Using Temporally and Spatially Resolved Measurements to Improve the Sensitivity of Fluorescence-Based Immunoassays
Abstract
1. Introduction
2. Materials and Methods
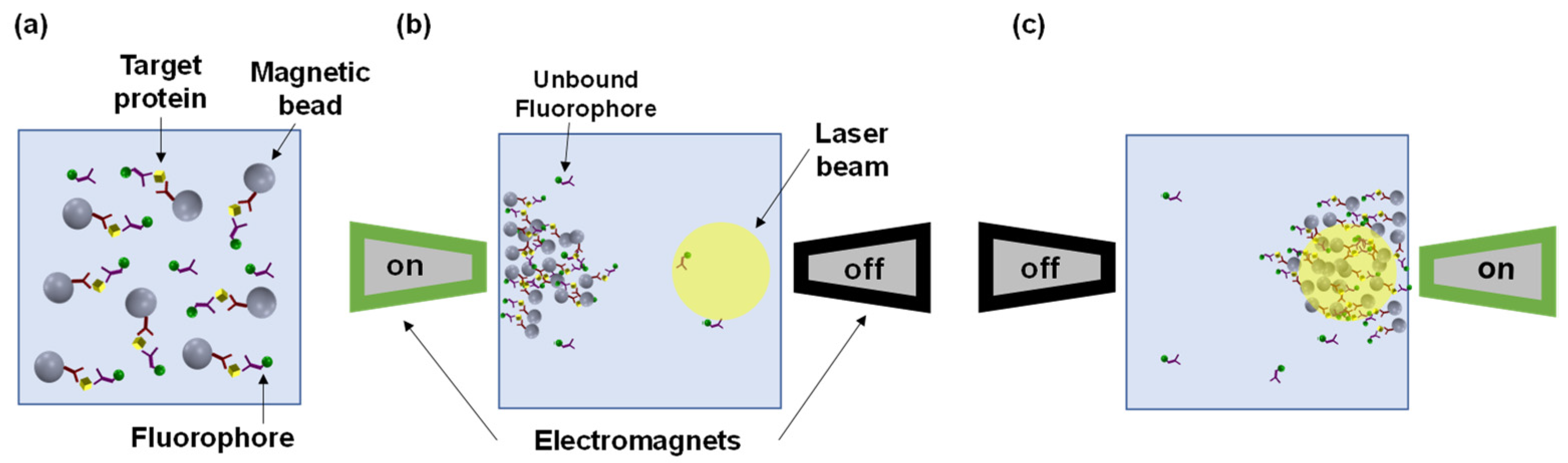
2.1. Magnetic Modulation Biosensing (MMB) System Description
2.2. Interleukin-8 Assay
2.3. Mathematical Model
2.4. Signal Processing Algorithms
2.4.1. Preliminary Process
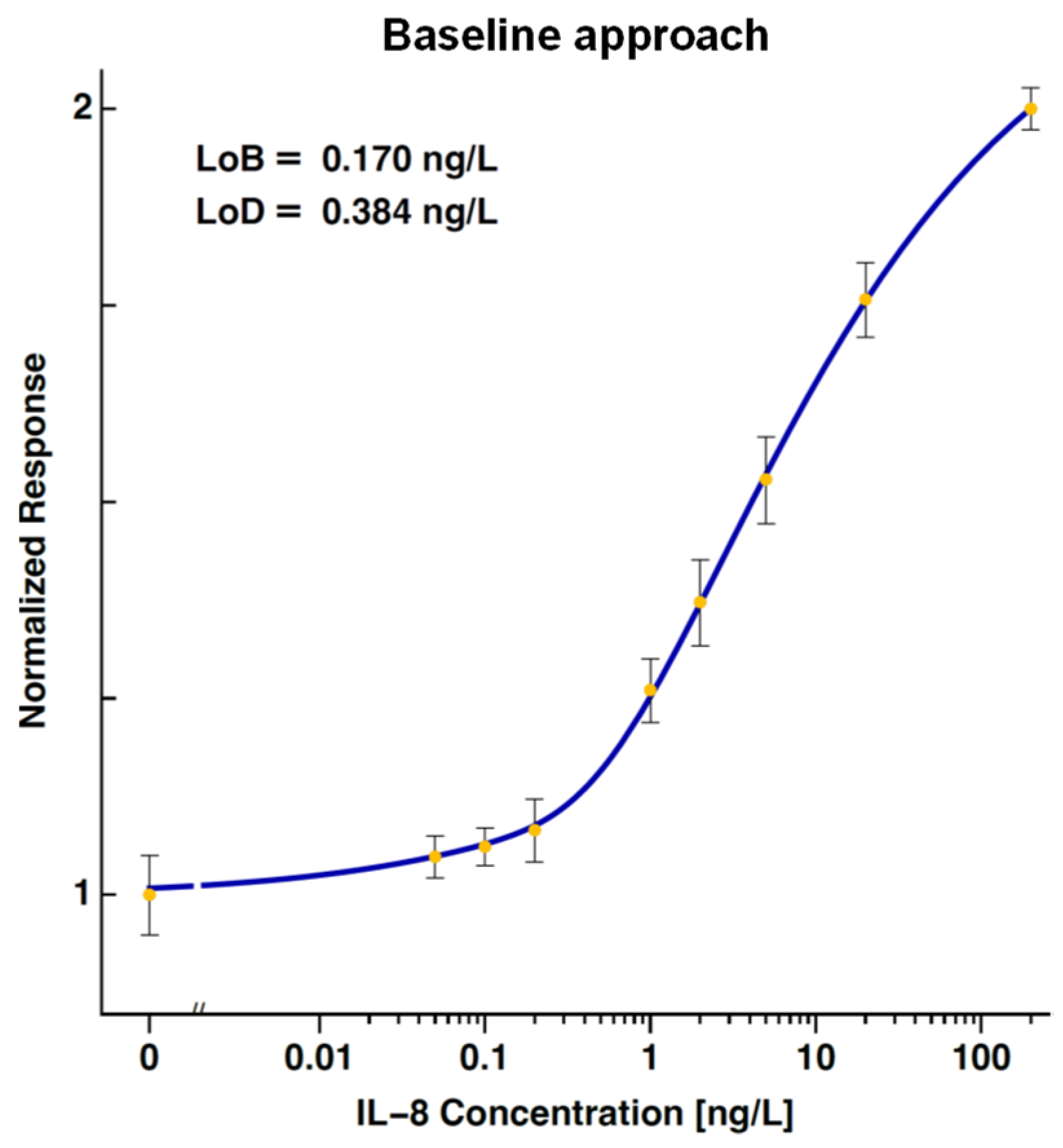
2.4.2. Baseline Approach
2.4.3. Feature 1
2.4.4. Feature 2
2.4.5. Homoscedasticity and Normality
2.4.6. Normalization
2.4.7. Dose–Response
2.4.8. Levels of Blank and Detection
2.4.9. Detection Procedure
- Apply a preliminary process (Section 2.4.1) by finding the center of mass, define a pinhole mask, and crop each frame to create , where
- Use either the Baseline approach (Section 2.4.2), Feature 1 (Section 2.4.3), or Feature 2 (Section 2.4.4) algorithms to convert the series of frames to a one-dimensional scalar time-series (, , or ), and then convert it to a scalar (, , or ).
- Use the natural logarithm function (Section 2.4.5) and normalize the result (Section 2.4.6).
- Determine the concentration using the inverse dose–response, , , or .
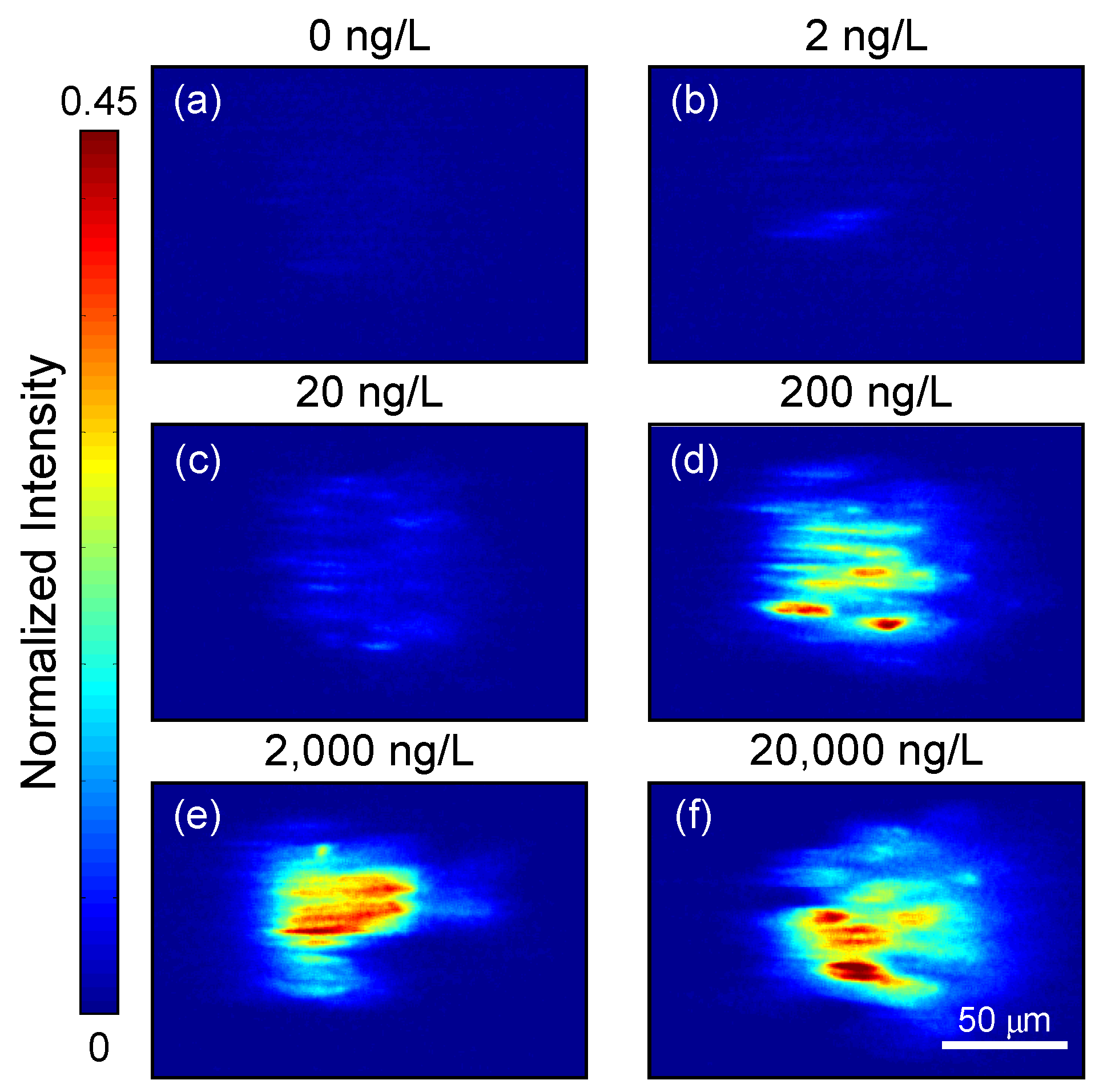
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pires, N.M.M.; Dong, T.; Hanke, U.; Hoivik, N. Recent Developments in Optical Detection Technologies in Lab-on-a-Chip Devices for Biosensing Applications. Sensors 2014, 14, 15458–15479. [Google Scholar] [CrossRef]
- Todd, J.; Freese, B.; Lu, A.; Held, D.; Morey, J.; Livingston, R.; Goix, P. Ultrasensitive Flow-Based Immunoassays Using Single-Molecule Counting. Clin. Chem. 2007, 53, 1990–1995. [Google Scholar] [CrossRef]
- Wu, A.H.B.; Fukushima, N.; Puskas, R.; Todd, J.; Goix, P. Development and Preliminary Clinical Validation of a High Sensitivity Assay for Cardiac Troponin Using a Capillary Flow (Single Molecule) Fluorescence Detector. Clin. Chem. 2006, 52, 2157–2159. [Google Scholar] [CrossRef]
- Rissin, D.M.; Kan, C.W.; Campbell, T.G.; Howes, S.C.; Fournier, D.R.; Song, L.; Piech, T.; Patel, P.P.; Chang, L.; Rivnak, A.J.; et al. Single-Molecule Enzyme-Linked Immunosorbent Assay Detects Serum Proteins at Subfemtomolar Concentrations. Nat. Biotech. 2010, 28, 595–599. [Google Scholar] [CrossRef]
- Dunbar, S.A. Applications of Luminex® xMAP™ Technology for Rapid, High-Throughput Multiplexed Nucleic Acid Detection. Clin. Chim. Acta 2006, 363, 71–82. [Google Scholar] [CrossRef]
- Forster, R.J.; Bertoncello, P.; Keyes, T.E. Electrogenerated Chemiluminescence. Annu. Rev. Anal. Chem. 2009, 2, 359–385. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51, 2415. [Google Scholar] [CrossRef]
- Llandro, J.; Palfreyman, J.J.; Ionescu, A.; Barnes, C.H. Magnetic Biosensor Technologies for Medical Applications: A Review. Med Biol. Eng. Comput. 2010, 48, 977–998. [Google Scholar] [CrossRef]
- Huang, X.; Li, Y.; Huang, X.; Xie, X.; Xu, Y.; Chen, Y.; Gao, W. A Novel Reverse Fluorescent Immunoassay Approach for Sensing Human Chorionic Gonadotropin Based on Silver-Gold Nano-Alloy and Magnetic Nanoparticles. Anal. Bioanal. Chem. 2016, 408, 619–627. [Google Scholar] [CrossRef]
- Hung, L.Y.; Huang, T.B.; Tsai, Y.C.; Yeh, C.S.; Lei, H.Y.; Lee, G.B. A Microfluidic Immunomagnetic Bead-Based System for the Rapid Detection of Influenza Infections: From Purified Virus Particles to Clinical Specimens. Biomed. Microdevices 2013, 15, 539–551. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, F.; Xia, X.; Ye, D.; Zhou, R.; Li, L.; Deng, T.; Ding, Z.; Liu, F. Developing a Fluorescence Substrate for Hrp-Based Diagnostic Assays with Superiorities over the Commercial ADHP. Chin. Chem. Lett. 2024, 35, 108970. [Google Scholar] [CrossRef]
- Peck, K.; Stryer, L.; Glazer, A.N.; Mathies, R.A. Single-Molecule Fluorescence Detection: Autocorrelation Criterion and Experimental Realization with Phycoerythrin. Proc. Natl. Acad. Sci. USA 1989, 86, 4087–4091. [Google Scholar] [CrossRef] [PubMed]
- Földes-Papp, Z. What It Means to Measure a Single Molecule in a Solution by Fluorescence Fluctuation Spectroscopy. Exp. Mol. Pathol. 2006, 80, 209–218. [Google Scholar] [CrossRef]
- Castro, A.; Williams, J.G. Single-Molecule Detection of Specific Nucleic Acid Sequences in Unamplified Genomic DNA. Anal. Chem. 1997, 69, 3915–3920. [Google Scholar] [CrossRef]
- Koffi, D.; Touré, A.O.; Varela, M.-L.; Vigan-Womas, I.; Béourou, S.; Brou, S.; Ehouman, M.-F.; Gnamien, L.; Richard, V.; Djaman, J.A.; et al. Analysis of Antibody Profiles in Symptomatic Malaria in Three Sentinel Sites of Ivory Coast by Using Multiplex, Fluorescent, Magnetic, Bead-Based Serological Assay (MAGPIX™). Malar. J. 2015, 14, 509. [Google Scholar] [CrossRef]
- Perraut, R.; Joos, C.; Sokhna, C.; Polson, H.E.; Trape, J.F.; Tall, A.; Marrama, L.; Mercereau-Puijalon, O.; Richard, V.; Longacre, S. Association of Antibody Responses to the Conserved Plasmodium falciparum Merozoite Surface Protein 5 with Protection Against Clinical Malaria. PLoS ONE 2014, 9, e101737. [Google Scholar] [CrossRef]
- Yeh, H.C.; Chao, S.Y.; Ho, Y.P.; Wang, T.H. Single-Molecule Detection and Probe Strategies for Rapid and Ultrasensitive Genomic Detection. Curr. Pharm. Biotechnol. 2005, 6, 453–461. [Google Scholar] [CrossRef]
- Moerner, W.E.; Fromm, D.P. Methods of Single-Molecule Fluorescence Spectroscopy and Microscopy. Rev. Sci. Instrum. 2003, 74, 3597–3619. [Google Scholar] [CrossRef]
- Barnes, M.D.; Whitten, W.B.; Ramsey, J.M. Detecting Single Molecules in Liquids. Anal. Chem. 1995, 67, A418–A423. [Google Scholar] [CrossRef]
- Perraut, R.; Richard, V.; Varela, M.L.; Trape, J.F.; Guillotte, M.; Tall, A. Comparative Analysis of IgG Responses to Plasmodium falciparum MSP1p19 and PF13-DBL1alpha1 Using ELISA and a Magnetic Bead-Based Duplex Assay (MAGPIX(R)-Luminex) in a Senegalese Meso-Endemic Community. Malar J. 2014, 13, 410. [Google Scholar] [CrossRef]
- Michelson, Y.; Lustig, Y.; Avivi, S.; Schwartz, E.; Danielli, A. Highly Sensitive and Specific Zika Virus Serological Assays Using a Magnetic Modulation Biosensing System. J. Infect. Dis. 2018, 219, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.; Margulis, M.; Danielli, A. Recent Advances in Rapid and Highly Sensitive Detection of Proteins and Specific DNA Sequences Using a Magnetic Modulation Biosensing System. Sensors 2022, 22, 4497. [Google Scholar] [CrossRef] [PubMed]
- Margulis, M.; Erster, O.; Roth, S.; Mandelboim, M.; Danielli, A. A Magnetic Modulation Biosensing-Based Molecular Assay for Rapid and Highly Sensitive Clinical Diagnosis of Coronavirus Disease 2019 (COVID-19). J. Mol. Diagn. 2021, 23, 1680–1690. [Google Scholar] [CrossRef] [PubMed]
- Verbarg, J.; Hadass, O.; Olivo, P.D.; Danielli, A. High Sensitivity Detection of a Protein Biomarker Interleukin-8 Utilizing a Magnetic Modulation Biosensing System. Sens. Actuators B Chem. 2017, 241, 614–618. [Google Scholar] [CrossRef]
- Roth, S.; Hadass, O.; Cohen, M.; Verbarg, J.; Wilsey, J.; Danielli, A. Improving the Sensitivity of Fluorescence-Based Immunoassays by Photobleaching the Autofluorescence of Magnetic Beads. Small 2018, 15, e1803751. [Google Scholar] [CrossRef] [PubMed]
- Borysiak, M.D.; Thompson, M.J.; Posner, J.D. Translating Diagnostic Assays from the Laboratory to the Clinic: Analytical and Clinical Metrics for Device Development and Evaluation. Lab Chip 2016, 16, 1293–1313. [Google Scholar] [CrossRef] [PubMed]
- Baltagi, B.H. Econometrics, 5th ed.; Springer Texts in Business and Economics; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Lee, D.K. Data Transformation: A Focus on the Interpretation. Korean J. Anesthesiol. 2020, 73, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2016, 10, e0146021. [Google Scholar] [CrossRef] [PubMed]
- Currie, L.A. Nomenclature in Evaluation of Analytical Methods Including Detection and Quantification Capabilities (IUPAC Recommendations 1995). Anal. Chim. Acta 1999, 391, 105–126. [Google Scholar] [CrossRef]
- Yang, K.; Tu, J.; Chen, T. Homoscedasticity: An Overlooked Critical Assumption for Linear Regression. Gen. Psychiatry 2019, 32, e100148. [Google Scholar] [CrossRef]

| Method | LoB | Gain | Gain (%) | LoD | Gain | Gain (%) |
|---|---|---|---|---|---|---|
| Baseline | 0.170 | - | - | 0.384 | - | - |
| Feature 1 | 0.082 | 2.1 | (52%) | 0.212 | 1.8 | (44%) |
| Feature 2 | 0.042 | 4.1 | (75%) | 0.158 | 2.4 | (58%) |
| Concentration [ng/L] | Baseline QR [ng/L] | Feature 1 QR [ng/L] | Gain (%) | Feature 2 QR [ng/L] | Gain (%) |
|---|---|---|---|---|---|
| 1 | 0.27 | 0.19 | 29.6% | 0.24 | 11.0% |
| 2 | 0.61 | 0.53 | 13.0% | 0.64 | −5.0% |
| 5 | 1.57 | 1.17 | 25.4% | 1.44 | 8.0% |
| 20 | 6.73 | 4.99 | 25.8% | 6.90 | −2.5% |
| Concentration [ng/L] | Baseline | Feature 1 | Feature 2 |
|---|---|---|---|
| 0 | 0.73 | 0.30 | 0.17 |
| 0.05 | 0.83 | 0.22 | 0.29 |
| 0.10 | 0.09 | 0.49 | 0.18 |
| 0.20 | 0.88 | 0.34 | 0.86 |
| 1.00 | 0.51 | 0.40 | 0.60 |
| Test | Baseline | Feature 1 | Feature 2 |
|---|---|---|---|
| Levene | 0.08 | 0.46 | 0.67 |
| Conover | 0.12 | 0.56 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kremer, R.; Roth, S.; Bross, A.; Danielli, A.; Noam, Y. Using Temporally and Spatially Resolved Measurements to Improve the Sensitivity of Fluorescence-Based Immunoassays. Biosensors 2024, 14, 220. https://doi.org/10.3390/bios14050220
Kremer R, Roth S, Bross A, Danielli A, Noam Y. Using Temporally and Spatially Resolved Measurements to Improve the Sensitivity of Fluorescence-Based Immunoassays. Biosensors. 2024; 14(5):220. https://doi.org/10.3390/bios14050220
Chicago/Turabian StyleKremer, Ran, Shira Roth, Avital Bross, Amos Danielli, and Yair Noam. 2024. "Using Temporally and Spatially Resolved Measurements to Improve the Sensitivity of Fluorescence-Based Immunoassays" Biosensors 14, no. 5: 220. https://doi.org/10.3390/bios14050220
APA StyleKremer, R., Roth, S., Bross, A., Danielli, A., & Noam, Y. (2024). Using Temporally and Spatially Resolved Measurements to Improve the Sensitivity of Fluorescence-Based Immunoassays. Biosensors, 14(5), 220. https://doi.org/10.3390/bios14050220






