Recent Advances in Aptamer-Based Biosensors for Bacterial Detection
Abstract
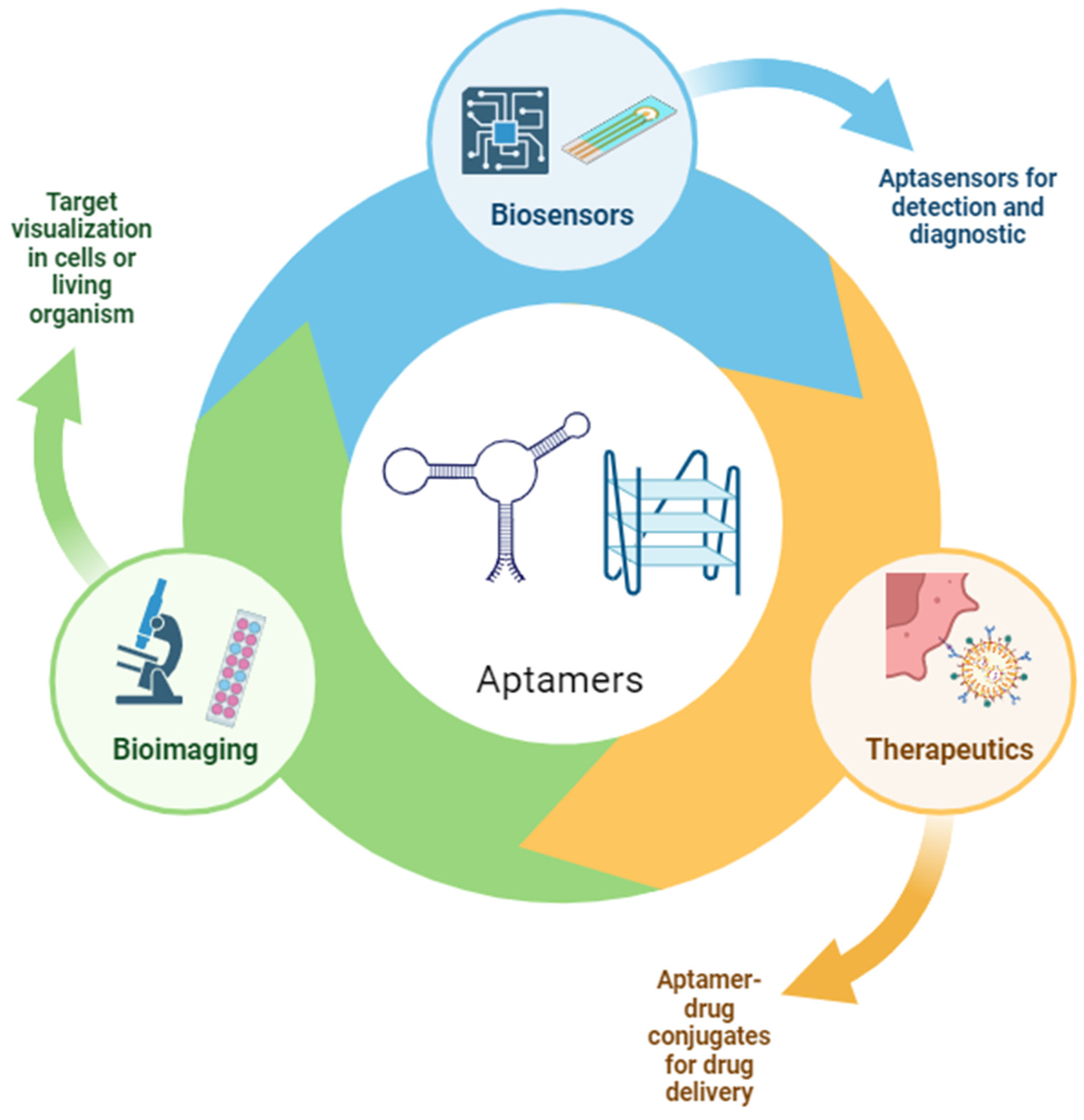
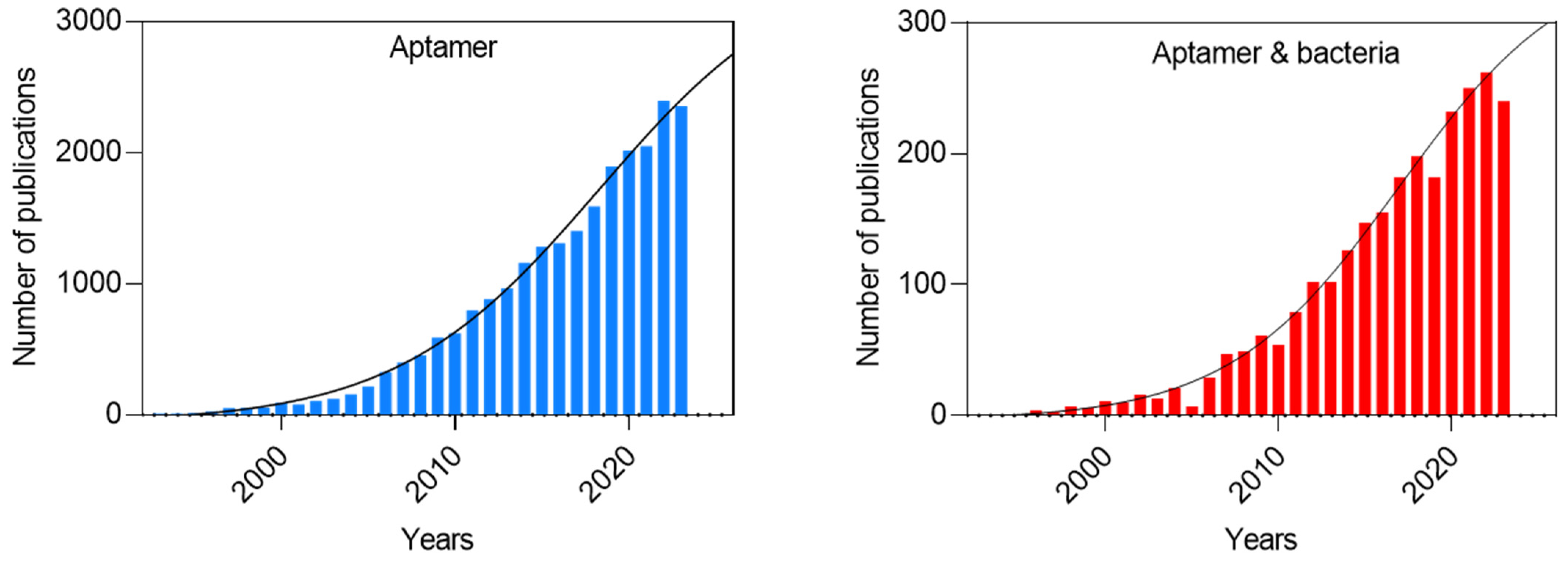
1. Introduction
2. Aptamer Selection Strategies for Bacterial Pathogen Detection
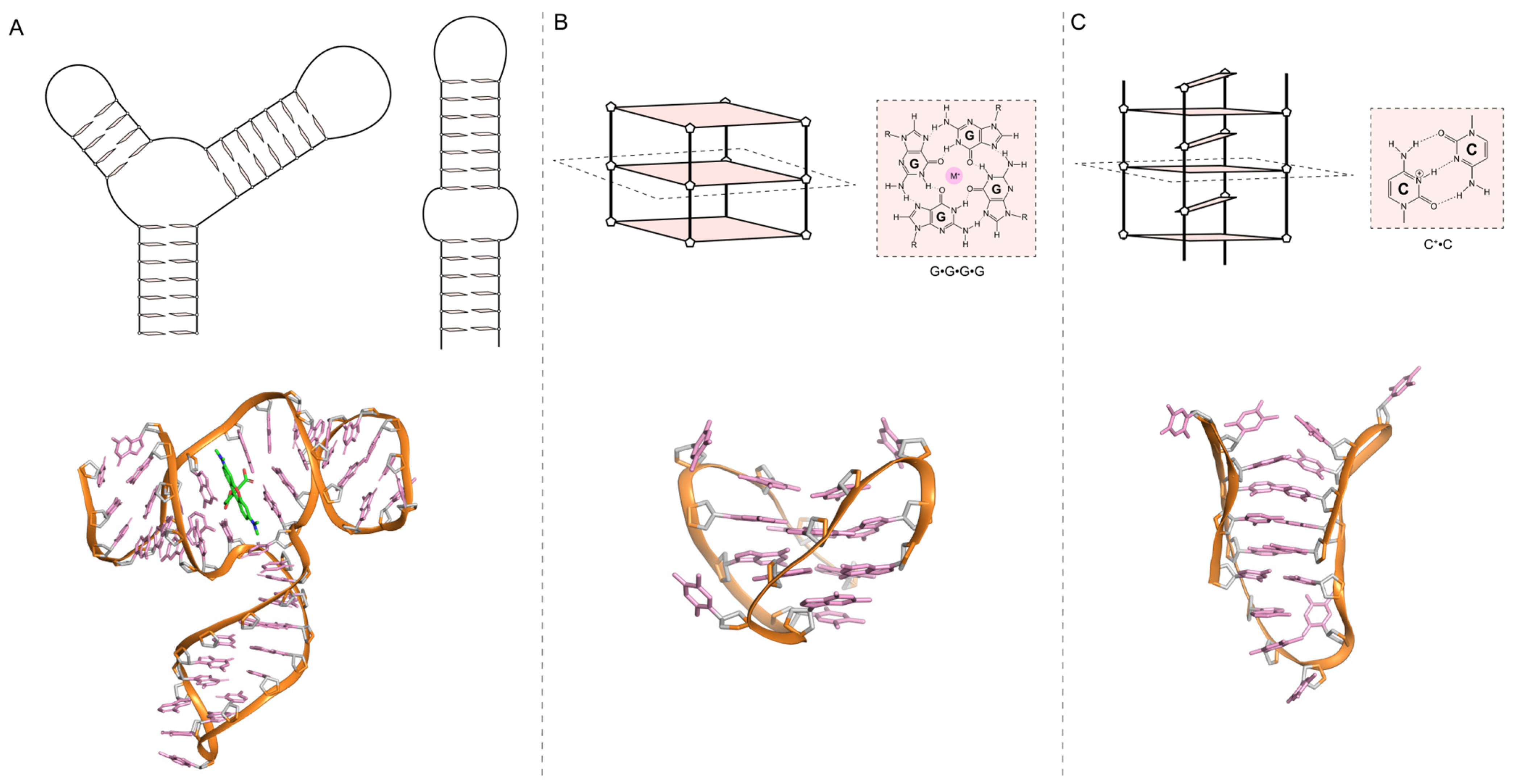
3. Aptamer Characterization: Affinity, Stability, and Structure
4. Aptasensors for Bacterial Detection
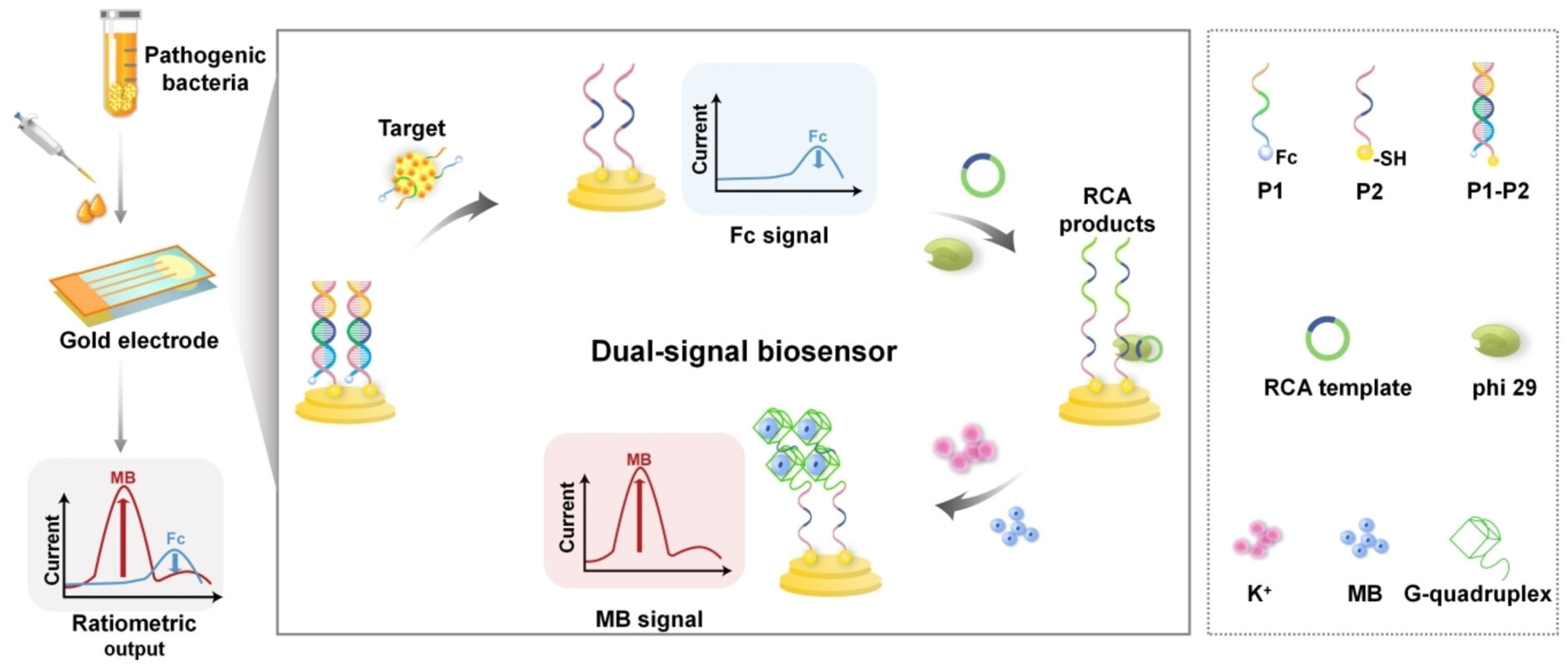
4.1. Electrochemical Aptasensors for Bacterial Detection
4.2. Optical Aptasensors for Bacterial Detection
4.3. Other Aptasensors for Bacterial Detection
5. Conclusions
6. Future Directions
| Bacterium | Aptamer | DNA or RNA | Target | Kd (nM) | LOD | Linear Range (CFU/mL) | Ref. |
|---|---|---|---|---|---|---|---|
| S. aureus | T1 T2 T3 A14 | RNA & DNA | IsdA protein | 2.2 ± 0.5 1.0 ± 0.3 0.7 ± 0.4 4 ± 2 | 113 pM 17 pM 11 pM 485 pM | / | [56] |
| S. aureus | SH-Apt2 | DNA | Whole cell | 210.7 | / | / | [61,201] |
| S. aureus | SA20 SA23 SA34 SA31 SA43 | DNA | Whole cell | 70.86 ± 39.22 61.50 ± 22.43 72.42 ± 35.23 82.86 ± 33.20 210.7 ± 135.9 | / | / | [65] |
| S. aureus | Apt1 Apt2 | DNA | Whole cell | 35 129 | 7.5–8.4 × 104 CFU/mL | 104–108 | [174,202] |
| S. aureus | H1 H2 cApt | DNA | / | / | 101 CFU/mL | 102–106 | [203] |
| S. aureus | A15 | DNA | Enterotoxin A protein | 48.57 | 8.7 × 10−3 µg/mL | 0.01–10 µg/mL | [204] |
| S. aureus | H1 H2 | DNA | Whole cell | / | 4–8 CFU/mL | 45–4.5 × 106 | [205] |
| E. coli | SH-Apt1 | DNA | Whole cell | 25.2 | / | / | [55,123] |
| E.coli | E1 E2 E3 | DNA | Whole cell | / | 3.7 × 102 CFU/mL | / | [162] |
| E.coli | / | DNA | Whole cell | / | 45 CFU/mL | 102–108 | [206] |
| E. coli | / | DNA | Whole cell | / | 0.05 CFU/mL | 0.1–104 | [207] |
| P. aeroginosa | F23 | DNA | Whole cell | 17.27 ± 5.00 | 104 CFU/mL | / | [208] |
| B. cereus | / | DNA | Whole cell | / | 22 CFU/mL | 49–49 × 106 | [209] |
| B. cereus | / | DNA | Whole Cell | / | 4 CFU/mL | 20–2 × 108 | [210] |
| Acinetobacter baumannii | AB K2 | DNA | Whole cell | 5.377 6.8 | 10 CFU/mL | 10–105 | [211] |
| Klebsiella pneumoniae | K2 | DNA | Whole cell | / | 10 CFU/mL | 10–105 | [211] |
| Leptospira interrogans | LAP3 | DNA | Outer, embrane protein | 133.13 | 57 CFU/mL | 60–6 × 105 | [212] |
| Listeria monocytogenes | A8 | DNA | Internalin A | / | 103 CFU/mL | 103–105 | [213] |
| Salmonella | Multi-apt | DNA | Multi | 11.72 | 7 CFU/mL | 10–107 | [214] |
| Yersinia enterocolitica | / | DNA | Whole cell | / | 3 CFU/mL | 10–109 | [215] |
| Bacillus cytotoxicus | BAS6R | DNA | Spore | / | 102–104 CFU/mL | 103–104 | [19] |
| Clostridium difficile | No name | DNA G-quadruplex | Toxin A protein (TOA) | / | 1 nM | 0–200 ng/mL | [216] |
| S. typhimurium | STA | DNA | Whole cell | / | 9 CFU/mL | 56–56 × 107 | [177] |
| S. typhimurium | H2 | DNA | Whole cell | / | 4–8 CFU/mL | 36–3.6 × 106 | [205] |
| S. Typhimurium | Apt ST | DNA | Whole cell | 10 | 30 CFU/mL | 102–106 | [217] |
| Vibrio parahaemolyticus | Apt VP | DNA | Whole cell | 16.88 | 10 CFU/mL | 102–106 | [217] |
| Campylobacter jejuni | ONS13 ONS-23TA | DNA | Whole cell | 292.8 ± 53.1 | 7.2 × 105 CFU/mL | / | [62,173] |
| Bacterium | Aptamer | DNA or RNA | Target | Kd (nM) | LOD | Linear Range (CFU/mL) | Ref. |
|---|---|---|---|---|---|---|---|
| S. aureus | Antibac1&2 | DNA | Peptidoglycan | 415 + 0.047 1261 + 0.280 | 82 pg/mL | / | [54,218] |
| S. aureus | Apt1 | DNA | Whole cell | 35 | 10–100 CFU/mL | 10–105 | [163,171] |
| S. aureus | P1 | DNA | Whole cell | / | 10 | 10–106 | [164] |
| S. aureus | A-SEB | DNA | SEB protein | 0.02 | 0.21 fM | 5.0–500 fM | [169] |
| E. coli | ECA I ECA II | DNA | Outer membrane proteins (OMPs) | / | / | 1 × 10−7–2 × 10−6 M | [219,220] |
| B. cereus | B15 B16 | DNA | Whole cell | 16.13 20.67 | 10 CFU/mL | / | [106] |
| B. cereus | 13–18 13–24 | DNA | Whole cell | 22.75 36.72 | 9.27 CFU/mL | / | [179] |
| Acinetobacterer baumanni | / | DNA | Whole cell | / | 150 CFU/mL | 1 × 103–1.0 × 108 | [221] |
| Clostridium difficile | / | DNA G-quadruplex | Toxin A protein (TOA) | 1 nM | 0–200 ng/mL | [216] | |
| Mycobacterium tuberculosis | / | DNA | MPT64 protein | 8.92 | 4.1 fMl | / | [222] |
| Bacterium | Aptamer | DNA or RNA | Target | Kd (nM) | LOD | Linear Range (CFU/mL) | Ref. |
|---|---|---|---|---|---|---|---|
| S. aureus | APTseb1 | DNA | Staphylococcal enterotoxin B (SEB) | / | / | / | [51] |
| S. aureus | G1 #2 #18 | RNA | Teichoic acid | / | / | / | [53] |
| S. aureus | H1 | DNA | / | / | 41 CFU/mL | 4.1 × 101 to 4.1 × 105 | [223] |
| S. aureus | AT-27 AT-33 AT-36 AT-49 | DNA | α-toxin protein | / | / | / | [224] |
| E. coli | GN6 GN12 | DNA | Outer membrane vesicles (OMV) | 29.94 20.36 | / | / | [194] |
| E. coli | 6-3 8-1 8-7 8-8 8-12 8-13 8-19 8-35 | RNA | Heme | 188 309 256 371 445 425 | / | / | [225] |
| E. coli | Stx1 stx2 | DNA | Shiga toxin Viz, stx1, and stx2 | 47 pM 29 pM | 44.5 pg/mL 41.3 pg/mg | 50 pg/mL 100 ng/mg | [226] |
| P. aeroginosa | F23 | DNA | Whole cell | 17.27 ± 5.00 | 104 CFU/mL | / | [208] |
| Vibrio cholerae | CT916 | Cholera toxin (CT) | 48.5 | 2.1–2.4 ng/ml | 0–10 ng/mL | [45] | |
| Clostridium perfringens | / | DNA | Whole cell | / | 1 CFU/mL | 1–108 | [155] |
Supplementary Materials
Funding
Conflicts of Interest
References
- Campos, J.C.D.M.; Antunes, L.C.M.; Ferreira, R.B.R. Global priority pathogens: Virulence, antimicrobial resistance and prospective treatment options. Future Microbiol. 2020, 15, 649–677. [Google Scholar] [CrossRef] [PubMed]
- WHO. 2022. Available online: https://cdn.who.int/media/docs/default-source/gho-documents/world-health-statistic-reports/worldhealthstatistics_2022.pdf (accessed on 27 November 2023).
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-Related Illness and Death in the United States. Emerg. Infect. Dis. 1999, 5, 607–625. [Google Scholar] [CrossRef] [PubMed]
- Marin, M.; Nikolic, M.V.; Vidic, J. Rapid point-of-need detection of bacteria and their toxins in food using gold nanoparticules. Comp. Rev. Food Sci. Food Saf. 2021, 20, 5880–5900. [Google Scholar] [CrossRef] [PubMed]
- Vidic, J.; Chaix, C.; Manzano, M.; Heyndrickx, M. Food Sensing: Detection of Bacillus cereus Spores in Dairy Products. Biosensors 2020, 10, 15. [Google Scholar] [CrossRef] [PubMed]
- Ramarao, N.; Tran, S.-L.; Marin, M.; Vidic, J. Advanced Methods for Detection of Bacillus cereus and Its Pathogenic Factors. Sensors 2020, 20, 2667. [Google Scholar] [CrossRef] [PubMed]
- Setlow, P.; Johnson, E.A. Spores and Their Significance. In Food Microbiology; Doyle, M.P., Diez-Gonzalez, F., Hill, C., Eds.; ASM Press: Washington, DC, USA, 2019; pp. 23–63. [Google Scholar] [CrossRef]
- Ikuta, K.S.; Swetschinski, L.R.; Aguilar, G.R.; Sharara, F.; Mestrovic, T.; Gray, A.P.; Weaver, N.D.; Wool, E.E.; Han, C.; Hayoon, A.G.; et al. Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2022, 400, 2221–2248. [Google Scholar] [CrossRef] [PubMed]
- Mullis, K.; Faloona, F.; Scharf, S.; Saiki, R.; Horn, G.; Erlich, H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harb. Symp. Quant. Biol. 1986, 51, 263–273. [Google Scholar] [CrossRef]
- Malorny, B.; Tassios, P.T.; Rådström, P.; Cook, N.; Wagner, M.; Hoorfar, J. Standardization of diagnostic PCR for the detection of foodborne pathogens. Int. J. Food Microbiol. 2003, 83, 39–48. [Google Scholar] [CrossRef]
- Toze, S. PCR and the detection of microbial pathogens in water and wastewater. Water Res. 1999, 33, 3545–3556. [Google Scholar] [CrossRef]
- Butler, J.E. Enzyme-Linked Immunosorbent Assay. J. Immunoass. 2000, 21, 165–209. [Google Scholar] [CrossRef]
- Thévenot, D.R.; Toth, K.; Durst, R.A.; Wilson, G.S. Electrochemical biosensors: Recommended definitions and classification*. Anal. Lett. 2001, 34, 635–659. [Google Scholar] [CrossRef]
- Wang, K.-Y.; Zeng, Y.-L.; Yang, X.-Y.; Li, W.-B.; Lan, X.-P. Utility of aptamer-fluorescence in situ hybridization for rapid detection of Pseudomonas aeruginosa. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 273–278. [Google Scholar] [CrossRef]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.; Tang, T.-H. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403. [Google Scholar] [CrossRef] [PubMed]
- Trunzo, N.E.; Hong, K.L. Recent Progress in the Identification of Aptamers Against Bacterial Origins and Their Diagnostic Applications. Int. J. Mol. Sci. 2020, 21, 5074. [Google Scholar] [CrossRef]
- Yi, J.; Xiao, W.; Li, G.; Wu, P.; He, Y.; Chen, C.; He, Y.; Ding, P.; Kai, T. The research of aptamer biosensor technologies for detection of microorganism. Appl. Microbiol. Biotechnol. 2020, 104, 9877–9890. [Google Scholar] [CrossRef]
- Rizzotto, F.; Marin, M.; Péchoux, C.; Auger, S.; Vidic, J. Colorimetric aptasensor for detection of Bacillus cytotoxicus spores in milk and ready-to-use food. Heliyon 2023, 9, e17562. [Google Scholar] [CrossRef]
- Bobrinetskiy, I.; Radovic, M.; Rizzotto, F.; Vizzini, P.; Jaric, S.; Pavlovic, Z.; Radonic, V.; Nikolic, M.V.; Vidic, J. Advances in Nanomaterials-Based Electrochemical Biosensors for Foodborne Pathogen Detection. Nanomaterials 2021, 11, 2700. [Google Scholar] [CrossRef]
- Sharifi, S.; Vahed, S.Z.; Ahmadian, E.; Dizaj, S.M.; Eftekhari, A.; Khalilov, R.; Ahmadi, M.; Hamidi-Asl, E.; Labib, M. Detection of pathogenic bacteria via nanomaterials-modified aptasensors. Biosens. Bioelectron. 2020, 150, 111933. [Google Scholar] [CrossRef]
- Liu, M.; Yue, F.; Kong, Q.; Liu, Z.; Guo, Y.; Sun, X. Aptamers against Pathogenic Bacteria: Selection Strategies and Apta-assay/Aptasensor Application for Food Safety. J. Agric. Food Chem. 2022, 70, 5477–5498. [Google Scholar] [CrossRef]
- Brown, A.; Brill, J.; Amini, R.; Nurmi, C.; Li, Y. Development of Better Aptamers: Structured Library Approaches, Selection Methods, and Chemical Modifications. Angew. Chem. Int. Ed. 2024, 63, e202318665. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Sampson, T. Aptamers and SELEX: The technology. World Pat. Inf. 2003, 25, 123–129. [Google Scholar] [CrossRef]
- Dausse, E.; Barré, A.; Aimé, A.; Groppi, A.; Rico, A.; Ainali, C.; Salgado, G.; Palau, W.; Daguerre, E.; Nikolski, M.; et al. Aptamer selection by direct microfluidic recovery and surface plasmon resonance evaluation. Biosens. Bioelectron. 2016, 80, 418–425. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.; Kuznetsov, A. Inside the Black Box: What Makes SELEX Better? Molecules 2019, 24, 3598. [Google Scholar] [CrossRef]
- Kohlberger, M.; Gadermaier, G. SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnol. Appl. Biochem. 2022, 69, 1771–1792. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Feng, Z.; Qin, H.; Chen, L.; Yan, M.; Li, L.; Qu, F. Recent progress of SELEX methods for screening nucleic acid aptamers. Talanta 2024, 266, 124998. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, M.; Ma, T.; Li, W.; Zhang, H. Review on the Selection of Aptamers and Application in Paper-Based Sensors. Biosensors 2022, 13, 39. [Google Scholar] [CrossRef]
- Cowperthwaite, M.C.; Ellington, A.D. Bioinformatic Analysis of the Contribution of Primer Sequences to Aptamer Structures. J. Mol. Evol. 2008, 67, 95–102. [Google Scholar] [CrossRef]
- Ellington, A.D.; Khrapov, M.; Shaw, C.A. The scene of a frozen accident. RNA 2000, 6, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Changayil, S.; Majerfeld, I.; Yarus, M. Selection of the simplest RNA that binds isoleucine. RNA 2003, 9, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Legiewicz, M.; Lozupone, C.; Knight, R.; Yarus, M. Size, constant sequences, and optimal selection. RNA 2005, 11, 1701–1709. [Google Scholar] [CrossRef] [PubMed]
- Shui, B.; Ozer, A.; Zipfel, W.; Sahu, N.; Singh, A.; Lis, J.T.; Shi, H.; Kotlikoff, M.I. RNA aptamers that functionally interact with green fluorescent protein and its derivatives. Nucleic Acids Res. 2012, 40, e39. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; McConnell, E.M.; Cruz-Toledo, J.; Bernard, E.D.; Pach, A.; Mastronardi, E.; Zhang, X.; Beking, M.; Francis, T.; Giamberardino, A.; et al. Analysis of In Vitro Aptamer Selection Parameters. J. Mol. Evol. 2015, 81, 150–161. [Google Scholar] [CrossRef] [PubMed]
- Pobanz, K.; Lupták, A. Improving the odds: Influence of starting pools on in vitro selection outcomes. Methods 2016, 106, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Sabeti, P.C.; Unrau, P.J.; Bartel, D.P. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 1997, 4, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Diafa, S.; Hollenstein, M. Generation of Aptamers with an Expanded Chemical Repertoire. Molecules 2015, 20, 16643–16671. [Google Scholar] [CrossRef] [PubMed]
- Meek, K.N.; Rangel, A.E.; Heemstra, J.M. Enhancing aptamer function and stability via in vitro selection using modified nucleic acids. Methods 2016, 106, 29–36. [Google Scholar] [CrossRef]
- Lapa, S.A.; Chudinov, A.V.; Timofeev, E.N. The Toolbox for Modified Aptamers. Mol. Biotechnol. 2016, 58, 79–92. [Google Scholar] [CrossRef]
- Mondal, B.; Ramlal, S.; Lavu, P.S.B.; Kingston, J. Highly Sensitive Colorimetric Biosensor for Staphylococcal Enterotoxin B by a Label-Free Aptamer and Gold Nanoparticles. Front. Microbiol. 2018, 9, 179. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhang, H.; Chen, X.; Wang, X.; Duan, N.; Wu, S.; Xu, B.; Wang, Z. A multicolor time-resolved fluorescence aptasensor for the simultaneous detection of multiplex Staphylococcus aureus enterotoxins in the milk. Biosens. Bioelectron. 2015, 74, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Frohnmeyer, E.; Frisch, F.; Falke, S.; Betzel, C.; Fischer, M. Highly affine and selective aptamers against cholera toxin as capture elements in magnetic bead-based sandwich ELAA. J. Biotechnol. 2018, 269, 35–42. [Google Scholar] [CrossRef]
- Bruno, J.G.; Richarte, A.M.; Carrillo, M.P.; Edge, A. An aptamer beacon responsive to botulinum toxins. Biosens. Bioelectron. 2012, 31, 240–243. [Google Scholar] [CrossRef] [PubMed]
- Subekin, A.; Alieva, R.; Kukushkin, V.; Oleynikov, I.; Zavyalova, E. Rapid SERS Detection of Botulinum Neurotoxin Type A. Nanomaterials 2023, 13, 2531. [Google Scholar] [CrossRef] [PubMed]
- Molejon, N.A.; Lapada, C.M.; Skouridou, V.; Rollon, A.P.; El-Shahawi, M.S.; Bashammakh, A.S.; O’Sullivan, C.K. Selection of G-rich ssDNA aptamers for the detection of enterotoxins of the cholera toxin family. Anal. Biochem. 2023, 669, 115118. [Google Scholar] [CrossRef] [PubMed]
- Bogomolova, A.; Aldissi, M. Real-time and label-free analyte detection in a flow-through mode using immobilized fluorescent aptamer/quantum dots molecular switches. Biosens. Bioelectron. 2015, 66, 290–296. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef] [PubMed]
- DeGrasse, J.A. A Single-Stranded DNA Aptamer That Selectively Binds to Staphylococcus aureus Enterotoxin B. PLoS ONE 2012, 7, e33410. [Google Scholar] [CrossRef]
- Chang, T.-W.; Blank, M.; Janardhanan, P.; Singh, B.R.; Mello, C.; Blind, M.; Cai, S. In vitro selection of RNA aptamers that inhibit the activity of type A botulinum neurotoxin. Biochem. Biophys. Res. Commun. 2010, 396, 854–860. [Google Scholar] [CrossRef]
- Han, S.R.; Lee, S.W. In vitro selection of RNA aptamer specific to Staphylococcus aureus. Ann. Microbiol. 2014, 64, 883–885. [Google Scholar] [CrossRef]
- Ferreira, I.M.; Lacerda, C.M.d.S.; de Faria, L.S.; Corrêa, C.R.; de Andrade, A.S.R. Selection of Peptidoglycan-Specific Aptamers for Bacterial Cells Identification. Appl. Biochem. Biotechnol. 2014, 174, 2548–2556. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.; Kim, G.; Park, S.B.; Lim, J.; Mo, C. Comparison of Whole-Cell SELEX Methods for the Identification of Staphylococcus Aureus-Specific DNA Aptamers. Sensors 2015, 15, 8884–8897. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, K.M.; Sabbih, G.; Algama, C.H.; Syed, R.; Danquah, M.K.; Dhakal, S. FRET-Based Single-Molecule Detection of Pathogen Protein IsdA Using Computationally Selected Aptamers. Anal. Chem. 2023, 95, 9839–9846. [Google Scholar] [CrossRef] [PubMed]
- Ohuchi, S. Cell-SELEX Technology. BioRes. Open Access 2012, 1, 265–272. [Google Scholar] [CrossRef]
- Sefah, K.; Shangguan, D.; Xiong, X.; O’Donoghue, M.B.; Tan, W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010, 5, 1169–1185. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-W.; Wang, H.-X.; Jia, G.-C.; Li, Z. Application of Aptamer-Based Biosensor for Rapid Detection of Pathogenic Escherichia coli. Sensors 2018, 18, 2518. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Yang, S.; Zhang, D.; Zhou, T.; Huang, J.; Gao, M.; Jiang, Y.; Liu, Y.; Yang, J. Ultrasensitive dual-enhanced sandwich strategy for simultaneous detection of Escherichia coli and Staphylococcus aureus based on optimized aptamers-functionalized magnetic capture probes and graphene oxide-Au nanostars SERS tags. J. Colloid Interface Sci. 2023, 634, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.-A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010, 87, 2323–2334. [Google Scholar] [CrossRef]
- Bruno, J.G.; Phillips, T.; Carrillo, M.P.; Crowell, R. Plastic-Adherent DNA Aptamer-Magnetic Bead and Quantum Dot Sandwich Assay for Campylobacter Detection. J. Fluoresc. 2009, 19, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Alibolandi, M.; Taghdisi, S.M.; Ramezani, P.; Hosseini Shamili, F.; Farzad, S.A.; Abnous, K.; Ramezani, M. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int. J. Pharm. 2017, 519, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.-Q.; Kim, E.R.; Gu, M.B. A new cognate aptamer pair-based sandwich-type electrochemical biosensor for sensitive detection of Staphylococcus aureus. Biosens. Bioelectron. 2022, 198, 113835. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-C.; Yang, C.-Y.; Sun, R.-L.; Cheng, Y.-F.; Kao, W.-C.; Yang, P.-C. Rapid single cell detection of Staphylococcus aureus by aptamer-conjugated gold nanoparticles. Sci. Rep. 2013, 3, 1863. [Google Scholar] [CrossRef]
- Lim, S.H.; Ryu, Y.C.; Hwang, B.H. Aptamer-immobilized Gold Nanoparticles Enable Facile and On-site Detection of Staphylococcus aureus. Biotechnol. Bioprocess Eng. 2021, 26, 107–113. [Google Scholar] [CrossRef]
- Manfredini, A.; Malusà, E.; Canfora, L. Aptamer-based technology for detecting Bacillus subtilis in soil. Appl. Microbiol. Biotechnol. 2023, 107, 6963–6972. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.-A. Selection of DNA aptamers for capture and detection of Salmonella typhimurium using a whole-cell SELEX approach in conjunction with cell sorting. Appl. Microbiol. Biotechnol. 2013, 97, 3677–3686. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.H.; Dwivedi, H.P.; Choi, S.J.; Jaykus, L.-A. Selection and characterization of DNA aptamers specific for Listeria species. Anal. Biochem. 2014, 459, 39–45. [Google Scholar] [CrossRef]
- Setlow, P. Resistance of spores of Bacillus species to ultraviolet light. Environ. Mol. Mutagen. 2001, 38, 97–104. [Google Scholar] [CrossRef]
- Setlow, P.; Christie, G. New Thoughts on an Old Topic: Secrets of Bacterial Spore Resistance Slowly Being Revealed. Microbiol. Mol. Biol. Rev. 2023, 87, e0008022. [Google Scholar] [CrossRef] [PubMed]
- Mazzaracchio, V.; Neagu, D.; Porchetta, A.; Marcoccio, E.; Pomponi, A.; Faggioni, G.; D’Amore, N.; Notargiacomo, A.; Pea, M.; Moscone, D.; et al. A label-free impedimetric aptasensor for the detection of Bacillus anthracis spore simulant. Biosens. Bioelectron. 2019, 126, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; You, T.; Jang, H.; Ryu, H.; Lee, E.-S.; Oh, M.-H.; Huh, Y.S.; Kim, S.M.; Jeon, T.-J. Aptamer-Conjugated Polydiacetylene Colorimetric Paper Chip for the Detection of Bacillus thuringiensis Spores. Sensors 2020, 20, 3124. [Google Scholar] [CrossRef] [PubMed]
- Ikanovic, M.; Rudzinski, W.E.; Bruno, J.G.; Allman, A.; Carrillo, M.P.; Dwarakanath, S.; Bhahdigadi, S.; Rao, P.; Kiel, J.L.; Andrews, C.J. Fluorescence Assay Based on Aptamer-Quantum Dot Binding to Bacillus thuringiensis Spores. J. Fluoresc. 2007, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Asif, A.; Mohsin, H.; Tanvir, R.; Rehman, Y. Revisiting the Mechanisms Involved in Calcium Chloride Induced Bacterial Transformation. Front. Microbiol. 2017, 8, 2169. [Google Scholar] [CrossRef]
- Raval, K.; Ganatra, T. Basics, types and applications of molecular docking: A review. IP Int. J. Compr. Adv. Pharmacol. 2022, 7, 12–16. [Google Scholar] [CrossRef]
- Dominguez, C.; Boelens, R.; Bonvin, A.M.J.J. HADDOCK: A Protein−Protein Docking Approach Based on Biochemical or Biophysical Information. J. Am. Chem. Soc. 2003, 125, 1731–1737. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Huey, R.; Olson, A.J. Using AutoDock for Ligand-Receptor Docking. Curr. Protoc. Bioinform. 2008, 24, 8.14.1–8.14.40. Available online: https://currentprotocols.onlinelibrary.wiley.com/doi/10.1002/0471250953.bi0814s24 (accessed on 21 November 2023). [CrossRef]
- Soon, S.; Nordin, N.A. In silico predictions and optimization of aptamers against Streptococcus agalactiae surface protein using computational docking. Mater. Today Proc. 2019, 16, 2096–2100. [Google Scholar] [CrossRef]
- Escamilla-Gutiérrez, A.; Córdova-Espinoza, M.G.; Sánchez-Monciváis, A.; Tecuatzi-Cadena, B.; Regalado-García, A.G.; Medina-Quero, K. In silico selection of aptamers for bacterial toxins detection. J. Biomol. Struct. Dyn. 2023, 41, 10909–10918. [Google Scholar] [CrossRef]
- Moradi, M.; Mohabatkar, H.; Behbahani, M.; Dini, G. Application of G-quadruplex aptamer conjugated MSNs to deliver ampicillin for suppressing S. aureus biofilm on mice bone. Arab. J. Chem. 2022, 15, 104274. [Google Scholar] [CrossRef]
- Selvam, R.; Lim, I.H.Y.; Lewis, J.C.; Lim, C.H.; Yap, M.K.K.; Tan, H.S. Selecting antibacterial aptamers against the BamA protein in Pseudomonas aeruginosa by incorporating genetic algorithm to optimise computational screening method. Sci. Rep. 2023, 13, 7582. [Google Scholar] [CrossRef] [PubMed]
- Yuan, K.; Meng, H.-M.; Wu, Y.; Chen, J.; Xu, H.; Qu, L.; Li, L.; Li, Z. Extracellular Milieu and Membrane Receptor Dual-Driven DNA Nanorobot for Accurate In Vivo Tumor Imaging. CCS Chem. 2022, 4, 1597–1609. [Google Scholar] [CrossRef]
- Ebrahimi, S.B.; Samanta, D.; Cheng, H.F.; Nathan, L.I.; Mirkin, C.A. Forced Intercalation (FIT)-Aptamers. J. Am. Chem. Soc. 2019, 141, 13744–13748. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, G.N.; Lee, M.P.H.; Neidle, S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature 2002, 417, 876–880. [Google Scholar] [CrossRef]
- Macaya, R.F.; Schultze, P.; Smith, F.W.; Roe, J.A.; Feigon, J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA 1993, 90, 3745–3749. [Google Scholar] [CrossRef]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and Modifications of Aptamers Selected from Live Cancer Cell Lines. ChemBioChem 2007, 8, 603–606. [Google Scholar] [CrossRef]
- Bing, T.; Yang, X.; Mei, H.; Cao, Z.; Shangguan, D. Conservative secondary structure motif of streptavidin-binding aptamers generated by different laboratories. Bioorganic Med. Chem. 2010, 18, 1798–1805. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Wang, C.; Yu, H.; Li, Y.; Zhao, Q.; Zhou, X.; Li, C.; Liu, M. Structural basis for high-affinity recognition of aflatoxin B1 by a DNA aptamer. Nucleic Acids Res. 2023, 51, 7666–7674. [Google Scholar] [CrossRef]
- Duchardt-Ferner, E.; Juen, M.; Bourgeois, B.; Madl, T.; Kreutz, C.; Ohlenschläger, O.; Wöhnert, J. Structure of an RNA aptamer in complex with the fluorophore tetramethylrhodamine. Nucleic Acids Res. 2020, 48, 949–961. [Google Scholar] [CrossRef]
- Schultze, P.; Macaya, R.F.; Feigon, J. Three-dimensional solution Structure of the Thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG. J. Mol. Biol. 1994, 235, 1532–1547. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Leroy, J.-L.; Guéron, M. An Intramolecular i-Motif: The Solution Structure and Base-pair Opening Kinetics of 9d(5mCCT3CCT3ACCT3CC). J. Mol. Biol. 1998, 278, 949–965. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.W.; Phan, A.T. Structural Basis of DNA Quadruplex–Duplex Junction Formation. Angew. Chem. Int. Ed. 2013, 52, 8566–8569. [Google Scholar] [CrossRef] [PubMed]
- Dolgosheina, E.V.; Jeng, S.C.Y.; Panchapakesan, S.S.S.; Cojocaru, R.; Chen, P.S.K.; Wilson, P.D.; Hawkins, N.; Wiggins, P.A.; Unrau, P.J. RNA Mango Aptamer-Fluorophore: A Bright, High-Affinity Complex for RNA Labeling and Tracking. ACS Chem. Biol. 2014, 9, 2412–2420. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Krafčiková, P.; Víglaský, V.; Strehlitz, B. G-quadruplex aptamer targeting Protein A and its capability to detect Staphylococcus aureus demonstrated by ELONA. Sci. Rep. 2016, 6, 33812. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, V.; Mir, B.; Alieva, R.; Arutyunyan, A.; Oleynikov, I.; Novikov, R.; Boravleva, E.; Kamzeeva, P.; Zatsepin, T.; Aralov, A.; et al. Unveiling the unusual i-motif-derived architecture of a DNA aptamer exhibiting high affinity for influenza A virus. ChemRxiv 2024. Available online: https://chemrxiv.org/engage/chemrxiv/article-details/65aac50c9138d231614d5daf (accessed on 6 March 2024).
- Lu, X.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef] [PubMed]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.; Neubock, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef]
- Biesiada, M.; Purzycka, K.J.; Szachniuk, M.; Blazewicz, J.; Adamiak, R.W. Automated RNA 3D Structure Prediction with RNAComposer. In RNA Structure Determination; Turner, D.H., Mathews, D.H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; Volume 1490, pp. 199–215. Available online: http://link.springer.com/10.1007/978-1-4939-6433-8_13 (accessed on 30 November 2023).
- Thevendran, R.; Citartan, M. Assays to Estimate the Binding Affinity of Aptamers. Talanta 2022, 238, 122971. [Google Scholar] [CrossRef]
- Kaur, H.; Yung, L.-Y.L. Probing High Affinity Sequences of DNA Aptamer against VEGF165. PLoS ONE 2012, 7, e31196. [Google Scholar] [CrossRef]
- Moon, J.; Kim, G.; Lee, S.; Park, S. Identification of Salmonella typhimurium-specific DNA aptamers developed using whole-cell SELEX and FACS analysis. J. Microbiol. Methods 2013, 95, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Ye, M.; Lu, M.; Chen, X.; Wu, S. DNA aptamers selection and characterization for development of impedimetric aptasensor for Bacillus cereus at different growing stages. Adv. Agrochem 2023, 2, 284–290. [Google Scholar] [CrossRef]
- Yılmaz, D.; Muslu, T.; Parlar, A.; Kurt, H.; Yüce, M. SELEX against whole-cell bacteria resulted in lipopolysaccharide binding aptamers. J. Biotechnol. 2022, 354, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.-L.; Lacroix, L. Analysis of Thermal Melting Curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef]
- Luo, Y.; Granzhan, A.; Verga, D.; Mergny, J. FRET-MC: A fluorescence melting competition assay for studying G4 structures in vitro. Biopolymers 2020, 112, e23415. [Google Scholar] [CrossRef]
- Esposito, V.; Scuotto, M.; Capuozzo, A.; Santamaria, R.; Varra, M.; Mayol, L.; Virgilio, A.; Galeone, A. A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance. Org. Biomol. Chem. 2014, 12, 8840–8843. [Google Scholar] [CrossRef]
- Mergny, J.-L. Thermal difference spectra: A specific signature for nucleic acid structures. Nucleic Acids Res. 2005, 33, e138. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, I.; Shafer, R.H. Effect of Loop Sequence and Size on DNA Aptamer Stability. Biochemistry 2000, 39, 1462–1468. [Google Scholar] [CrossRef]
- Reilly, S.M.; Morgan, R.K.; Brooks, T.A.; Wadkins, R.M. Effect of Interior Loop Length on the Thermal Stability and pKa of i-Motif DNA. Biochemistry 2015, 54, 1364–1370. [Google Scholar] [CrossRef]
- Ahmed, S.; Kaushik, M.; Chaudhary, S.; Kukreti, S. Structural polymorphism of a cytosine-rich DNA sequence forming i-motif structure: Exploring pH based biosensors. Int. J. Biol. Macromol. 2018, 111, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Kankia, B.I.; Marky, L.A. Folding of the Thrombin Aptamer into a G-Quadruplex with Sr2+: Stability, Heat, and Hydration. J. Am. Chem. Soc. 2001, 123, 10799–10804. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Chen, Z.; Liu, D.; Jiang, H.; Zhang, Z.-K.; Lu, A.; Zhang, B.-T.; Yu, Y.; Zhang, G. Structural Biology for the Molecular Insight between Aptamers and Target Proteins. Int. J. Mol. Sci. 2021, 22, 4093. [Google Scholar] [CrossRef] [PubMed]
- Heredia, F.L.; Roche-Lima, A.; Parés-Matos, E.I. A novel artificial intelligence-based approach for identification of deoxynucleotide aptamers. PLoS Comput. Biol. 2021, 17, e1009247. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Pei, W.; Han, Y.; Jayaseelan, S.; Shekhtman, A.; Shi, H.; Niu, L. One RNA aptamer sequence, two structures: A collaborating pair that inhibits AMPA receptors. Nucleic Acids Res. 2009, 37, 4022–4032. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.J.; Favorov, O.; Li, K.; Nuthanakanti, A.; Hussein, D.; Michaud, A.; Lafontaine, D.A.; Busan, S.; Serganov, A.; Aubé, J.; et al. SHAPE-enabled fragment-based ligand discovery for RNA. Proc. Natl. Acad. Sci. USA 2022, 119, e2122660119. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zheng, Y.-T.; Su, A.-M.; Sun, B.; Xi, X.-G.; Hou, X.-M. Remodeling the conformational dynamics of I-motif DNA by helicases in ATP-independent mode at acidic environment. iScience 2022, 25, 103575. [Google Scholar] [CrossRef] [PubMed]
- Kypr, J.; Kejnovska, I.; Renciuk, D.; Vorlickova, M. Circular dichroism and conformational polymorphism of DNA. Nucleic Acids Res. 2009, 37, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, P.; Dembska, A.; Juskowiak, B. Monitoring of pH Using an i-Motif-Forming Sequence Containing a Fluorescent Cytosine Analogue, tC. Molecules 2019, 24, 952. [Google Scholar] [CrossRef]
- Chen, C.; Li, M.; Xing, Y.; Li, Y.; Joedecke, C.C.; Jin, J.; Yang, Z.; Liu, D. Study of pH-Induced Folding and Unfolding Kinetics of the DNA i-Motif by Stopped-Flow Circular Dichroism. Langmuir 2012, 28, 17743–17748. [Google Scholar] [CrossRef]
- Santos, T.; Lopes-Nunes, J.; Alexandre, D.; Miranda, A.; Figueiredo, J.; Silva, M.S.; Mergny, J.-L.; Cruz, C. Stabilization of a DNA aptamer by ligand binding. Biochimie 2022, 200, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.-H.; Chen, R.-H.; Lee, C.-H.; Chang, Y.; Chen, C.-S.; Chen, W.-Y. Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry. Colloids Surf. B Biointerfaces 2011, 88, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Balasco, N.; Autiero, I.; Vitagliano, L.; Sica, F. Structural Insights into Protein–Aptamer Recognitions Emerged from Experimental and Computational Studies. Int. J. Mol. Sci. 2023, 24, 16318. [Google Scholar] [CrossRef] [PubMed]
- Wijmenga, S.S.; Van Buuren, B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998, 32, 287–387. [Google Scholar] [CrossRef]
- Someya, T.; Baba, S.; Fujimoto, M.; Kawai, G.; Kumasaka, T.; Nakamura, K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: Insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012, 40, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Menichelli, E.; Lam, B.J.; Wang, Y.; Wang, V.S.; Shaffer, J.; Tjhung, K.F.; Bursulaya, B.; Nguyen, T.N.; Vo, T.; Alper, P.B.; et al. Discovery of small molecules that target a tertiary-structured RNA. Proc. Natl. Acad. Sci. USA 2022, 119, e2213117119. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Napolitano, V.; Rossitto, E.; Osman, W.; Nagano, M.; Wakui, K.; Popowicz, G.M.; Yoshimoto, K.; Sica, F. Steric hindrance and structural flexibility shape the functional properties of a guanine-rich oligonucleotide. Nucleic Acids Res. 2023, 51, 8880–8890. [Google Scholar] [CrossRef] [PubMed]
- Kratschmer, C.; Levy, M. Effect of Chemical Modifications on Aptamer Stability in Serum. Nucleic Acid Ther. 2017, 27, 335–344. [Google Scholar] [CrossRef]
- Peng, C.G.; Damha, M.J. G-quadruplex induced stabilization by 2′-deoxy-2′-fluoro-d-arabinonucleic acids (2′F-ANA). Nucleic Acids Res. 2007, 35, 4977–4988. [Google Scholar]
- Elskens, J.P.; Elskens, J.M.; Madder, A. Chemical Modification of Aptamers for Increased Binding Affinity in Diagnostic Applications: Current Status and Future Prospects. Int. J. Mol. Sci. 2020, 21, 4522. [Google Scholar] [CrossRef]
- Dunn, M.R.; Jimenez, R.M.; Chaput, J.C. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Kasahara, Y.; Kuwahara, M. Artificial Specific Binders Directly Recovered from Chemically Modified Nucleic Acid Libraries. J. Nucleic Acids 2012, 2012, 156482. [Google Scholar] [CrossRef] [PubMed]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Al Bawab, A.; Ismail, S.I. Aptamers Chemistry: Chemical Modifications and Conjugation Strategies. Molecules 2019, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R. Nuclease resistance of DNA nanostructures. Nat. Rev. Chem. 2021, 5, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, G.; Wang, T.; Fu, J.; Li, R.; Song, L.; Wang, Z.; Ding, B.; Chen, F. Enhanced Stability of DNA Nanostructures by Incorporation of Unnatural Base Pairs. ChemPhysChem 2017, 18, 2977–2980. [Google Scholar] [CrossRef]
- Tabuchi, Y.; Yang, J.; Taki, M. Relative Nuclease Resistance of a DNA Aptamer Covalently Conjugated to a Target Protein. Int. J. Mol. Sci. 2022, 23, 7778. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, A.; Edwardson, T.G.W.; Hancock, M.A.; Dore, M.D.; Sleiman, H.F. Development of DNA Nanostructures for High-Affinity Binding to Human Serum Albumin. J. Am. Chem. Soc. 2017, 139, 7355–7362. [Google Scholar] [CrossRef] [PubMed]
- Ponnuswamy, N.; Bastings, M.M.C.; Nathwani, B.; Ryu, J.H.; Chou, L.Y.T.; Vinther, M.; Li, W.A.; Anastassacos, F.M.; Mooney, D.J.; Shih, W.M. Oligolysine-based coating protects DNA nanostructures from low-salt denaturation and nuclease degradation. Nat. Commun. 2017, 8, 15654. [Google Scholar] [CrossRef]
- Keum, J.-W.; Bermudez, H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. 2009, 45, 7036–7038. [Google Scholar] [CrossRef]
- Gerling, T.; Kube, M.; Kick, B.; Dietz, H. Sequence-programmable covalent bonding of designed DNA assemblies. Sci. Adv. 2018, 4, eaau1157. [Google Scholar] [CrossRef] [PubMed]
- El-Khoury, R.; Damha, M.J. End-ligation can dramatically stabilize i-motifs at neutral pH. Chem. Commun. 2023, 59, 3715–3718. [Google Scholar] [CrossRef]
- Hahn, J.; Wickham, S.F.J.; Shih, W.M.; Perrault, S.D. Addressing the Instability of DNA Nanostructures in Tissue Culture. ACS Nano 2014, 8, 8765–8775. [Google Scholar] [CrossRef]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2018, 4, 46–54. [Google Scholar] [CrossRef]
- Dausse, E.; Da Rocha Gomes, S.; Toulmé, J.J. Aptamers: A new class of oligonucleotides in the drug discovery pipeline? Curr. Opin. Pharmacol. 2009, 9, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Röthlisberger, P.; Hollenstein, M. Aptamer chemistry. Adv. Drug Deliv. Rev. 2018, 134, 3–21. [Google Scholar] [CrossRef]
- Chan, K.Y.; Kinghorn, A.B.; Hollenstein, M.; Tanner, J.A. Chemical Modifications for a Next Generation of Nucleic Acid Aptamers. ChemBioChem 2022, 23, e202200006. [Google Scholar] [CrossRef]
- Chowdary Akkina, R.; Payala, V.; Sushma Maganti, S. Tools for Rapid Detection and Control of Foodborne Microbial Pathogens. In Foodborne Pathogens—Recent Advances in Control and Detection; Lamas, A., Manuel Franco, C., Regal, P., Eds.; IntechOpen: Visakhapatnam, India, 2023; Available online: https://www.intechopen.com/chapters/81425 (accessed on 14 November 2023).
- Wandtke, T.; Wędrowska, E.; Szczur, M.; Przybylski, G.; Libura, M.; Kopiński, P. Aptamers—Diagnostic and Therapeutic Solution in SARS-CoV-2. Int. J. Mol. Sci. 2022, 23, 1412. [Google Scholar] [CrossRef] [PubMed]
- Wandtke, T.; Woźniak, J.; Kopiński, P. Aptamers in Diagnostics and Treatment of Viral Infections. Viruses 2015, 7, 751–780. [Google Scholar] [CrossRef]
- Wang, T.; Chen, C.; Larcher, L.M.; Barrero, R.A.; Veedu, R.N. Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnol. Adv. 2019, 37, 28–50. [Google Scholar] [CrossRef]
- Wang, W.; Yuan, W.; Wang, D.; Mai, X.; Wang, D.; Zhu, Y.; Liu, F.; Sun, Z. Dual-mode sensor based on the synergy of magnetic separation and functionalized probes for the ultrasensitive detection of Clostridium perfringens. RSC Adv. 2022, 12, 25744–25752. [Google Scholar] [CrossRef]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Electrochemical Detection of Protein Using a Double Aptamer Sandwich. Anal. Lett. 2004, 37, 2901–2909. [Google Scholar] [CrossRef]
- Rizzotto, F.; Khalife, M.; Hou, Y.; Chaix, C.; Lagarde, F.; Scaramozzino, N.; Vidic, J. Recent Advances in Electrochemical Biosensors for Food Control. Micromachines 2023, 14, 1412. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Lai, R.Y.; Plaxco, K.W. Preparation of electrode-immobilized, redox-modified oligonucleotides for electrochemical DNA and aptamer-based sensing. Nat. Protoc. 2007, 2, 2875–2880. [Google Scholar] [CrossRef]
- Vidic, J.; Vizzini, P.; Manzano, M.; Kavanaugh, D.; Ramarao, N.; Zivkovic, M.; Radonic, V.; Knezevic, N.; Giouroudi, I.; Gadjanski, I. Point-of-Need DNA Testing for Detection of Foodborne Pathogenic Bacteria. Sensors 2019, 19, 1100. [Google Scholar] [CrossRef] [PubMed]
- Majdinasab, M.; Hayat, A.; Marty, J.L. Aptamer-based assays and aptasensors for detection of pathogenic bacteria in food samples. TrAC Trends Anal. Chem. 2018, 107, 60–77. [Google Scholar] [CrossRef]
- Fischer, C.; Hünniger, T.; Jarck, J.-H.; Frohnmeyer, E.; Kallinich, C.; Haase, I.; Hahn, U.; Fischer, M. Food Sensing: Aptamer-Based Trapping of Bacillus cereus Spores with Specific Detection via Real Time PCR in Milk. J. Agric. Food Chem. 2015, 63, 8050–8057. [Google Scholar] [CrossRef]
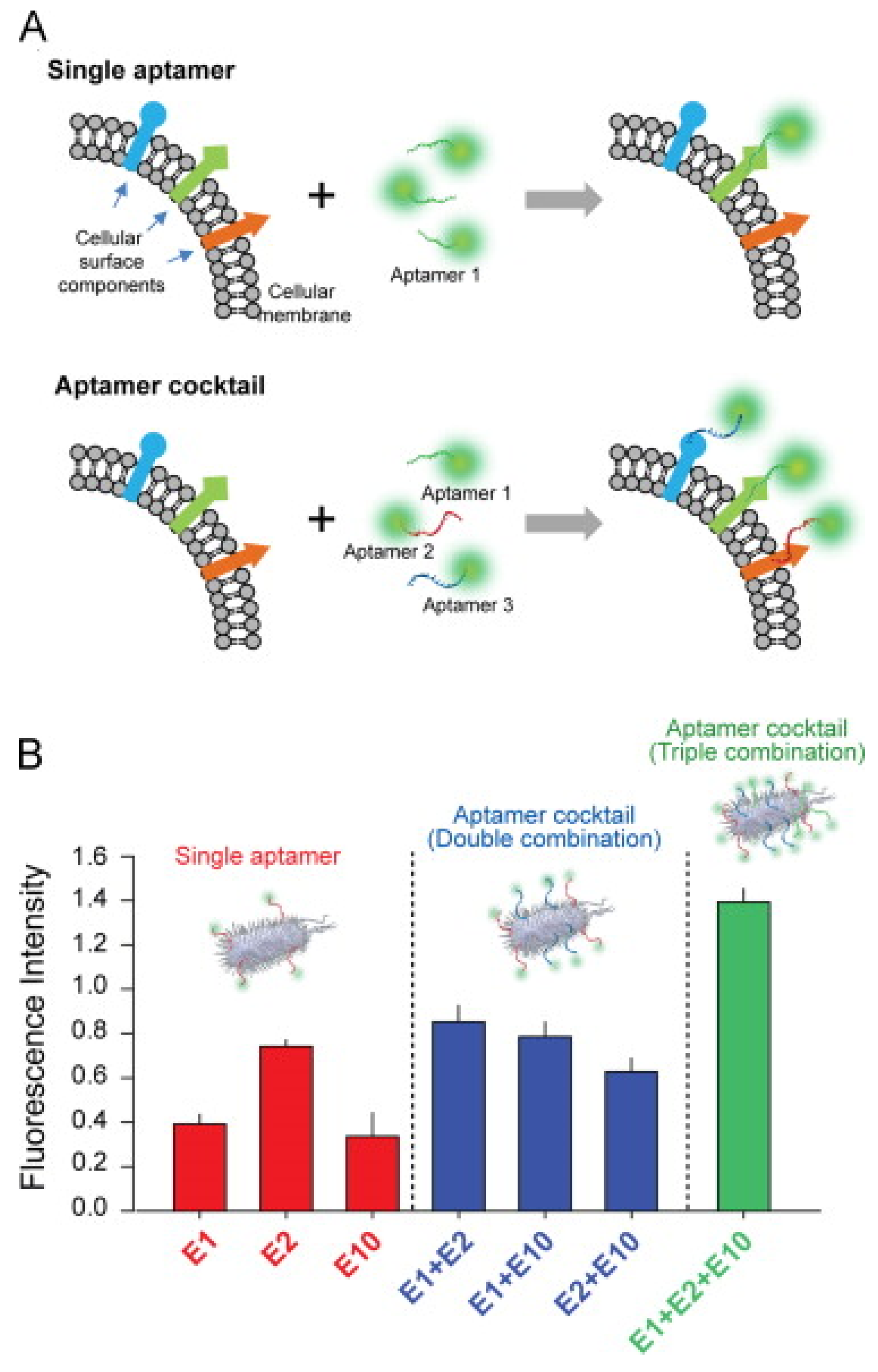
- Kim, Y.S.; Chung, J.; Song, M.Y.; Jurng, J.; Kim, B.C. Aptamer cocktails: Enhancement of sensing signals compared to single use of aptamers for detection of bacteria. Biosens. Bioelectron. 2014, 54, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Liu, P.P.; Yan, J.; Luan, D.; Sun, T.; Bian, X. Dual Synthetic Receptor-Based Sandwich Electrochemical Sensor for Highly Selective and Ultrasensitive Detection of Pathogenic Bacteria at the Single-Cell Level. Anal. Chem. 2023, 95, 5561–5567. [Google Scholar] [CrossRef]
- Ding, Z.; Yang, S.; Wang, J.; Zhao, Z.; Xu, H.; Chen, Z.; Liu, Z.; Wang, Y.; Bao, J.; Chang, K.; et al. Rolling Circle Amplification/G-Quadruplex-Based Dual Signal Ratiometric Electrochemical Aptasensor for Ultrasensitive Detection of Pathogenic Bacteria. ChemElectroChem 2023, 10, e202300257. [Google Scholar] [CrossRef]
- Demidov, V.V. Rolling-circle amplification in DNA diagnostics: The power of simplicity. Expert Rev. Mol. Diagn. 2002, 2, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Ali, M.M.; Brook, M.A.; Li, Y. Rolling Circle Amplification: Applications in Nanotechnology and Biodetection with Functional Nucleic Acids. Angew. Chem. Int. Ed. 2008, 47, 6330–6337. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.-T.; Nie, J.; Zhang, D.-W.; Chen, J.-T.; Zhou, Y.-L.; Zhang, X.-X. Methylene Blue as a G-Quadruplex Binding Probe for Label-Free Homogeneous Electrochemical Biosensing. Anal. Chem. 2014, 86, 9489–9495. [Google Scholar] [CrossRef] [PubMed]
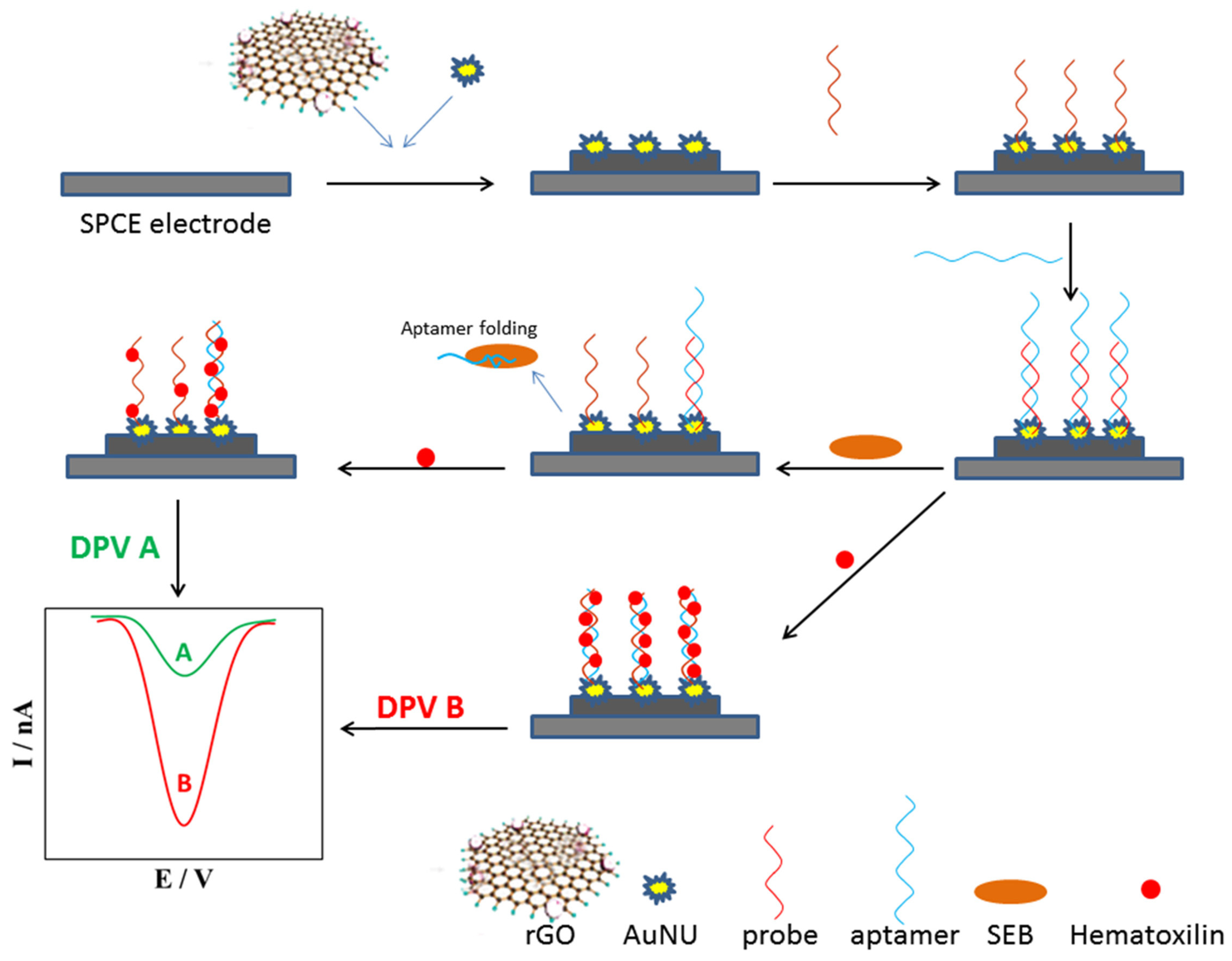
- Nodoushan, S.M.; Nasirizadeh, N.; Amani, J.; Halabian, R.; Fooladi, A.A.I. An electrochemical aptasensor for staphylococcal enterotoxin B detection based on reduced graphene oxide and gold nano-urchins. Biosens. Bioelectron. 2019, 127, 221–228. [Google Scholar] [CrossRef]
- Gholamzad, M.; Khatami, M.R.; Ghassemi, S.; Vaise Malekshahi, Z.; Shooshtari, M.B. Detection of Staphylococcus Enterotoxin B (SEB) Using an Immunochromatographic Test Strip. Jundishapur J. Microbiol. 2015, 8, e26793. Available online: https://brieflands.com/articles/jjm-56578.html (accessed on 16 November 2023). [CrossRef] [PubMed]
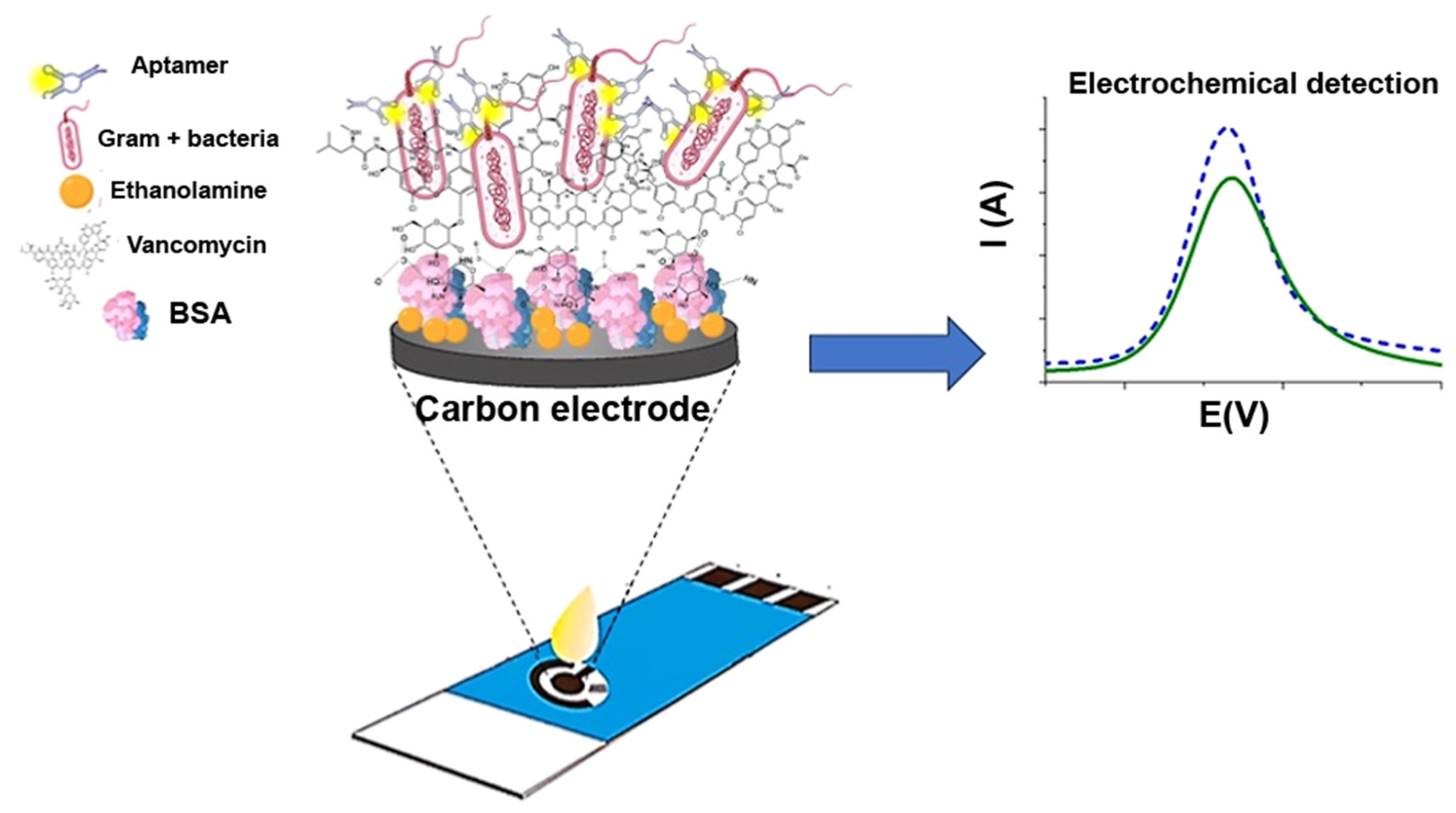
- Novakovic, Z.; Khalife, M.; Costache, V.; Camacho, M.J.; Cardoso, S.; Martins, V.; Gadjanski, I.; Radovic, M.; Vidic, J. Rapid Detection and Identification of Vancomycin-Sensitive Bacteria Using an Electrochemical Aptsa-Sensor. ACS Omega 2024, 9, 2841–2849. [Google Scholar] [CrossRef] [PubMed]
- Bajaj, A.; Abutoama, M.; Isaacs, S.; Abuleil, M.J.; Yaniv, K.; Kushmaro, A.; Modic, M.; Cvelbar, U.; Abdulhalim, I. Biofilm growth monitoring using guided wave ultralong-range Surface Plasmon Resonance: A proof of concept. Biosens. Bioelectron. 2023, 228, 115204. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-J.; Kim, H.-S.; Chon, J.-W.; Kim, D.-H.; Hyeon, J.-Y.; Seo, K.-H. New colorimetric aptasensor for rapid on-site detection of Campylobacter jejuni and Campylobacter coli in chicken carcass samples. Anal. Chim. Acta 2018, 1029, 78–85. [Google Scholar] [CrossRef]
- Marin, M.; Rizzotto, F.; Léguillier, V.; Péchoux, C.; Borezee-Durant, E.; Vidic, J. Naked-eye detection of Staphylococcus aureus in powdered milk and infant formula using gold nanoparticles. J. Microbiol. Methods 2022, 201, 106578. [Google Scholar] [CrossRef]
- Balbinot, S.; Srivastav, A.M.; Vidic, J.; Abdulhalim, I.; Manzano, M. Plasmonic biosensors for food control. Trends Food Sci. Technol. 2021, 111, 128–140. [Google Scholar] [CrossRef]
- Ko, J.; Park, S.-G.; Lee, S.; Wang, X.; Mun, C.; Kim, S.; Kim, D.-H.; Choo, J. Culture-Free Detection of Bacterial Pathogens on Plasmonic Nanopillar Arrays Using Rapid Raman Mapping. ACS Appl. Mater. Interfaces 2018, 10, 6831–6840. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Ouyang, Q.; Zhao, J. Fabricating a Novel Raman Spectroscopy-Based Aptasensor for Rapidly Sensing Salmonella typhimurium. Food Anal. Methods 2017, 10, 3032–3041. [Google Scholar] [CrossRef]
- Bruno, J.G.; Carrillo, M.P. Development of Aptamer Beacons for Rapid Presumptive Detection of Bacillus Spores. J. Fluoresc. 2012, 22, 915–924. [Google Scholar] [CrossRef]
- Zhou, Z.; Lan, X.; Zhu, L.; Zhang, Y.; Chen, K.; Zhang, W.; Xu, W. Portable dual-aptamer microfluidic chip biosensor for Bacillus cereus based on aptamer tailoring and dumbbell-shaped probes. J. Hazard. Mater. 2023, 445, 130545. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Yan, H.; Zheng, Y.; Zu, Y.; Yang, S.; Hu, H.; Shi, S.; Liang, H.; Niu, X. Joint concanavalin A-aptamer enabled dual recognition for anti-interference visual detection of Salmonella typhimurium in complex food matrices. Food Chem. 2023, 426, 136581. [Google Scholar] [CrossRef]
- Martinović, T.; Andjelković, U.; Gajdošik, M.; Rešetar, D.; Josić, D. Foodborne pathogens and their toxins. J. Proteom. 2016, 147, 226–235. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Staphylococcus aureus toxins. Curr. Opin. Microbiol. 2014, 17, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Tătaru, A.-M.; Canciu, A.; Tertiș, M.; Cristea, C.; Cernat, A. Staphylococcus aureus—Review on potential targets for sensors development. Bioelectrochemistry 2023, 153, 108492. [Google Scholar] [CrossRef]
- Mainil, J. Escherichia coli virulence factors. Vet. Immunol. Immunopathol. 2013, 152, 2–12. [Google Scholar] [CrossRef]
- Smits, W.K.; Lyras, D.; Lacy, D.B.; Wilcox, M.H.; Kuijper, E.J. Clostridium difficile infection. Nat. Rev. Dis. Primers 2016, 2, 16020. [Google Scholar] [CrossRef]
- Pickett, C.L.; Pesci, E.C.; Cottle, D.L.; Russell, G.; Erdem, A.N.; Zeytin, H. Prevalence of cytolethal distending toxin production in Campylobacter jejuni and relatedness of Campylobacter sp. cdtB gene. Infect. Immun. 1996, 64, 2070–2078. [Google Scholar] [CrossRef] [PubMed]
- Churchill, R.L.; Lee, H.; Hall, J.C. Detection of Listeria monocytogenes and the toxin listeriolysin O in food. J. Microbiol. Methods 2006, 64, 141–170. [Google Scholar] [CrossRef] [PubMed]
- Platt-Samoraj, A. Toxigenic Properties of Yersinia enterocolitica Biotype 1A. Toxins 2022, 14, 118. [Google Scholar] [CrossRef]
- Sakaguchi, G. Clostridium botulinum toxin. Pharmacol. Ther. 1983, 19, 165–194. [Google Scholar] [CrossRef]
- Rajkovic, A. Microbial toxins and low level of foodborne exposure. Trends Food Sci. Technol. 2014, 38, 149–157. [Google Scholar] [CrossRef]
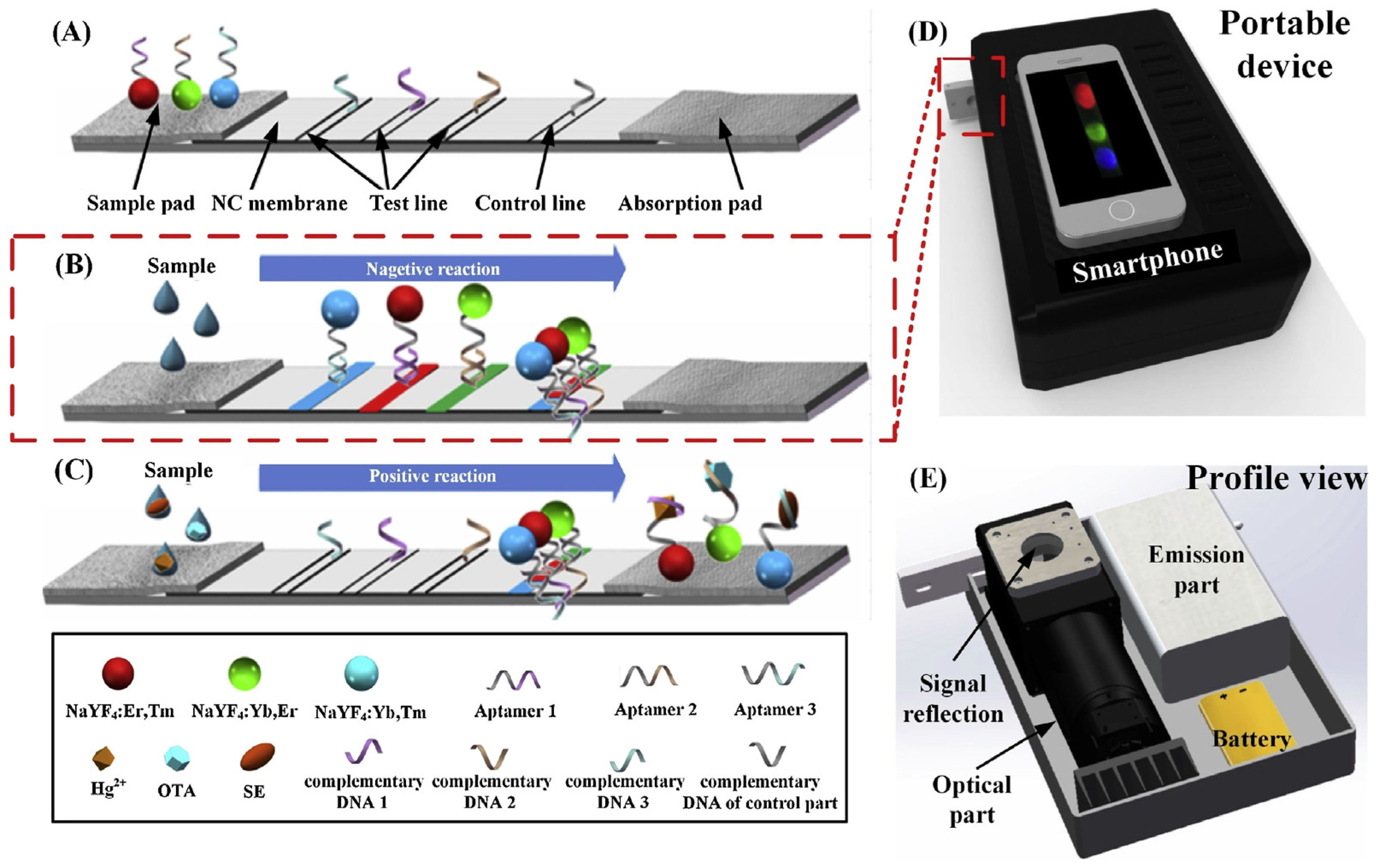
- Jin, B.; Yang, Y.; He, R.; Park, Y.I.; Lee, A.; Bai, D.; Li, F.; Lu, T.J.; Xu, F.; Lin, M. Lateral flow aptamer assay integrated smartphone-based portable device for simultaneous detection of multiple targets using upconversion nanoparticles. Sens. Actuators B Chem. 2018, 276, 48–56. [Google Scholar] [CrossRef]
- Roca, C.; Avalos-Padilla, Y.; Prieto-Simón, B.; Iglesias, V.; Ramírez, M.; Imperial, S.; Fernàndez-Busquets, X. Selection of an Aptamer against the Enzyme 1-deoxy-D-xylulose-5-phosphate Reductoisomerase from Plasmodium falciparum. Pharmaceutics 2022, 14, 2515. [Google Scholar] [CrossRef]
- Wang, K.; Gan, L.; Jiang, L.; Zhang, X.; Yang, X.; Chen, M.; Lan, X. Neutralization of Staphylococcal Enterotoxin B by an Aptamer Antagonist. Antimicrob. Agents Chemother. 2015, 59, 2072–2077. [Google Scholar] [CrossRef]
- Shin, H.-S.; Gedi, V.; Kim, J.-K.; Lee, D.-K. Detection of Gram-negative bacterial outer membrane vesicles using DNA aptamers. Sci. Rep. 2019, 9, 13167. [Google Scholar] [CrossRef]
- Rotariu, L.; Lagarde, F.; Jaffrezic-Renault, N.; Bala, C. Electrochemical biosensors for fast detection of food contaminants—Trends and perspective. TrAC Trends Anal. Chem. 2016, 79, 80–87. [Google Scholar] [CrossRef]
- Xu, L.; Li, D.; Ramadan, S.; Li, Y.; Klein, N. Facile biosensors for rapid detection of COVID-19. Biosens. Bioelectron. 2020, 170, 112673. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shi, Z.; Liu, Q. Smartphone-based biosensors for portable food evaluation. Curr. Opin. Food Sci. 2019, 28, 74–81. [Google Scholar] [CrossRef]
- Choi, J.R. Development of Point-of-Care Biosensors for COVID-19. Front. Chem. 2020, 8, 517. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. TrAC Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Dejeu, J.; Van der Heyden, A.; Spinelli, N.; Defrancq, E.; Coche-Guérente, L. Recent progress in the design of G-quadruplex–based electrochemical aptasensors. Curr. Opin. Electrochem. 2021, 30, 100812. [Google Scholar] [CrossRef]
- Kim, Y.S.; Song, M.Y.; Jurng, J.; Kim, B.C. Isolation and characterization of DNA aptamers against Escherichia coli using a bacterial cell–systematic evolution of ligands by exponential enrichment approach. Anal. Biochem. 2013, 436, 22–28. [Google Scholar] [CrossRef]
- Abbaspour, A.; Norouz-Sarvestani, F.; Noori, A.; Soltani, N. Aptamer-conjugated silver nanoparticles for electrochemical dual-aptamer-based sandwich detection of staphylococcus aureus. Biosens. Bioelectron. 2015, 68, 149–155. [Google Scholar] [CrossRef]
- Kang, Q.; Xing, X.; Zhang, S.; He, L.; Li, J.; Jiao, J.; Du, X.; Wang, S. A novel Aptamer-induced CHA amplification strategy for ultrasensitive detection of Staphylococcus aureus and NIR-triggered photothermal bactericidal Activity based on aptamer-modified magnetic Fe3O4@AuNRs. Sens. Actuators B Chem. 2023, 382, 133554. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, X.; Xia, Y.; Wu, S.; Duan, N.; Ma, X.; Wang, Z. Selection, identification and application of a DNA aptamer against Staphylococcus aureus enterotoxin A. Anal. Methods 2014, 6, 690–697. [Google Scholar] [CrossRef]
- Chen, J.; Huang, Z.; Luo, Z.; Yu, Q.; Xu, Y.; Wang, X.; Li, Y.; Duan, Y. Multichannel-Structured Three-Dimensional Chip for Highly Sensitive Pathogenic Bacteria Detection Based on Fast DNA-Programmed Signal Polymerization. Anal. Chem. 2018, 90, 12019–12026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Yang, Y.; Li, L.; Tao, X.; Song, E. Rapid detection of pathogenic bacteria based on a universal dual-recognition FRET sensing system constructed with aptamer-quantum dots and lectin-gold nanoparticles. Chin. Chem. Lett. 2023, 34, 108102. [Google Scholar] [CrossRef]
- Jiang, D.; Yang, C.; Fan, Y.; Polly Leung, H.M.; Inthavong, K.; Zhang, Y.; Li, Z.; Yang, M. Ultra-sensitive photoelectrochemical aptamer biosensor for detecting E. coli O157:H7 based on nonmetallic plasmonic two-dimensional hydrated defective tungsten oxide nanosheets coupling with nitrogen-doped graphene quantum dots (dWO3•H2O@N-GQDs). Biosens. Bioelectron. 2021, 183, 113214. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, F.R.W.; Cesca, K.; Valério, A.; de Oliveira, D.; Hotza, D. Colorimetric detection of Pseudomonas aeruginosa by aptamer-functionalized gold nanoparticles. Appl. Microbiol. Biotechnol. 2023, 107, 71–80. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Sheng, R.; Li, H.; Ahmad, W.; Chen, Q. Rapid and selective detection of Bacillus cereus in food using cDNA-based up-conversion fluorescence spectrum copy and aptamer modified magnetic separation. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 267, 120618. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Chen, L.; Teng, M.; Hao, M.; Feng, B.; Yang, F.; Shen, H.; Yu, S.; Wang, L. Dual recognition strategy for the rapid and precise detection of Bacillus cereus using post-modified nano-MOF and aptamer. Sens. Actuators B Chem. 2023, 386, 133745. [Google Scholar] [CrossRef]
- Effah, C.Y.; Ding, L.; Tan, L.; He, S.; Li, X.; Yuan, H.; Li, Y.; Liu, S.; Sun, T.; Wu, Y. A SERS bioassay based on vancomycin-modified PEI-interlayered nanocomposite and aptamer-functionalized SERS tags for synchronous detection of Acinetobacter baumannii and Klebsiella pneumoniae. Food Chem. 2023, 423, 136242. [Google Scholar] [CrossRef]
- Vishwakarma, A.; Meganathan, Y.; Ramya, M. Aptamer-based assay for rapid detection, surveillance, and screening of pathogenic Leptospira in water samples. Sci. Rep. 2023, 13, 13379. [Google Scholar] [CrossRef] [PubMed]
- Ohk, S.H.; Koo, O.K.; Sen, T.; Yamamoto, C.M.; Bhunia, A.K. Antibody-aptamer functionalized fibre-optic biosensor for specific detection of Listeria monocytogenes from food: Antibody-aptamer fibre-optic sensor for L. monocytogenes. J. Appl. Microbiol. 2010, 109, 808–817. [Google Scholar] [CrossRef]
- Sun, M.; Ma, N.; Shi, H.; Cheong, L.-Z.; Yang, W.; Qiao, Z. A HCR based multivalent aptamer amplifier for ultrasensitive detection of Salmonella. Sens. Actuators B Chem. 2023, 375, 132860. [Google Scholar] [CrossRef]
- Tavassoli, M.; Khezerlou, A.; Hamishehkar, H.; Ehsani, A.; Khalilzadeh, B. An ultrasensitive aptamer-based fluorescent on/off system for trace amount evaluation of Yersinia enterocolitica in food samples. Microchim. Acta 2023, 190, 253. [Google Scholar] [CrossRef] [PubMed]
- Luo, P.; Liu, Y.; Xia, Y.; Xu, H.; Xie, G. Aptamer biosensor for sensitive detection of toxin A of Clostridium difficile using gold nanoparticles synthesized by Bacillus stearothermophilus. Biosens. Bioelectron. 2014, 54, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Shen, Z.; Tan, L.; Yuan, J.; Gan, N. Electrochemical aptasensor for simultaneous detection of foodborne pathogens based on a double stirring bars-assisted signal amplification strategy. Sens. Actuators B Chem. 2021, 345, 130337. [Google Scholar] [CrossRef]
- Bakhshandeh, F.; Saha, S.; Sen, P.; Sakib, S.; MacLachlan, R.; Kanji, F.; Osman, E.; Soleymani, L. A universal bacterial sensor created by integrating a light modulating aptamer complex with photoelectrochemical signal readout. Biosens. Bioelectron. 2023, 235, 115359. [Google Scholar] [CrossRef] [PubMed]
- Bruno, J.G.; Carrillo, M.P.; Phillips, T.; Andrews, C.J. A Novel Screening Method for Competitive FRET-Aptamers Applied to E. coli Assay Development. J. Fluoresc. 2010, 20, 1211–1223. [Google Scholar] [CrossRef] [PubMed]
- Queirós, R.B.; De-Los-Santos-Álvarez, N.; Noronha, J.; Sales, M. A label-free DNA aptamer-based impedance biosensor for the detection of E. coli outer membrane proteins. Sens. Actuators B Chem. 2013, 181, 766–772. [Google Scholar] [CrossRef]
- Abedi, R.; Raoof, J.B.; Mohseni, M.; Hashkavayi, A.B. Development of a label-free impedimetric aptasensor for the detection of Acinetobacter baumannii bacteria. Anal. Biochem. 2023, 679, 115288. [Google Scholar] [CrossRef] [PubMed]
- Sypabekova, M.; Dukenbayev, K.; Tsepke, A.; Akisheva, A.; Oralbayev, N.; Kanayeva, D. An aptasensor for the detection of Mycobacterium tuberculosis secreted immunogenic protein MPT64 in clinical samples towards tuberculosis detection. Sci. Rep. 2019, 9, 16273. [Google Scholar] [CrossRef] [PubMed]
- Lian, Y.; He, F.; Wang, H.; Tong, F. A new aptamer/graphene interdigitated gold electrode piezoelectric sensor for rapid and specific detection of Staphylococcus aureus. Biosens. Bioelectron. 2015, 65, 314–319. [Google Scholar] [CrossRef]
- Alizadeh, N.; Memar, M.Y.; Mehramuz, B.; Abibiglou, S.S.; Hemmati, F.; Kafil, H.S. Current advances in aptamer-assisted technologies for detecting bacterial and fungal toxins. J. Appl. Microbiol. 2018, 124, 644–651. [Google Scholar] [CrossRef]
- Niles, J.C.; Marletta, M.A. Utilizing RNA Aptamers To Probe a Physiologically Important Heme-Regulated Cellular Network. ACS Chem. Biol. 2006, 1, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Shorie, M.; Sabherwal, P. Biolayer interferometry-SELEX for Shiga toxin antigenic-peptide aptamers & detection via chitosan-WSe2 aptasensor. Biosens. Bioelectron. 2020, 167, 112498. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Léguillier, V.; Heddi, B.; Vidic, J. Recent Advances in Aptamer-Based Biosensors for Bacterial Detection. Biosensors 2024, 14, 210. https://doi.org/10.3390/bios14050210
Léguillier V, Heddi B, Vidic J. Recent Advances in Aptamer-Based Biosensors for Bacterial Detection. Biosensors. 2024; 14(5):210. https://doi.org/10.3390/bios14050210
Chicago/Turabian StyleLéguillier, Vincent, Brahim Heddi, and Jasmina Vidic. 2024. "Recent Advances in Aptamer-Based Biosensors for Bacterial Detection" Biosensors 14, no. 5: 210. https://doi.org/10.3390/bios14050210
APA StyleLéguillier, V., Heddi, B., & Vidic, J. (2024). Recent Advances in Aptamer-Based Biosensors for Bacterial Detection. Biosensors, 14(5), 210. https://doi.org/10.3390/bios14050210






