Rong, Z.; Liu, X.; Gu, B.; Xiao, R.; Wang, S. Layer-by-layer assembly of magnetic-core dual quantum dot-shell nanocomposites for fluorescence lateral flow detection of bacteria. Nanoscale 2020, 12, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, Y.; Dorsel, P.P.; Wu, C. Efficient visual adsorption of Pb(2+) by nanocellulose/sodium alginate microspheres with fluorescence sensitivity. Int. J. Biol. Macromol. 2023, 228, 13–22. [Google Scholar] [CrossRef] [PubMed]
- San Jose, L.; Garcia, O.; Quijada-Garrido, I.; Lopez-Gonzalez, M. RAFT Hydroxylated Polymers as Templates and Ligands for the Synthesis of Fluorescent ZnO Quantum Dots. Nanomaterials 2022, 12, 3441. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xu, H.; Shao, Y.; Liu, J.; Fan, L.J. Preparation and evaluation of fluorescent poly(p-phenyleneethylene) covalently coated microspheres with reactive sites for bioconjugation. J. Colloid Interface Sci. 2019, 540, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Zhang, T.; Yu, B.; Ma, R.; Yue, Q.; Alghamdi, A.A.; Deng, Y. A facile construction of bifunctional core-shell magnetic fluorescent Fe3O4@YVO4:Eu3+ microspheres for latent fingerprint detection. J. Colloid Interface Sci. 2022, 605, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Chen, Y.; Hong, X.; Liu, Z.; Yuan, W. Porous microsphere and its applications. Int. J. Nanomed. 2013, 8, 1111–1120. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Yu, H.; Yang, L.; He, Z.; Zhou, L.; Sun, J.; Gu, X.; Yang, W.; Tang, B.Z. Core-Shell Fluorescent Polymeric Particles with Tunable White Light Emission Based on Aggregation Microenvironment Manipulation. Angew. Chem. Int. Ed. Engl. 2021, 60, 25246–25251. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Hu, Y.; Cao, Z.; Xiao, L.; Lou, J.; Liu, L.; Wang, Y.; Zhao, Z.; Qi, D.; Cui, Q. Efficient synthesis of high solid content emulsions of AIE polymeric nanoparticles with tunable brightness and surface functionalization through miniemulsion polymerization. Dyes Pigments 2019, 163, 371–380. [Google Scholar] [CrossRef]
- Bicak, T.C.; Garnier, M.; Sabbah, M.; Griffete, N. One-Step Synthesis of Fluorescent Poly(divinylbenzene) Particles without Fluorescent Monomers. Macromol. Rapid Commun. 2023, 44, e2200966. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.D.; Wang, Y.; Han, G.Z. Synthesis of carboxylated silicon phthalocyanine photosensitive microspheres with controllable etching. Des. Monomers Polym. 2019, 22, 98–105. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Huang, H.; Jiang, R.; Mao, L.; Liu, M.; Chen, J.; Deng, F.; Zhou, N.; Zhang, X.; Wei, Y. Click multiwalled carbon nanotubes: A novel method for preparation of carboxyl groups functionalized carbon quantum dots. Mater. Sci. Eng. C 2020, 108, 110376. [Google Scholar] [CrossRef] [PubMed]
- Argudo, P.G.; Carril, M.; Martin-Romero, M.T.; Giner-Casares, J.J.; Carrillo-Carrion, C. Surface-Active Fluorinated Quantum Dots for Enhanced Cellular Uptake. Chemistry 2019, 25, 195–199. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Z.; Han, Z.; Fan, L.; Liu, S.; Yang, H.; Chen, Z.; Sun, T.; Ning, B. A highly sensitive and dual-readout immunoassay for norfloxacin in milk based on QDs-FM@ALP-SA and click chemistry. Talanta 2021, 234, 122703. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Chen, B.; He, M.; Hu, B. A dual-functional magnetic microsphere for ICP-MS quantification and fluorescence imaging of matrix metalloproteinase 2 in cell secretion. Anal. Chim. Acta 2021, 1161, 338479. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Guan, J.; Liu, X.; Shi, Y.; Wu, Q.; Luo, M.; Zhu, Y.; Wang, Z.; Wang, L.; Pan, Y. Rapid detection of avian leukosis virus using a fluorescent microsphere immunochromatographic test strip assay. Poult. Sci. 2019, 98, 6492–6496. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, Z.Z.; Huang, Z.Y. Rapid detection of trace malachite green using a fluorescence probe based on signal amplification through electrostatic self-assembly of CdTe QDs and polystyrene microsphere. Mar. Pollut. Bull. 2020, 151, 110812. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.; Enomoto, K.; Ohshiro, K.; Inoue, D.; Kikitsu, T.; Hyeon-Deuk, K.; Pu, Y.J.; Kim, D. Controlling the dimension of the quantum resonance in CdTe quantum dot superlattices fabricated via layer-by-layer assembly. Nat. Commun. 2020, 11, 5471. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Li, B.; Tao, J.; Cheng, J.; Liu, H. The Complex Co-infections of Multiple Porcine Diarrhea Viruses in Local Area Based on the Luminex xTAG Multiplex Detection Method. Front. Vet. Sci. 2021, 8, 602866. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Fu, Q.; Wang, Y.; Long, L.; Jiang, W.; Chen, M.; Xia, H.; Zhang, P.; Tan, F. CRISPR-based quantum dot nanobead lateral flow assay for facile detection of varicella-zoster virus. Appl. Microbiol. Biotechnol. 2023, 107, 3319–3328. [Google Scholar] [CrossRef] [PubMed]
- Renuka, R.M.; Maroli, N.; Achuth, J.; Ponmalai, K.; Kadirvelu, K. Highly adaptable and sensitive FRET-based aptamer assay for the detection of Salmonella paratyphi A. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 243, 118662. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Yan, H.; Zheng, B.; Faheem, A.; Guo, A.; Hu, C.; Hu, Y. Light-Regulated Natural Fluorescence of the PCC 6803@ZIF-8 Composite as an Encoded Microsphere for the Detection of Multiple Biomarkers. ACS Sens. 2021, 6, 2574–2583. [Google Scholar] [CrossRef] [PubMed]
- Seder, I.; Coronel-Tellez, R.; Helalat, S.H.; Sun, Y. Fully integrated sample-in-answer-out platform for viral detection using digital reverse transcription recombinase polymerase amplification (dRT-RPA). Biosens. Bioelectron. 2023, 237, 115487. [Google Scholar] [CrossRef] [PubMed]
- Noppakuadrittidej, P.; Charlermroj, R.; Makornwattana, M.; Kaew-Amdee, S.; Waditee-Sirisattha, R.; Vilaivan, T.; Praneenararat, T.; Karoonuthaisiri, N. Development of peptide nucleic acid-based bead array technology for Bacillus cereus detection. Sci. Rep. 2023, 13, 12482. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Zhai, J.; Wang, Y.; Du, X.; Wang, Z.; Xie, X. Photoswitch-Based Fluorescence Encoding of Microspheres in a Limited Spectral Window for Multiplexed Detection. Anal. Chem. 2022, 94, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Hinney, B.; Wiedermann, S.; Bosco, A.; Rinaldi, L.; Hofer, M.; Joachim, A.; Krucken, J.; Steinborn, R. Development of a three-colour digital PCR for early and quantitative detection of benzimidazole resistance-associated single nucleotide polymorphisms in Haemonchus contortus. Int. J. Parasitol. Drugs Drug Resist. 2023, 22, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Hu, O.; Li, Z.; Wu, J.; Tan, Y.; Chen, Z.; Tong, Y. A Multicomponent Nucleic Acid Enzyme-Cleavable Quantum Dot Nanobeacon for Highly Sensitive Diagnosis of Tuberculosis with the Naked Eye. ACS Sens. 2023, 8, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Eze, N.A.; Milam, V.T. Quantitative Analysis of In Situ Locked Nucleic Acid and DNA Competitive Displacement Events on Microspheres. Langmuir 2022, 38, 6871–6881. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, S.; Barzon, V.; Buxens, A.; Gorrini, M.; Larruskain, A.; El Hamss, R.; Balderacchi, A.M.; Corsico, A.G.; Ferrarotti, I. Molecular diagnosis of alpha1-antitrypsin deficiency: A new method based on Luminex technology. J. Clin. Lab. Anal. 2020, 34, e23279. [Google Scholar] [CrossRef] [PubMed]
- Kibriya, M.G.; Raza, M.; Kamal, M.; Haq, Z.; Paul, R.; Mareczko, A.; Pierce, B.L.; Ahsan, H.; Jasmine, F. Relative Telomere Length Change in Colorectal Carcinoma and Its Association with Tumor Characteristics, Gene Expression and Microsatellite Instability. Cancers 2022, 14, 2250. [Google Scholar] [CrossRef] [PubMed]
- Ashby, M.; Rajko-Nenow, P.; Batten, C.; Flannery, J. Simultaneous Detection of Bluetongue Virus Serotypes Using xMAP Technology. Microorganisms 2020, 8, 1564. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, J.; Liang, J.; Liu, Y.; Liu, C.; Liu, Y.; Xu, T.; Zhang, X. Ultrasound-enhanced catalytic hairpin assembly capable of ultrasensitive microRNA biosensing for the early screening of Alzheimer’s disease. Biosens. Bioelectron. 2023, 242, 115746. [Google Scholar] [CrossRef] [PubMed]
- Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T., Jr. Nucleotide Identification in DNA Using Dielectrophoresis Spectroscopy. Micromachines 2019, 11, 39. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Yi, X.; Yu, X.; Wang, Y.; Zhang, C.; Qin, L.; Guo, D.; Zhou, S.; Zhang, G.; Deng, Y.; et al. High-Throughput Strategies for the Discovery of Anticancer Drugs by Targeting Transcriptional Reprogramming. Front. Oncol. 2021, 11, 762023. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e1417. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C. DDX3X is Epigenetically Repressed in Renal Cell Carcinoma and Serves as a Prognostic Indicator and Therapeutic Target in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2881. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Licklider, L.J.; Gygi, S.P.; Reed, R. Comprehensive proteomic analysis of the human spliceosome. Nature 2002, 419, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.H.; Chen, C.M.; Cheng, P.L.; Shih, J.W.; Tsou, A.P.; Lee, Y.H. DDX3, a DEAD box RNA helicase with tumor growth-suppressive property and transcriptional regulation activity of the p21waf1/cip1 promoter, is a candidate tumor suppressor. Cancer Res. 2006, 66, 6579–6588. [Google Scholar] [CrossRef] [PubMed]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.M.; Cheng, C.H.; Pan, S.L.; Yang, P.M.; Lin, D.Y.; Lee, K.H. Gene Expression Signature-Based Approach Identifies Antifungal Drug Ciclopirox as a Novel Inhibitor of HMGA2 in Colorectal Cancer. Biomolecules 2019, 9, 688. [Google Scholar] [CrossRef] [PubMed]
- Wen, K.C.; Sung, P.L.; Wu, A.T.H.; Chou, P.C.; Lin, J.H.; Huang, C.F.; Yeung, S.J.; Lee, M.H. Neoadjuvant metformin added to conventional chemotherapy synergizes anti-proliferative effects in ovarian cancer. J. Ovarian Res. 2020, 13, 95. [Google Scholar] [CrossRef] [PubMed]
- Pushparaj, P.N.; Abdulkareem, A.A.; Naseer, M.I. Identification of Novel Gene Signatures using Next-Generation Sequencing Data from COVID-19 Infection Models: Focus on Neuro-COVID and Potential Therapeutics. Front. Pharmacol. 2021, 12, 688227. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.P.; Hsieh, Y.Y.; Chou, C.J.; Yang, P.M. Systematic polypharmacology and drug repurposing via an integrated L1000-based Connectivity Map database mining. R. Soc. Open Sci. 2018, 5, 181321. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, L.; Zeng, W.; Zhang, L.; He, N.; Lu, Z. High-throughput quantitative detection of triple-negative breast cancer-associated expressed miRNAs by rolling circle amplification on fluorescence-encoded microspheres. Chin. Chem. Lett. 2023, 34, 108141. [Google Scholar] [CrossRef]
- Wang, N.; Song, L.; Deng, T.; Li, J. Microsphere-based suspension array for simultaneous recognition and quantification of multiple cancer-associated miRNA via DNAzyme-Mediated signal amplification. Anal. Chim. Acta 2020, 1140, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, J.; Shen, S.; Geng, Z.; Ling, Y.; Peng, B. Evaluation of Different Blood Circulating miRNAs for Hepatocellular Carcinoma Diagnosis. J. Nanosci. Nanotechnol. 2020, 20, 1983–1988. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Wu, L.R.; Yan, Y.H.; Zhang, J.X.; Chu, T.; Kwong, L.N.; Patel, A.A.; Zhang, D.Y. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 2022, 6, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Li, X.; Xie, K.P. Coupled liquid biopsy and bioinformatics for pancreatic cancer early detection and precision prognostication. Mol. Cancer 2021, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- Dasari, A.; Morris, V.K.; Allegra, C.J.; Atreya, C.; Benson, A.B., 3rd; Boland, P.; Chung, K.; Copur, M.S.; Corcoran, R.B.; Deming, D.A.; et al. ctDNA applications and integration in colorectal cancer: An NCI Colon and Rectal-Anal Task Forces whitepaper. Nat. Rev. Clin. Oncol. 2020, 17, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Hapsianto, B.N.; Kojima, N.; Kurita, R.; Yamagata, H.; Fujita, H.; Fujii, T.; Kim, S.H. Direct Capture and Amplification of Small Fragmented DNAs Using Nitrogen-Mustard-Coated Microbeads. Anal. Chem. 2022, 94, 7594–7600. [Google Scholar] [CrossRef] [PubMed]
- Kerachian, M.A.; Azghandi, M.; Javadmanesh, A.; Ghaffarzadegan, K.; Mozaffari-Jovin, S. Selective capture of plasma cell-free tumor DNA on magnetic beads: A sensitive and versatile tool for liquid biopsy. Cell. Oncol. 2020, 43, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Malla, M.; Loree, J.M.; Kasi, P.M.; Parikh, A.R. Using Circulating Tumor DNA in Colorectal Cancer: Current and Evolving Practices. J. Clin. Oncol. 2022, 40, 2846–2857. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Ma, P.; Chen, X.; Cheng, Z.; Lin, J. High-sensitivity fluorescence detection for lung cancer CYFRA21-1 DNA based on accumulative hybridization of quantum dots. J. Mater. Chem. B 2022, 10, 1386–1392. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, J.B.; Hou, L.K.; Yu, F.; Zhang, J.; Wu, W.; Tang, X.M.; Sun, F.; Lu, H.M.; Deng, J.; et al. Liquid biopsy in lung cancer: Significance in diagnostics, prediction, and treatment monitoring. Mol. Cancer 2022, 21, 25. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.; Delabat, S.; Carattini, Y.L.; Andrews, D.M. SARS-CoV-2 and Variant Diagnostic Testing Approaches in the United States. Viruses 2021, 13, 2492. [Google Scholar] [CrossRef] [PubMed]
- Artika, I.M.; Dewi, Y.P.; Nainggolan, I.M.; Siregar, J.E.; Antonjaya, U. Real-Time Polymerase Chain Reaction: Current Techniques, Applications, and Role in COVID-19 Diagnosis. Genes 2022, 13, 2387. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, L.; Deng, X.; Liang, R.; Su, M.; He, C.; Hu, L.; Su, Y.; Ren, J.; Yu, F.; et al. Recent advances in the detection of respiratory virus infection in humans. J. Med. Virol. 2020, 92, 408–417. [Google Scholar] [CrossRef]
- Falzarano, M.S.; Scotton, C.; Passarelli, C.; Ferlini, A. Duchenne Muscular Dystrophy: From Diagnosis to Therapy. Molecules 2015, 20, 18168–18184. [Google Scholar] [CrossRef] [PubMed]
- Hammitt, L.L.; Feikin, D.R.; Scott, J.A.G.; Zeger, S.L.; Murdoch, D.R.; O’Brien, K.L.; Deloria Knoll, M. Addressing the Analytic Challenges of Cross-Sectional Pediatric Pneumonia Etiology Data. Clin. Infect. Dis. 2017, 64, S197–S204. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, J.E.; Stratton, C.W.; Persing, D.H.; Tang, Y.W. Forty Years of Molecular Diagnostics for Infectious Diseases. J. Clin. Microbiol. 2022, 60, e0244621. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, S.A. Nucleic acid sample preparation techniques for bead-based suspension arrays. Methods 2023, 219, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Reslova, N.; Michna, V.; Kasny, M.; Mikel, P.; Kralik, P. xMAP Technology: Applications in Detection of Pathogens. Front. Microbiol. 2017, 8, 55. [Google Scholar] [CrossRef] [PubMed]

| Method | Advantages | Disadvantages |
|---|---|---|
| Adsorption method |
|
|
| Embedding method |
|
|
| Self-assembly method |
|
|
| Chemical bonding method |
|
|
| Copolymerization method |
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Luo, G.; Ren, J.; Wang, Q.; Zhao, X.; Wei, L.; Wang, Y.; Liu, Y.; Deng, Y.; Li, S. Recent Advances in and Application of Fluorescent Microspheres for Multiple Nucleic Acid Detection. Biosensors 2024, 14, 265. https://doi.org/10.3390/bios14060265
Chen Z, Luo G, Ren J, Wang Q, Zhao X, Wei L, Wang Y, Liu Y, Deng Y, Li S. Recent Advances in and Application of Fluorescent Microspheres for Multiple Nucleic Acid Detection. Biosensors. 2024; 14(6):265. https://doi.org/10.3390/bios14060265
Chicago/Turabian StyleChen, Zhu, Gaoming Luo, Jie Ren, Qixuan Wang, Xinping Zhao, Linyu Wei, Yue Wang, Yuan Liu, Yan Deng, and Song Li. 2024. "Recent Advances in and Application of Fluorescent Microspheres for Multiple Nucleic Acid Detection" Biosensors 14, no. 6: 265. https://doi.org/10.3390/bios14060265
APA StyleChen, Z., Luo, G., Ren, J., Wang, Q., Zhao, X., Wei, L., Wang, Y., Liu, Y., Deng, Y., & Li, S. (2024). Recent Advances in and Application of Fluorescent Microspheres for Multiple Nucleic Acid Detection. Biosensors, 14(6), 265. https://doi.org/10.3390/bios14060265




