Catalytic Hairpin Assembly-Based Self-Ratiometric Gel Electrophoresis Detection Platform for Reliable Nucleic Acid Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
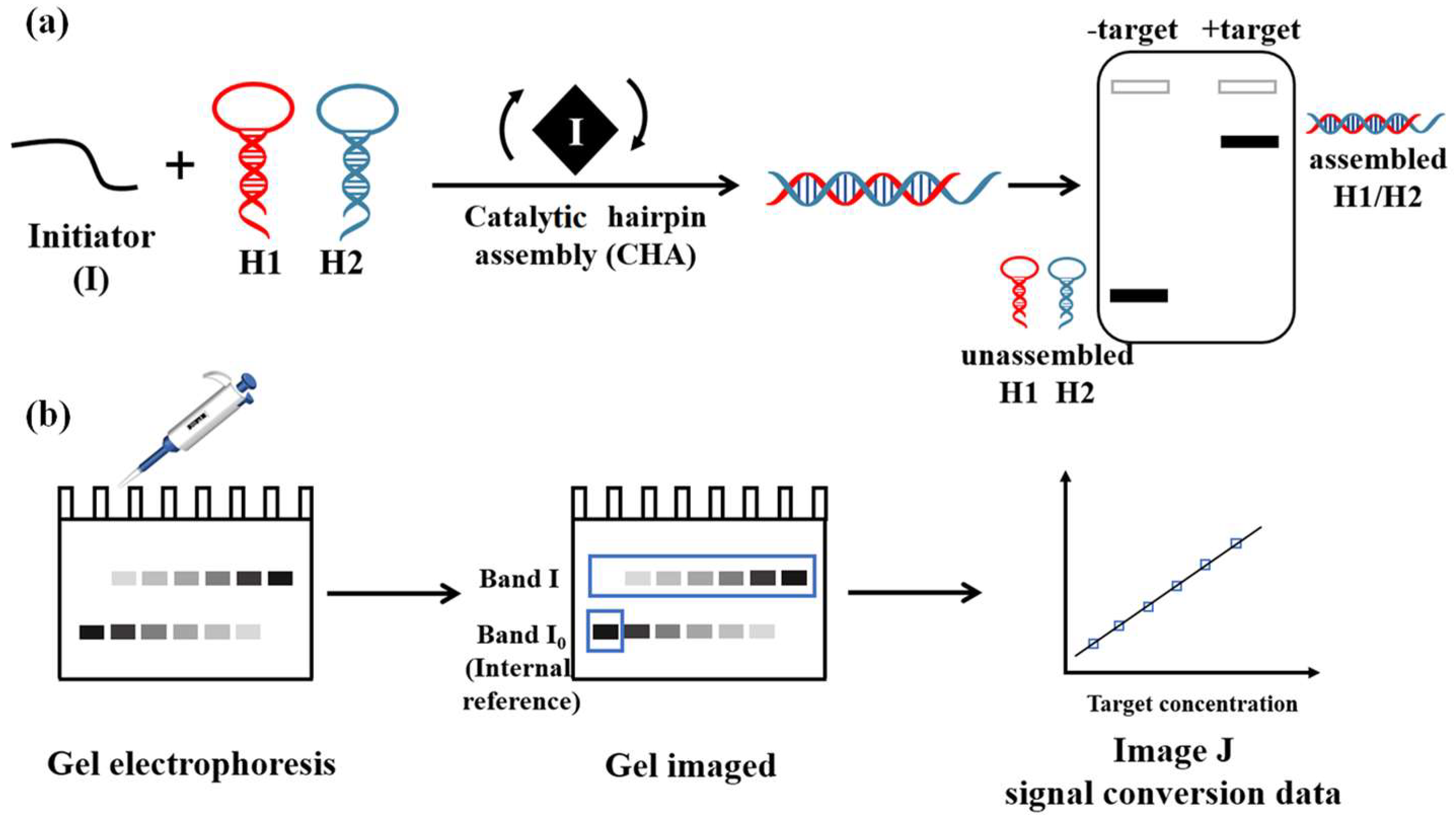
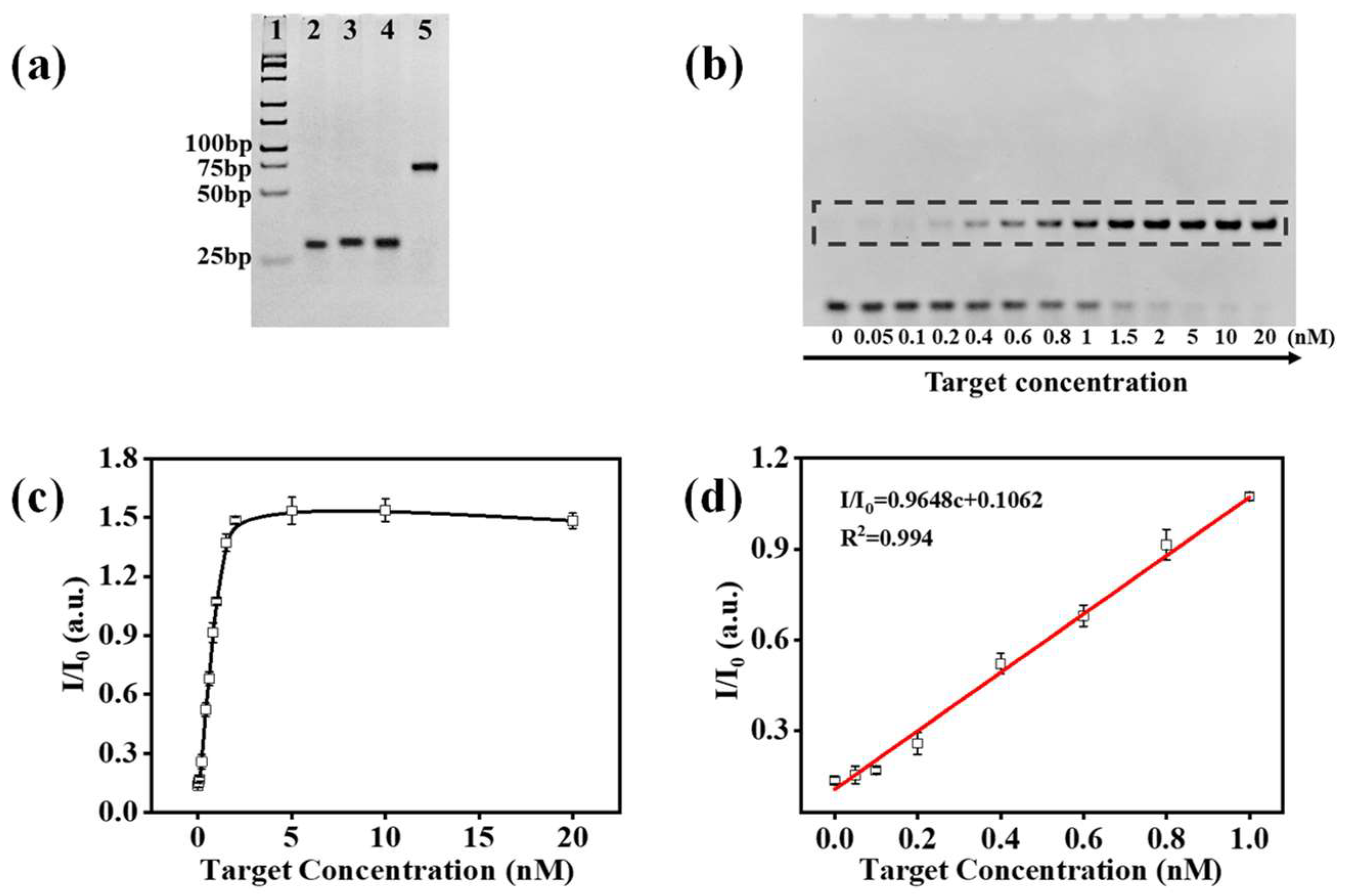
2.2. Catalytic Hairpin Assembly
2.3. Gel Analysis
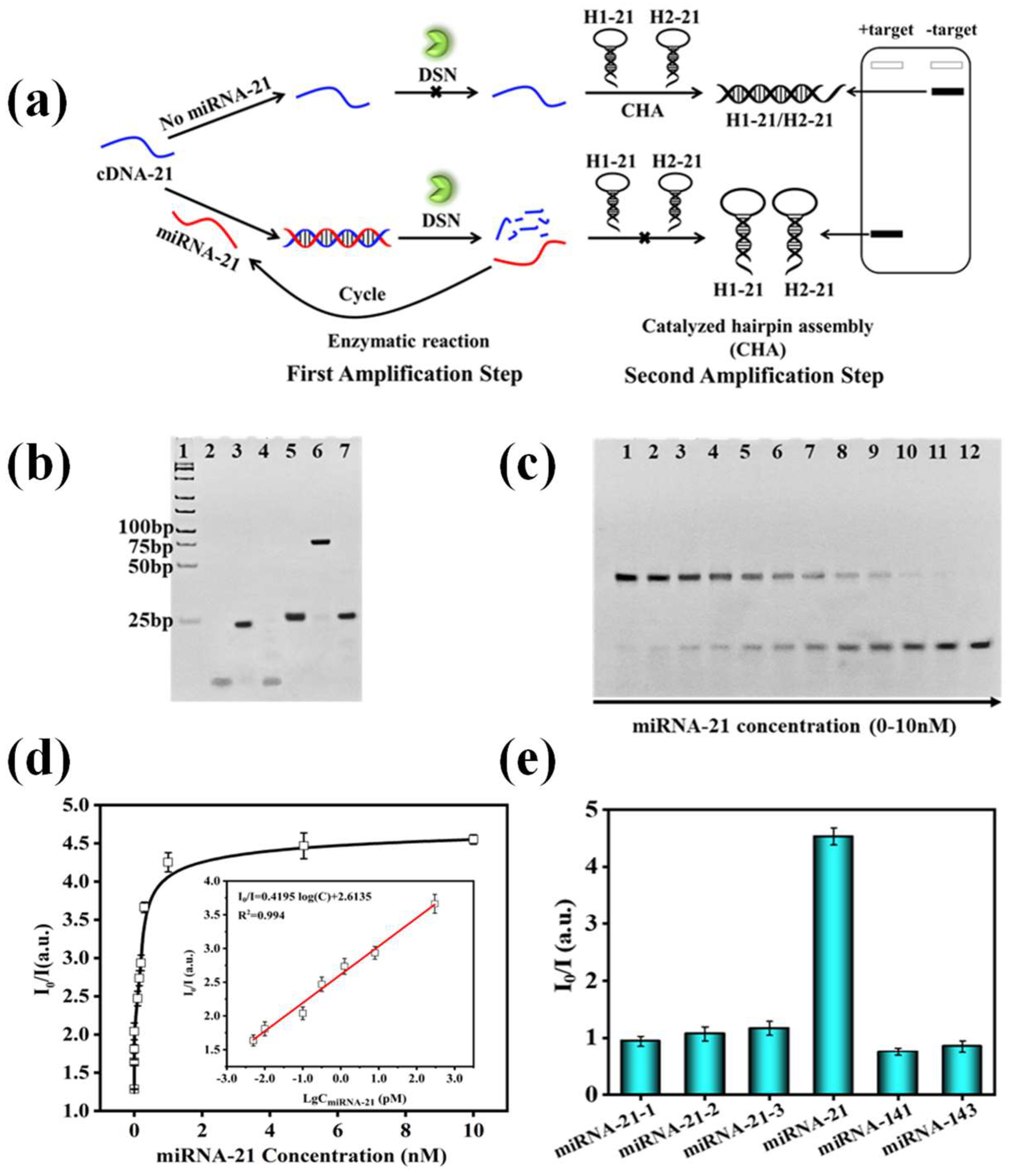
2.4. Protocol for miRNA-21 and HBV Detection
2.5. HBV Detection
2.6. MiRNA-21 and HBV Quantification
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, Y.; Zuo, X.; Li, Q.; Chen, F.; Chen, Y.R.; Deng, J.; Han, D.; Hao, C.; Huang, F.; Huang, Y.; et al. Nucleic Acids Analysis. Sci. China Chem. 2021, 64, 171–203. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal amplification of nucleic acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Seeman, N.C.; Sleiman, H.F. DNA nanotechnology. Nat. Rev. Mater. 2017, 3, 1–23. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Q.; Zhang, X.; Huang, H.; Tang, S.; Chai, Y.; Xu, Z.; Li, M.; Chen, X.; Liu, J.; et al. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J. Nanobiotechnol. 2022, 20, 279. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D.; Mirkin, C.A. DNA-based nanostructures for live-cell analysis. J. Am. Chem. Soc. 2020, 142, 11343–11356. [Google Scholar] [CrossRef]
- Sackmann, E.K.; Fulton, A.L.; Beebe, D.J. The present and future role of microfluidics in biomedical research. Nature 2014, 507, 181–189. [Google Scholar] [CrossRef]
- Karim, K.; Lamaoui, A.; Amine, A. Paper-based optical sensors paired with smartphones for biomedical analysis. J. Pharm. Biomed. Anal. 2023, 225, 115207. [Google Scholar] [CrossRef]
- Thorne, H. Electrophoretic separation of polyoma virus DNA from host cell DNA. Virology 1966, 29, 234–239. [Google Scholar] [CrossRef]
- Bishop, D.H.L.; Claybrook, J.R.; Spiegelman, S. Electrophoretic separation of viral nucleic acids on polyacrylamide gels. J. Mol. Biol. 1967, 26, 373–387. [Google Scholar] [CrossRef]
- Smithies, O. Zone electrophoresis in starch gels: Group variations in the serum proteins of normal human adults. Biochem. J. 1955, 61, 629–641. [Google Scholar]
- Liu, Y.; Yu, Y.; Meng, Q.; Jia, X.; Zhu, J.; Tang, C.; Zhao, Q.; Feng, X.; Zhang, J. A Fluorescent Probe for the Specific Staining of Cysteine Containing Proteins and Thioredoxin Reductase in SDS-PAGE. Biosensors 2021, 11, 132. [Google Scholar] [CrossRef] [PubMed]
- Koussa, M.A.; Halvorsen, K.; Ward, A.; Wong, W.P. DNA nanoswitches: A quantitative platform for gel-based biomolecular interaction analysis. Nat. Methods 2015, 12, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Ranallo, S.; Amodio, A.; Idili, A.; Porchetta, A.; Ricci, F. Electronic control of DNA-based nanoswitches and nanodevices. Chem. Sci. 2016, 7, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Hansen, C.H.; Yang, D.; Koussa, M.A.; Wong, W.P. Nanoswitch-linked immunosorbent assay (NLISA) for fast, sensitive, and specific protein detection. Proc. Natl. Acad. Sci. USA 2017, 114, 10367–10372. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Shi, L.; Wang, H.; Li, T. A DNA nanoswitch-controlled reversible nanosensor. Nucleic Acids Res. 2017, 45, 541–546. [Google Scholar] [CrossRef]
- Chandrasekaran, A.R.; MacIsaac, M.; Dey, P.; Levchenko, O.; Zhou, L.; Andres, M.; Dey, B.K.; Halvorsen, K. Cellular microRNA detection with miRacles: microRNA-activated conditional looping of engineered switches. Sci. Adv. 2019, 5, eaau9443. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Li, H.; Jia, Y.; Mak, P.I.; Martins, R.P. SARS-CoV-2 RNA Detection with Duplex-Specific Nuclease Signal Amplification. Micromachines 2021, 12, 197. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Chandrasekaran, A.R.; Punnoose, J.A.; Bonenfant, G.; Charles, S.; Levchenko, O.; Badu, P.; Cavaliere, C.; Pager, C.T.; Halvorsen, K. Programmable low-cost DNA-based platform for viral RNA detection. Sci. Adv. 2020, 6, eabc6246. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R.; Zavala, J.; Halvorsen, K. Programmable DNA nanoswitches for detection of nucleic acid sequences. ACS Sens. 2016, 1, 120–123. [Google Scholar] [CrossRef]
- Wang, H.B.; Bai, H.Y.; Dong, G.L.; Liu, Y.M. DNA-templated Au nanoclusters coupled with proximity-dependent hybridization and guanine-rich DNA induced quenching: A sensitive fluorescent biosensing platform for DNA detection. Nanoscale Adv. 2019, 1, 1482–1488. [Google Scholar] [CrossRef]
- Zhang, P.; Zandieh, M.; Ding, Y.; Wu, L.; Wang, X.; Liu, J.; Li, Z. A Label-Free, Mix-and-Detect ssDNA-Binding Assay Based on Cationic Conjugated Polymers. Biosensors 2023, 13, 122. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R.; Trivedi, R.; Halvorsen, K. Ribonuclease-responsive DNA nanoswitches. Cell Rep. Phys. Sci. 2020, 1, 100117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, P.; Hou, M.; Chen, L.; Wang, J.; Yang, H.; Feng, W. An electrochemical biosensor based on ARGET ATRP with DSN-assisted target recycling for sensitive detection of tobacco mosaic virus RNA. Bioelectrochemistry 2022, 144, 108037. [Google Scholar] [CrossRef] [PubMed]
- Kachwala, M.J.; Smith, C.W.; Nandu, N.; Yigit, M.V. Reprogrammable gel electrophoresis detection assay using CRISPR-Cas12a and hybridization chain reaction. Anal. Chem. 2021, 93, 1934–1938. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Zhang, J.; Meng, X.; He, J.; Zhang, K.; Cao, Y.; Wang, D.; Dong, H.; Zhang, X. Catalytic hairpin assembly gel assay for multiple and sensitive microRNA detection. Theranostics 2018, 8, 2646–2656. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.L.; Lu, H.J.; Xu, J.J.; Zhou, H.; Chen, H.Y. Recent advances of ratiometric electrochemiluminescence biosensors. J. Mater. Chem. B 2019, 7, 6469–6475. [Google Scholar] [CrossRef]
- Chen, L.G.; Li, J.; Sun, L.; Wang, H.B. Ratiometric fluorometric assay triggered by alkaline phosphatase: Proof-of-concept toward a split-type biosensing strategy for DNA detection. Talanta 2024, 271, 125703. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Jia, Z.; Yu, M.; Zhang, M.; Xu, C. A Ratiometric Fluorescent Sensor Based on Chelation-Enhanced Fluorescence of Carbon Dots for Zinc Ion Detection. Molecules 2023, 28, 7818. [Google Scholar] [CrossRef]
- Luo, Y.; Wu, N.; Wang, L.; Song, Y.; Du, Y.; Ma, G. Biosensor Based on Covalent Organic Framework Immobilized Acetylcholinesterase for Ratiometric Detection of Carbaryl. Biosensors 2022, 12, 625. [Google Scholar] [CrossRef]
- HeeáLee, M.; SeungáKim, J. Small molecule-based ratiometric fluorescence probes for cations, anions, and biomolecules. Chem. Soc. Rev. 2015, 44, 4185–4191. [Google Scholar]
- Loas, A.; Lippard, S.J. Direct ratiometric detection of nitric oxide with Cu (II)-based fluorescent probes. J. Mater. Chem. B 2017, 5, 8929–8933. [Google Scholar] [CrossRef]
- Aron, A.T.; Loehr, M.O.; Bogena, J.; Chang, C.J. An endoperoxide reactivity-based FRET probe for ratiometric fluorescence imaging of labile iron pools in living cells. J. Am. Chem. Soc. 2016, 138, 14338–14346. [Google Scholar] [CrossRef]
- Viviani, V.R.; Pelentir, G.F.; Bevilaqua, V.R. Bioluminescence Color-Tuning Firefly Luciferases: Engineering and Prospects for Real-Time Intracellular pH Imaging and Heavy Metal Biosensing. Biosensors 2022, 12, 400. [Google Scholar] [CrossRef]
- Hao, N.; Hua, R.; Zhang, K.; Lu, J.; Wang, K. A sunlight powered portable photoelectrochemical biosensor based on a potentiometric resolve ratiometric principle. Anal. Chem. 2018, 90, 13207–13211. [Google Scholar] [CrossRef]
- Zheng, Y.N.; Liang, W.B.; Xiong, C.Y.; Zhuo, Y.; Chai, Y.Q.; Yuan, R. Universal ratiometric photoelectrochemical bioassay with target-nucleotide transduction-amplification and electron-transfer tunneling distance regulation strategies for ultrasensitive determination of microRNA in cells. Anal. Chem. 2017, 89, 9445–9451. [Google Scholar] [CrossRef]
- Yin, P.; Choi, H.M.; Calvert, C.R.; Pierce, N.A. Programming biomolecular self-assembly pathways. Nature 2008, 451, 318–322. [Google Scholar] [CrossRef]
- Li, B.; Ellington, A.D.; Chen, X. Rational, modular adaptation of enzyme-free DNA circuits to multiple detection methods. Nucleic Acids Res. 2011, 39, e110. [Google Scholar] [CrossRef]
- Deng, R.; Zhang, K.; Li, J. Isothermal amplification for microRNA detection: From the test tube to the cell. Acc. Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, Q.; Wang, S.-Y.; Deng, X.-B.; Zhang, C.-H. Catalytic Hairpin Assembly-Based Self-Ratiometric Gel Electrophoresis Detection Platform for Reliable Nucleic Acid Analysis. Biosensors 2024, 14, 232. https://doi.org/10.3390/bios14050232
Xi Q, Wang S-Y, Deng X-B, Zhang C-H. Catalytic Hairpin Assembly-Based Self-Ratiometric Gel Electrophoresis Detection Platform for Reliable Nucleic Acid Analysis. Biosensors. 2024; 14(5):232. https://doi.org/10.3390/bios14050232
Chicago/Turabian StyleXi, Qiang, Si-Yi Wang, Xiao-Bing Deng, and Chong-Hua Zhang. 2024. "Catalytic Hairpin Assembly-Based Self-Ratiometric Gel Electrophoresis Detection Platform for Reliable Nucleic Acid Analysis" Biosensors 14, no. 5: 232. https://doi.org/10.3390/bios14050232
APA StyleXi, Q., Wang, S.-Y., Deng, X.-B., & Zhang, C.-H. (2024). Catalytic Hairpin Assembly-Based Self-Ratiometric Gel Electrophoresis Detection Platform for Reliable Nucleic Acid Analysis. Biosensors, 14(5), 232. https://doi.org/10.3390/bios14050232




