Unraveling the Dynamics of SARS-CoV-2 Mutations: Insights from Surface Plasmon Resonance Biosensor Kinetics
Abstract
1. Introduction
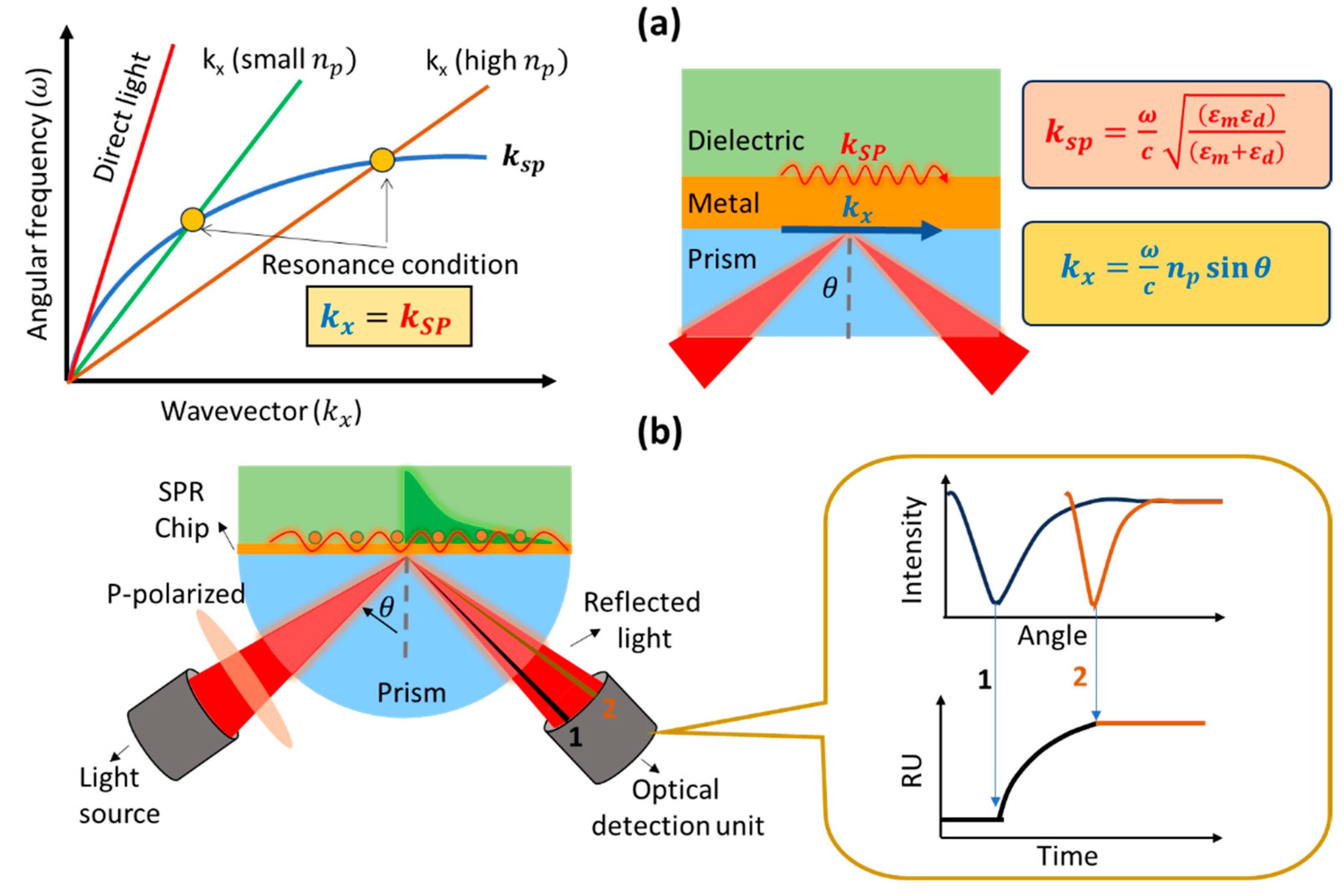
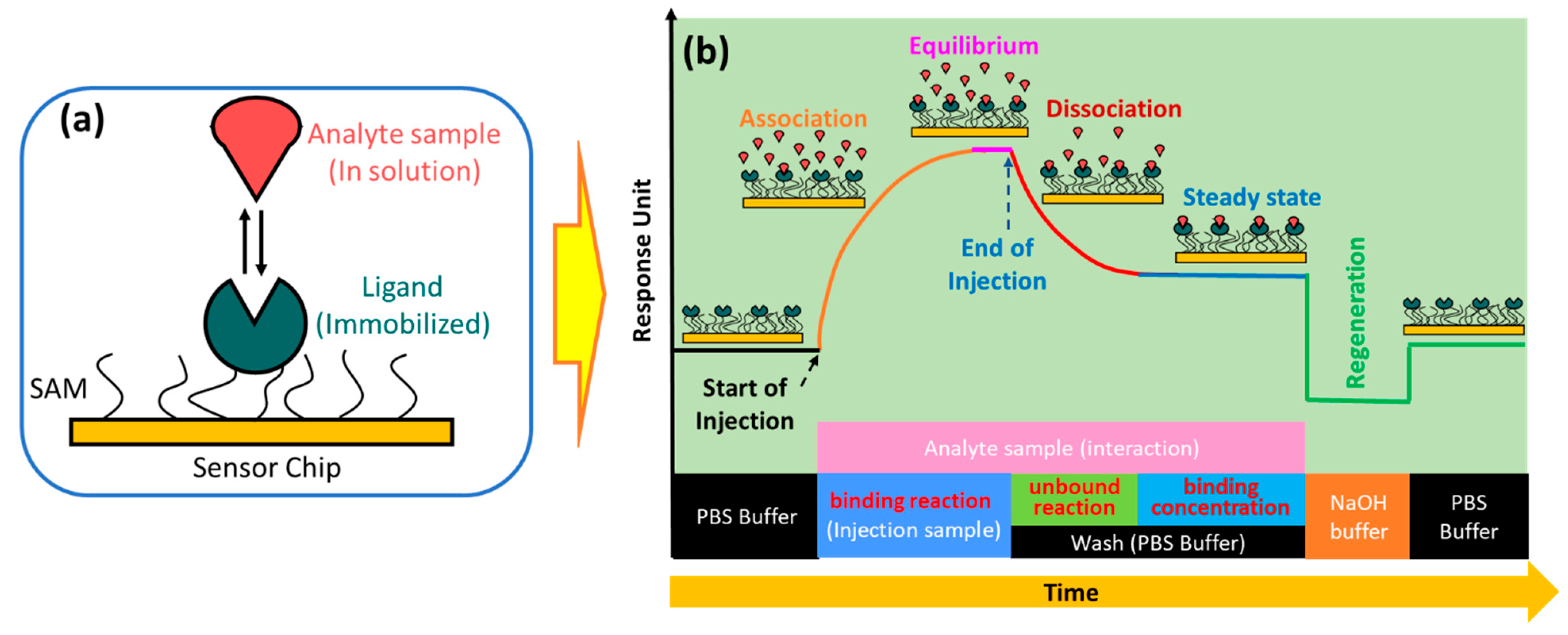
2. Working Principle and Development of a Prism-Based SPR Biosensor
3. Principle of SPR Kinetics
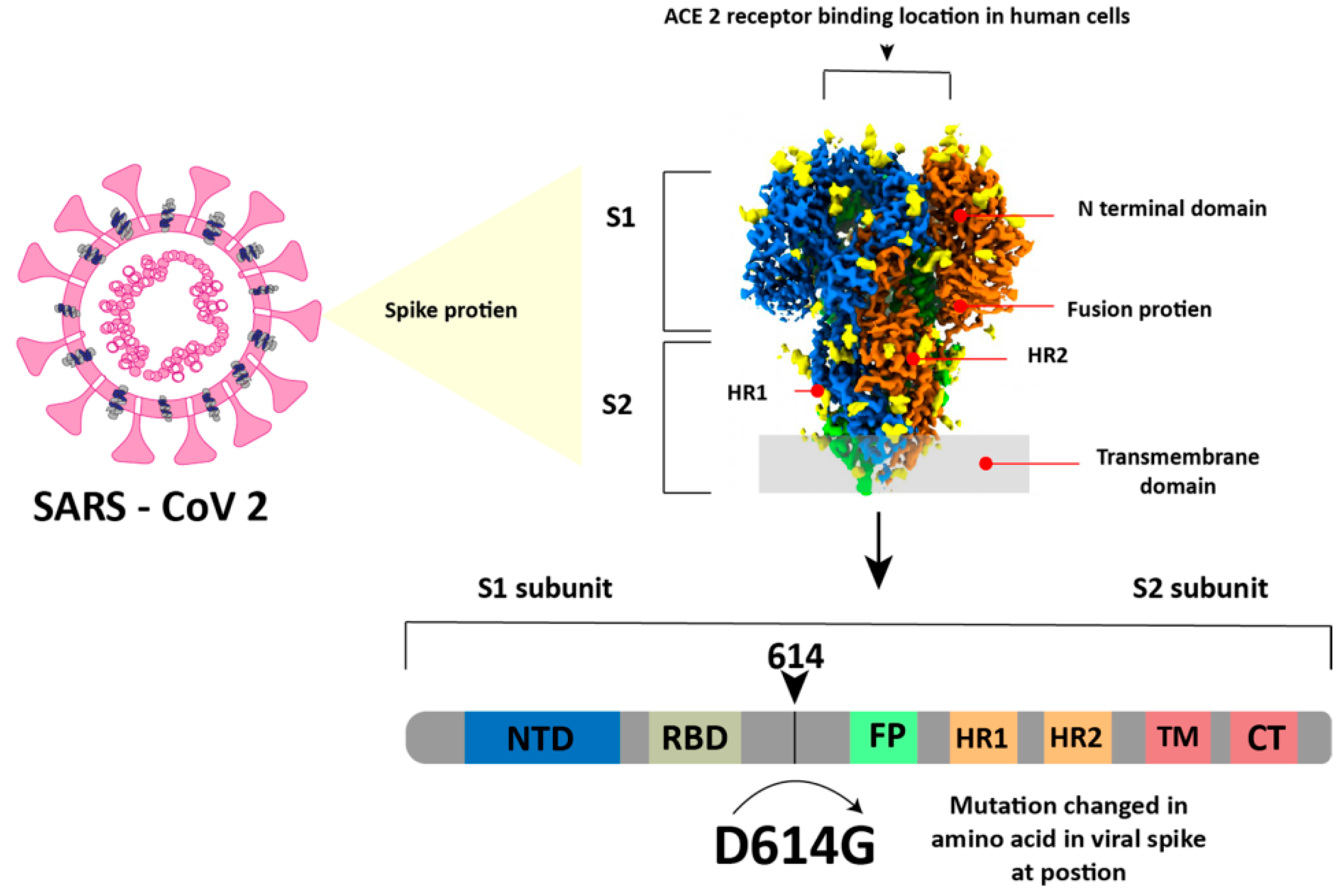
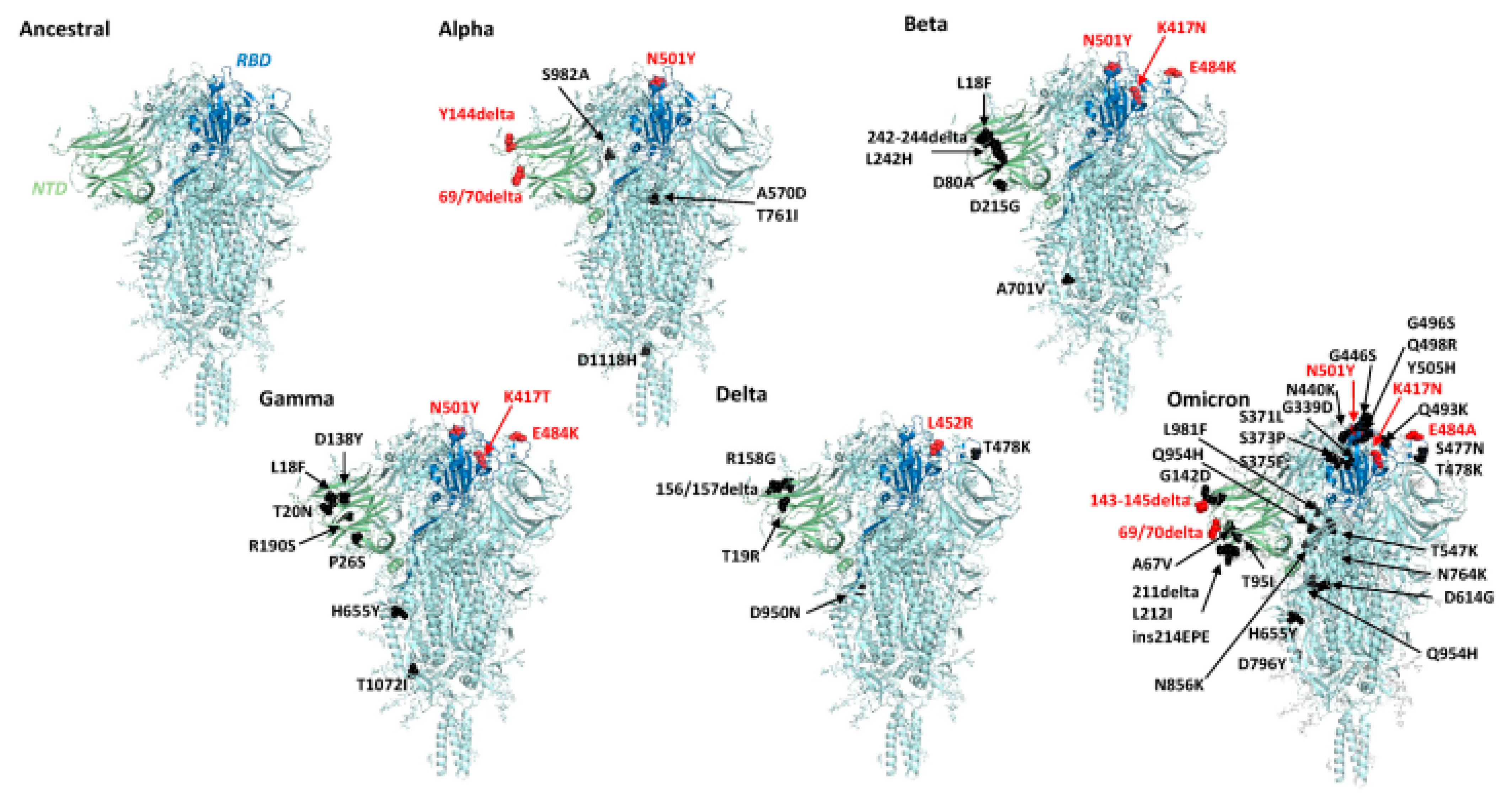
4. COVID-19 Virus and Its Mutation
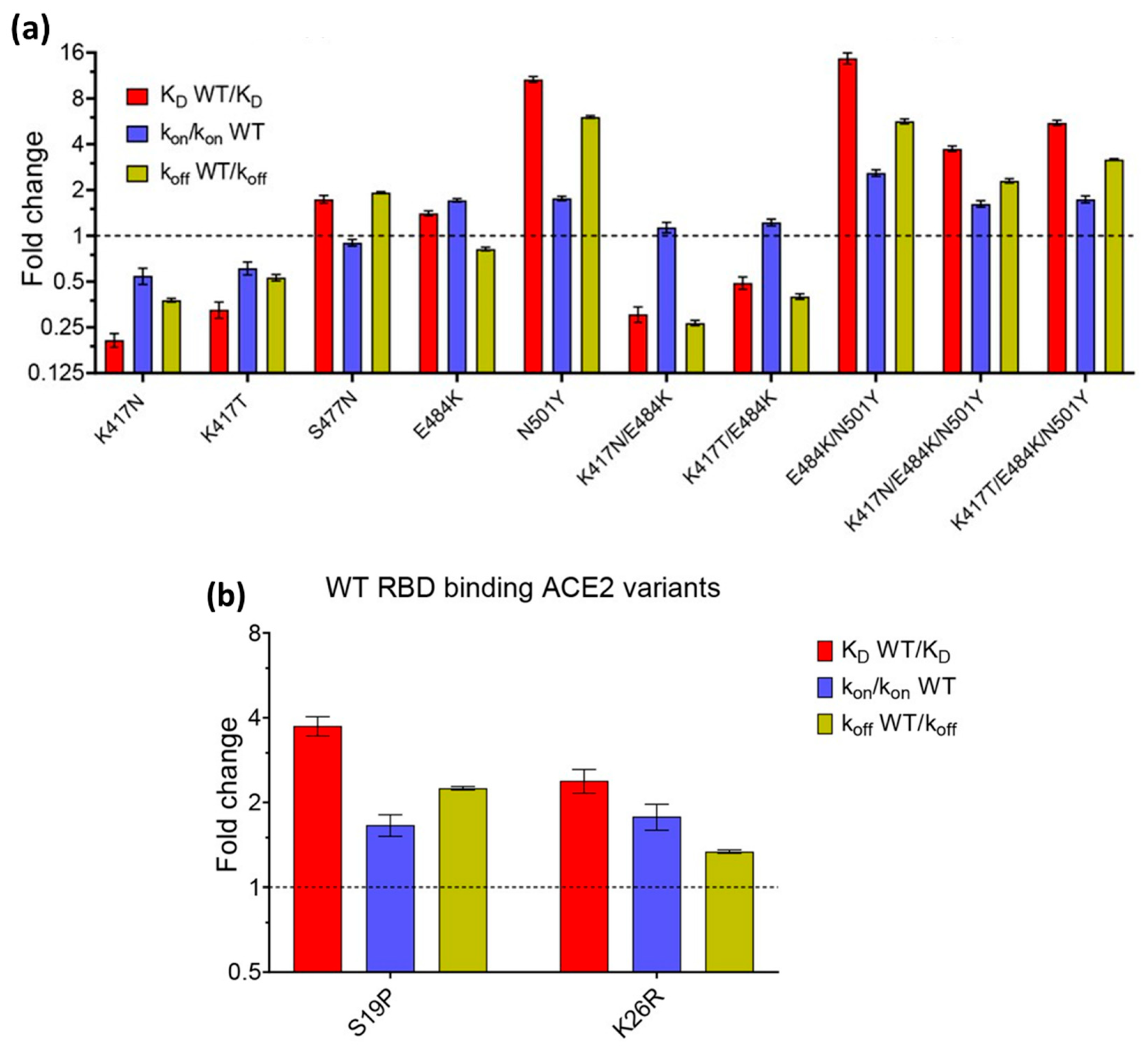
5. Application of SPR Technology for SARS-CoV-2 Detection and Analysis of Its Binding
6. Conclusions and Future Prospective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ciotti, M.; Angeletti, S.; Minieri, M.; Giovannetti, M.; Benvenuto, D.; Pascarella, S.; Sagnelli, C.; Bianchi, M.; Bernardini, S.; Ciccozzi, M. COVID-19 Outbreak: An Overview. Chemotherapy 2020, 64, 215–223. [Google Scholar] [CrossRef]
- Hui, D.S.; I Azhar, E.; Madani, T.A.; Ntoumi, F.; Kock, R.; Dar, O.; Ippolito, G.; Mchugh, T.D.; Memish, Z.A.; Drosten, C.; et al. The Continuing 2019-NCoV Epidemic Threat of Novel Coronaviruses to Global Health—The Latest 2019 Novel Coronavirus Outbreak in Wuhan, China. Int. J. Infect. Dis. 2020, 91, 264–266. [Google Scholar] [CrossRef]
- Wu, J.T.; Leung, K.; Leung, G.M. Nowcasting and Forecasting the Potential Domestic and International Spread of the 2019-NCoV Outbreak Originating in Wuhan, China: A Modelling Study. Lancet 2020, 395, 689–697. [Google Scholar] [CrossRef]
- Cosar, B.; Karagulleoglu, Z.Y.; Unal, S.; Ince, A.T.; Uncuoglu, D.B.; Tuncer, G.; Kilinc, B.R.; Ozkan, Y.E.; Ozkoc, H.C.; Demir, I.N.; et al. SARS-CoV-2 Mutations and Their Viral Variants. Cytokine Growth Factor Rev. 2022, 63, 10–22. [Google Scholar] [CrossRef]
- He, C.; Lin, C.; Mo, G.; Xi, B.; Li, A.; Huang, D.; Wan, Y.; Chen, F.; Liang, Y.; Zuo, Q.; et al. Rapid and Accurate Detection of SARS-CoV-2 Mutations Using a Cas12a-Based Sensing Platform. Biosens. Bioelectron. 2022, 198, 113857. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Naiyer, S.; Mansuri, S.; Soni, N.; Singh, V.; Bhat, K.H.; Singh, N.; Arora, G.; Mansuri, M.S. COVID-19 Diagnosis: A Comprehensive Review of the RT-QPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12, 1503. [Google Scholar] [CrossRef] [PubMed]
- Adnan, N.; Khandker, S.S.; Haq, A.; Chaity, M.A.; Khalek, A.; Nazim, A.Q.; Kaitsuka, T.; Tomizawa, K.; Mie, M.; Kobatake, E.; et al. Detection of SARS-CoV-2 by Antigen ELISA Test Is Highly Swayed by Viral Load and Sample Storage Condition. Expert Rev. Anti. Infect. Ther. 2022, 20, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Hui, T.C.H.; Khoo, H.W.; Young, B.E.; Mohideen, S.M.H.; Lee, Y.S.; Lim, C.J.; Leo, Y.S.; Kaw, G.J.L.; Lye, D.C.; Tan, C.H. Clinical Utility of Chest Radiography for Severe COVID-19. Quant. Imaging Med. Surg. 2020, 10, 1540–1550. [Google Scholar] [CrossRef] [PubMed]
- Sharif, P.M.; Nematizadeh, M.; Saghazadeh, M.; Saghazadeh, A.; Rezaei, N. Computed Tomography Scan in COVID-19: A Systematic Review and Meta-Analysis. Pol. J. Radiol. 2022, 87, e1–e23. [Google Scholar] [CrossRef]
- Oliveira, M.C.; Scharan, K.O.; Thomés, B.I.; Bernardelli, R.S.; Reese, F.B.; Kozesinski-Nakatani, A.C.; Martins, C.C.; Lobo, S.M.A.; Réa-Neto, Á. Diagnostic Accuracy of a Set of Clinical and Radiological Criteria for Screening of COVID-19 Using RT-PCR as the Reference Standard. BMC Pulm. Med. 2023, 23, 81. [Google Scholar] [CrossRef] [PubMed]
- Filchakova, O.; Dossym, D.; Ilyas, A.; Kuanysheva, T.; Abdizhamil, A.; Bukasov, R. Review of COVID-19 Testing and Diagnostic Methods. Talanta 2022, 244, 123409. [Google Scholar] [CrossRef]
- Mariani, S.; Minunni, M. Surface Plasmon Resonance Applications in Clinical Analysis. Anal. Bioanal. Chem. 2014, 406, 2303–2323. [Google Scholar] [CrossRef]
- Huo, Z.; Li, Y.; Chen, B.; Zhang, W.; Yang, X.; Yang, X. Recent Advances in Surface Plasmon Resonance Imaging and Biological Applications. Talanta 2023, 255, 124213. [Google Scholar] [CrossRef]
- Pandey, P.S.; Raghuwanshi, S.K.; Shadab, A.; Ansari, M.T.I.; Tiwari, U.K.; Kumar, S. SPR Based Biosensing Chip for COVID-19 Diagnosis—A Review. IEEE Sens. J. 2022, 22, 13800–13810. [Google Scholar] [CrossRef]
- Syed Nor, S.N.; Rasanang, N.S.; Karman, S.; Zaman, W.S.W.K.; Harun, S.W.; Arof, H. A Review: Surface Plasmon Resonance-Based Biosensor for Early Screening of SARS-CoV2 Infection. IEEE Access 2022, 10, 1228–1244. [Google Scholar] [CrossRef]
- Mauriz, E.; Lechuga, L.M. Current Trends in Spr Biosensing of SARS-CoV-2 Entry Inhibitors. Chemosensors 2021, 9, 330. [Google Scholar] [CrossRef]
- Wei, H.; Zhang, C.; Du, X.; Zhang, Z. Research Progress of Biosensors for Detection of SARS-CoV-2 Variants Based on ACE2. Talanta 2023, 251, 123813. [Google Scholar] [CrossRef] [PubMed]
- Gul, I.; Zhai, S.; Zhong, X.; Chen, Q.; Yuan, X.; Du, Z.; Chen, Z.; Raheem, M.A.; Deng, L.; Leeansyah, E.; et al. Angiotensin-Converting Enzyme 2-Based Biosensing Modalities and Devices for Coronavirus Detection. Biosensors 2022, 12, 984. [Google Scholar] [CrossRef] [PubMed]
- Ratswohl, C.; Vázquez García, C.; Ahmad, A.U.W.; Gonschior, H.; Lebedin, M.; Silvis, C.E.; Spatt, L.; Gerhard, C.; Lehmann, M.; Sander, L.E.; et al. A Design Strategy to Generate a SARS-CoV-2 RBD Vaccine That Abrogates ACE2 Binding and Improves Neutralizing Antibody Responses. Eur. J. Immunol. 2023, 53, e2350408. [Google Scholar] [CrossRef] [PubMed]
- Damborský, P.; Švitel, J.; Katrlík, J. Optical Biosensors. Essays Biochem. 2016, 60, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Eddin, F.B.K.; Fen, Y.W. The Principle of Nanomaterials Based Surface Plasmon Resonance Biosensors and Its Potential for Dopamine Detection. Molecules 2020, 25, 2769. [Google Scholar] [CrossRef]
- Sharma, A.K.; Jha, R.; Gupta, B.D. Fiber-Optic Sensors Based on Surface Plasmon Resonance: A Comprehensive Review. IEEE Sens. J. 2007, 7, 1118–1129. [Google Scholar] [CrossRef]
- Nurrohman, D.T.; Chiu, N.-F. Surface Plasmon Resonance Biosensor Performance Analysis on 2D Material Based on Graphene and Transition Metal Dichalcogenides. ECS J. Solid State Sci. Technol. 2020, 9, 115023. [Google Scholar] [CrossRef]
- Das, C.M.; Yang, F.; Yang, Z.; Liu, X.; Hoang, Q.T.; Xu, Z.; Neermunda, S.; Kong, K.V.; Ho, H.P.; Ju, L.A.; et al. Computational Modeling for Intelligent Surface Plasmon Resonance Sensor Design and Experimental Schemes for Real-Time Plasmonic Biosensing: A Review. Adv. Theory Simul. 2023, 6, 2200886. [Google Scholar] [CrossRef]
- D’Agata, R.; Bellassai, N.; Jungbluth, V.; Spoto, G. Recent Advances in Antifouling Materials for Surface Plasmon Resonance Biosensing in Clinical Diagnostics and Food Safety. Polymers 2021, 13, 1929. [Google Scholar] [CrossRef]
- Daniyal, W.M.E.M.M.; Fen, Y.W.; Fauzi, N.I.M.; Hashim, H.S.; Ramdzan, N.S.M.; Omar, N.A.S. Recent Advances in Surface Plasmon Resonance Optical Sensors for Potential Application in Environmental Monitoring. Sens. Mater. 2020, 32, 4191–4200. [Google Scholar] [CrossRef]
- Nurrohman, D.T.; Wang, Y.-H.; Chiu, N.-F. Exploring Graphene and MoS2 Chips Based Surface Plasmon Resonance Biosensors for Diagnostic Applications. Front. Chem. 2020, 8, 728. [Google Scholar] [CrossRef]
- Chiu, N.F.; Kuo, C.T.; Lin, T.L.; Chang, C.C.; Chen, C.Y. Ultra-High Sensitivity of the Non-Immunological Affinity of Graphene Oxide-Peptide-Based Surface Plasmon Resonance Biosensors to Detect Human Chorionic Gonadotropin. Biosens. Bioelectron. 2017, 94, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Chiu, N.F.; Tai, M.J.; Nurrohman, D.T.; Lin, T.L.; Wang, Y.H.; Chen, C.Y. Immunoassay-Amplified Responses Using a Functionalized Mos2-Based Spr Biosensor to Detect Papp-A2 in Maternal Serum Samples to Screen for Fetal down’s Syndrome. Int. J. Nanomed. 2021, 16, 2715–2733. [Google Scholar] [CrossRef]
- Murali, S.; Rustandi, R.R.; Zheng, X.; Payne, A.; Shang, L. Applications of Surface Plasmon Resonance and Biolayer Interferometry for Virus–Ligand Binding. Viruses 2022, 14, 717. [Google Scholar] [CrossRef]
- Kausaite-Minkstimiene, A.; Popov, A.; Ramanaviciene, A. Ultra-Sensitive SPR Immunosensors: A Comprehensive Review of Labeling and Interface Modification Using Nanostructures. TrAC-Trends Anal. Chem. 2024, 170, 117468. [Google Scholar] [CrossRef]
- Vachali, P.P.; Li, B.; Bartschi, A.; Bernstein, P.S. Surface Plasmon Resonance (SPR)-Based Biosensor Technology for the Quantitative Characterization of Protein-Carotenoid Interactions. Arch. Biochem. Biophys. 2015, 572, 66–72. [Google Scholar] [CrossRef]
- Sparks, R.P.; Jenkins, J.L.; Fratti, R. Use of Surface Plasmon Resonance (SPR) to Determine Binding Affinities and Kinetic Parameters Between Components Important in Fusion Machinery. In Methods in Molecular Biology; Fratti, R., Ed.; Springer New York: New York, NY, USA, 2019; Volume 1860, pp. 199–210. ISBN 978-1-4939-8759-7. [Google Scholar]
- Dorozinska, H.V.; Turu, T.A.; Markina, O.M.; Dorozinsky, G.V.; Maslov, V.P. Influence of Temperature on the Measuring Accuracy of Devices Based on Surface Plasmon Resonance Phenomenon. Mod. Instrum. 2018, 7, 1–10. [Google Scholar] [CrossRef]
- Ritzefeld, M.; Sewald, N. Real-Time Analysis of Specific Protein-DNA Interactions with Surface Plasmon Resonance. J. Amino Acids 2012, 2012, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Hahnefeld, C.; Drewianka, S.; Herberg, F.W. Determination of Kinetic Data Using Surface Plasmon Resonance Biosensors. In Molecular Diagnosis of Infectious Diseases; Humana Press: Totowa, NJ, USA, 2004; Volume 94, pp. 299–320. [Google Scholar]
- Chiu, N.F.; Huang, T.Y.; Lai, H.C.; Liu, K.C. Graphene Oxide-Based SPR Biosensor Chip for Immunoassay Applications. Nanoscale Res. Lett. 2014, 9, 445. [Google Scholar] [CrossRef] [PubMed]
- Trabucchi, A.; Iacono, R.F.; Guerra, L.L.; Faccinetti, N.I.; Krochik, A.G.; Arriazu, M.C.; Poskus, E.; Valdez, S.N. Characterization of Insulin Antibodies by Surface Plasmon Resonance in Two Clinical Cases: Brittle Diabetes and Insulin Autoimmune Syndrome. PLoS ONE 2013, 8, e84099. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Yang, H.S.; Karbaschi, M.; Racine-Brzostek, S.E.; Li, P.; Zuk, R.; Yang, Y.J.; Klasse, P.J.; Shi, Y.; Zhao, Z. Measurements of SARS-CoV-2 Antibody Dissociation Rate Constant by Chaotrope-Free Biolayer Interferometry in Serum of COVID-19 Convalescent Patients. Biosens. Bioelectron. 2022, 209, 114237. [Google Scholar] [CrossRef] [PubMed]
- Maros, H.; Juniar, S. Handbook of Surface Plasmon Resonance; Schasfoort, R.B.M., Ed.; Royal Society of Chemistry: Cambridge, UK, 2017; ISBN 978-1-78262-730-2. [Google Scholar]
- Ezzati Nazhad Dolatabadi, J.; de la Guardia, M. Tips on Ligand Immobilization and Kinetic Study Using Surface Plasmon Resonance. BioImpacts 2016, 6, 117–118. [Google Scholar] [CrossRef] [PubMed]
- Healthcare, G.E. Biacore Sensor Surface Handbook; Biacore: Uppsala, Sweden, 2008; Volume BR-1005-71. [Google Scholar]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q.; Shi, X.; Wang, Q.; Zhang, L.; et al. Structure of the SARS-CoV-2 Spike Receptor-Binding Domain Bound to the ACE2 Receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.M. Progress in Electrochemical Biosensing of SARS-CoV-2 Virus for COVID-19 Management. Chemosensors 2022, 10, 287. [Google Scholar] [CrossRef]
- Taha, B.A.; Al-Jubouri, Q.; Al Mashhadany, Y.; Zan, M.S.D.B.; Bakar, A.A.A.; Fadhel, M.M.; Arsad, N. Photonics Enabled Intelligence System to Identify SARS-CoV-2 Mutations. Appl. Microbiol. Biotechnol. 2022, 106, 3321–3336. [Google Scholar] [CrossRef]
- Van Vo, G.; Bagyinszky, E.; An, S.S.A. COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development. Microorganisms 2022, 10, 598. [Google Scholar] [CrossRef]
- Mohammadi, M.; Shayestehpour, M.; Mirzaei, H. The Impact of Spike Mutated Variants of SARS-CoV-2 [Alpha, Beta, Gamma, Delta, and Lambda] on the Efficacy of Subunit Recombinant Vaccines. Braz. J. Infect. Dis. 2021, 25, 101606. [Google Scholar] [CrossRef] [PubMed]
- Xi, H.; Jiang, H.; Juhas, M.; Zhang, Y. Multiplex Biosensing for Simultaneous Detection of Mutations in SARS-CoV-2. ACS Omega 2021, 6, 25846–25859. [Google Scholar] [CrossRef]
- Zhang, L.; Jackson, C.B.; Mou, H.; Ojha, A.; Peng, H.; Quinlan, B.D.; Rangarajan, E.S.; Pan, A.; Vanderheiden, A.; Suthar, M.S.; et al. SARS-CoV-2 Spike-Protein D614G Mutation Increases Virion Spike Density and Infectivity. Nat. Commun. 2020, 11, 6013. [Google Scholar] [CrossRef] [PubMed]
- McLean, G.; Kamil, J.; Lee, B.; Moore, P.; Schulz, T.F.; Muik, A.; Sahin, U.; Türeci, Ö.; Pather, S. The Impact of Evolving SARS-CoV-2 Mutations and Variants on COVID-19 Vaccines. MBio 2022, 13, e02979-21. [Google Scholar] [CrossRef] [PubMed]
- Ghoula, M.; Deyawe Kongmeneck, A.; Eid, R.; Camproux, A.C.; Moroy, G. Comparative Study of the Mutations Observed in the SARS-CoV-2 RBD Variants of Concern and Their Impact on the Interaction with the ACE2 Protein. J. Phys. Chem. B 2023, 127, 8586–8602. [Google Scholar] [CrossRef]
- Lee, J.H.; Lee, Y.; Lee, S.K.; Kim, J.; Lee, C.S.; Kim, N.H.; Kim, H.G. Versatile Role of ACE2-Based Biosensors for Detection of SARS-CoV-2 Variants and Neutralizing Antibodies. Biosens. Bioelectron. 2022, 203, 114034. [Google Scholar] [CrossRef] [PubMed]
- Drobysh, M.; Liustrovaite, V.; Kanetski, Y.; Brasiunas, B.; Zvirbliene, A.; Rimkute, A.; Gudas, D.; Kucinskaite-Kodze, I.; Simanavicius, M.; Ramanavicius, S.; et al. Electrochemical Biosensing Based Comparative Study of Monoclonal Antibodies against SARS-CoV-2 Nucleocapsid Protein. Sci. Total Environ. 2024, 908, 168154. [Google Scholar] [CrossRef]
- Zhang, Y.; Juhas, M.; Kwok, C.K. Aptamers Targeting SARS-CoV-2: A Promising Tool to Fight against COVID-19. Trends Biotechnol. 2023, 41, 528–544. [Google Scholar] [CrossRef]
- Kumar, T.H.V.; Srinivasan, S.; Krishnan, V.; Vaidyanathan, R.; Babu, K.A.; Natarajan, S.; Veerapandian, M. Peptide-Based Direct Electrochemical Detection of Receptor Binding Domains of SARS-CoV-2 Spike Protein in Pristine Samples. Sensors Actuators B Chem. 2023, 377, 133052. [Google Scholar] [CrossRef]
- Masterson, A.N.; Muhoberac, B.B.; Gopinadhan, A.; Wilde, D.J.; Deiss, F.T.; John, C.C.; Sardar, R. Multiplexed and High-Throughput Label-Free Detection of RNA/Spike Protein/IgG/IgM Biomarkers of SARS-CoV-2 Infection Utilizing Nanoplasmonic Biosensors. Anal. Chem. 2021, 93, 8754–8763. [Google Scholar] [CrossRef]
- Liu, Y.; Qin, Z.; Jia, X.; Zhou, J.; Li, H.; Wang, X.; Chen, Y.; Deng, J.; Jin, Z.; Wang, G. Directly and Ultrasensitivity Detecting SARS-CoV-2 Spike Protein in Pharyngeal Swab Solution by Using SERS-Based Biosensor. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2023, 303, 123275. [Google Scholar] [CrossRef]
- Hao, X.; St-Pierre, J.P.; Zou, S.; Cao, X. Localized Surface Plasmon Resonance Biosensor Chip Surface Modification and Signal Amplifications toward Rapid and Sensitive Detection of COVID-19 Infections. Biosens. Bioelectron. 2023, 236, 115421. [Google Scholar] [CrossRef]
- Wu, Q.; Wu, W.; Chen, F.; Ren, P. Highly Sensitive and Selective Surface Plasmon Resonance Biosensor for the Detection of SARS-CoV-2 Spike S1 Protein. Analyst 2022, 147, 2809–2818. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Xu, X.; Wang, Y.; Li, M.; Zhou, K.; Zhang, L.; Tan, Y. Surface Plasmon Resonance Biosensor with Laser Heterodyne Feedback for Highly-Sensitive and Rapid Detection of COVID-19 Spike Antigen. Biosens. Bioelectron. 2022, 206, 114163. [Google Scholar] [CrossRef]
- Bae, M.; Choi, S.; Kim, J.; Seo, G.; Lee, Y.W. Temperature-Insensitive Label-Free SARS-CoV-2 Spike Protein Detection Based on Complementary Refractive Index and Temperature Dependence of Multi-Mode Interference and Grating Resonance. Talanta 2024, 266, 125091. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, X.; Pan, J.; Zhang, Y.; Zhang, L.; Wang, C.; Yan, X.; Liu, X.; Lu, G. Ultrasensitive Detection of SARS-CoV-2 Spike Protein in Untreated Saliva Using SERS-Based Biosensor. Biosens. Bioelectron. 2021, 190, 113421. [Google Scholar] [CrossRef] [PubMed]
- Bistaffa, M.J.; Camacho, S.A.; Pazin, W.M.; Constantino, C.J.L.; Oliveira, O.N.; Aoki, P.H.B. Immunoassay Platform with Surface-Enhanced Resonance Raman Scattering for Detecting Trace Levels of SARS-CoV-2 Spike Protein. Talanta 2022, 244, 123381. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Sun, A.Y.; Huang, Y.C.; Wang, C.H.; Wang, S.C.; Chau, L.K. Integration of Power-Free and Self-Contained Microfluidic Chip with Fiber Optic Particle Plasmon Resonance Aptasensor for Rapid Detection of SARS-CoV-2 Nucleocapsid Protein. Biosensors 2022, 12, 785. [Google Scholar] [CrossRef] [PubMed]
- Gheware, A.; Ray, A.; Rana, D.; Bajpai, P.; Nambirajan, A.; Arulselvi, S.; Mathur, P.; Trikha, A.; Arava, S.; Das, P.; et al. ACE2 Protein Expression in Lung Tissues of Severe COVID-19 Infection. Sci. Rep. 2022, 12, 4058. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Chopra, P.; Li, X.; Bouwman, K.M.; Tompkins, S.M.; Wolfert, M.A.; de Vries, R.P.; Boons, G.-J. Heparan Sulfate Proteoglycans as Attachment Factor for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef] [PubMed]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM Structure of the 2019-NCoV Spike in the Prefusion Conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef]
- Glycoprotein, C.-S.; Walls, A.C.; Park, Y.; Tortorici, M.A.; Wall, A.; Mcguire, A.T.; Veesler, D.; Walls, A.C.; Park, Y.; Tortorici, M.A.; et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
- Xue, T.; Wu, W.; Guo, N.; Wu, C.; Huang, J.; Lai, L.; Liu, H.; Li, Y.; Wang, T.; Wang, Y. Single Point Mutations Can Potentially Enhance Infectivity of SARS-CoV-2 Revealed by: In Silico Affinity Maturation and SPR Assay. RSC Adv. 2021, 11, 14737–14745. [Google Scholar] [CrossRef]
- Barton, M.I.; Macgowan, S.; Kutuzov, M.; Dushek, O.; Barton, G.J.; Anton Van Der Merwe, P. Effects of Common Mutations in the Sars-Cov-2 Spike Rbd and Its Ligand the Human Ace2 Receptor on Binding Affinity and Kinetics. Elife 2021, 10, e70658. [Google Scholar] [CrossRef] [PubMed]
- Fratev, F. R346K Mutation in the Mu Variant of SARS-CoV-2 Alters the Interactions with Monoclonal Antibodies from Class 2: A Free Energy Perturbation Study. J. Chem. Inf. Model. 2022, 62, 627–631. [Google Scholar] [CrossRef]
- Aggarwal, A.; Naskar, S.; Maroli, N.; Gorai, B.; Dixit, N.M.; Maiti, P.K. Mechanistic Insights into the Effects of Key Mutations on SARS-CoV-2 RBD-ACE2 Binding. Phys. Chem. Chem. Phys. 2021, 23, 26451–26458. [Google Scholar] [CrossRef]
- Taft, J.M.; Weber, C.R.; Gao, B.; Ehling, R.A.; Han, J.; Frei, L.; Metcalfe, S.W.; Overath, M.D.; Yermanos, A.; Kelton, W.; et al. Deep Mutational Learning Predicts ACE2 Binding and Antibody Escape to Combinatorial Mutations in the SARS-CoV-2 Receptor-Binding Domain. Cell 2022, 185, 4008–4022.e14. [Google Scholar] [CrossRef]
- Chen, C.; Boorla, V.S.; Banerjee, D.; Chowdhury, R.; Cavener, V.S.; Nissly, R.H.; Gontu, A.; Boyle, N.R.; Vandegrift, K.; Nair, M.S.; et al. Computational Prediction of the Effect of Amino Acid Changes on the Binding Affinity between SARS-CoV-2 Spike RBD and Human ACE2. Proc. Natl. Acad. Sci. USA 2021, 118, e2106480118. [Google Scholar] [CrossRef] [PubMed]
- Barnes, J.E.; Lund-Andersen, P.K.; Patel, J.S.; Ytreberg, F.M. The Effect of Mutations on Binding Interactions between the SARS-CoV-2 Receptor Binding Domain and Neutralizing Antibodies B38 and CB6. Sci. Rep. 2022, 12, 18819. [Google Scholar] [CrossRef] [PubMed]
- Leonard, A.C.; Weinstein, J.J.; Steiner, P.J.; Erbse, A.H.; Fleishman, S.J.; Whitehead, T.A. Stabilization of the SARS-CoV-2 Receptor Binding Domain by Protein Core Redesign and Deep Mutational Scanning. Protein Eng. Des. Sel. 2022, 35, gzac002. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, A.P.; Katsamba, P.S.; Liao, J.; Sampson, J.M.; Bahna, F.; Mannepalli, S.; Morano, N.C.; Shapiro, L.; Friesner, R.A.; Honig, B. Free Energy Perturbation Calculations of Mutation Effects on SARS-CoV-2 RBD::ACE2 Binding Affinity. J. Mol. Biol. 2023, 435, 168187. [Google Scholar] [CrossRef] [PubMed]
- Makowski, E.K.; Schardt, J.S.; Smith, M.D.; Tessier, P.M. Mutational Analysis of SARS-CoV-2 Variants of Concern Reveals Key Tradeoffs between Receptor Affinity and Antibody Escape. PLoS Comput. Biol. 2022, 18, e1010160. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.K.; Hsu, C.H.; Lai, C.S. Enhancement of the Au/ZnO-NA Plasmonic SERS Signal Using Principal Component Analysis as a Machine Learning Approach. IEEE Photonics J. 2020, 12, 2200611. [Google Scholar] [CrossRef]
- Paryanti, G.; Faig, H.; Rokach, L.; Sadot, D. A Direct Learning Approach for Neural Network Based Pre-Distortion for Coherent Nonlinear Optical Transmitter. J. Light. Technol. 2020, 38, 3883–3896. [Google Scholar] [CrossRef]
- Williamson, I.A.D.; Hughes, T.W.; Minkov, M.; Bartlett, B.; Pai, S.; Fan, S. Tunable Nonlinear Activation Functions for Optical Neural Networks. Opt. InfoBase Conf. Pap. 2020. [Google Scholar] [CrossRef]
- Arano-Martinez, J.A.; Martínez-González, C.L.; Salazar, M.I.; Torres-Torres, C. A Framework for Biosensors Assisted by Multiphoton Effects and Machine Learning. Biosensors 2022, 12, 710. [Google Scholar] [CrossRef]
- Qatamin, A.H.; Ghithan, J.H.; Moreno, M.; Nunn, B.M.; Jones, K.B.; Zamborini, F.P.; Keynton, R.S.; O’toole, M.G.; Mendes, S.B. Detection of Influenza Virus by Electrochemical Surface Plasmon Resonance under Potential Modulation. Appl. Opt. 2019, 58, SW4C.4. [Google Scholar] [CrossRef]
- Song, C.; Jiang, X.; Yang, Y.; Zhang, J.; Larson, S.; Zhao, Y.; Wang, L. High-Sensitive Assay of Nucleic Acid Using Tetrahedral DNA Probes and DNA Concatamers with a Surface-Enhanced Raman Scattering/Surface Plasmon Resonance Dual-Mode Biosensor Based on a Silver Nanorod-Covered Silver Nanohole Array. ACS Appl. Mater. Interfaces 2020, 12, 31242–31254. [Google Scholar] [CrossRef] [PubMed]
- Hasler, R.; Steger-Polt, M.H.; Reiner-Rozman, C.; Fossati, S.; Lee, S.; Aspermair, P.; Kleber, C.; Ibáñez, M.; Dostalek, J.; Knoll, W. Optical and Electronic Signal Stabilization of Plasmonic Fiber Optic Gate Electrodes: Towards Improved Real-Time Dual-Mode Biosensing. Front. Phys. 2023, 11, 1202132. [Google Scholar] [CrossRef]
- Caucheteur, C.; Guo, T.; Albert, J. Review of Plasmonic Fiber Optic Biochemical Sensors: Improving the Limit of Detection. Anal. Bioanal. Chem. 2015, 407, 3883–3897. [Google Scholar] [CrossRef] [PubMed]

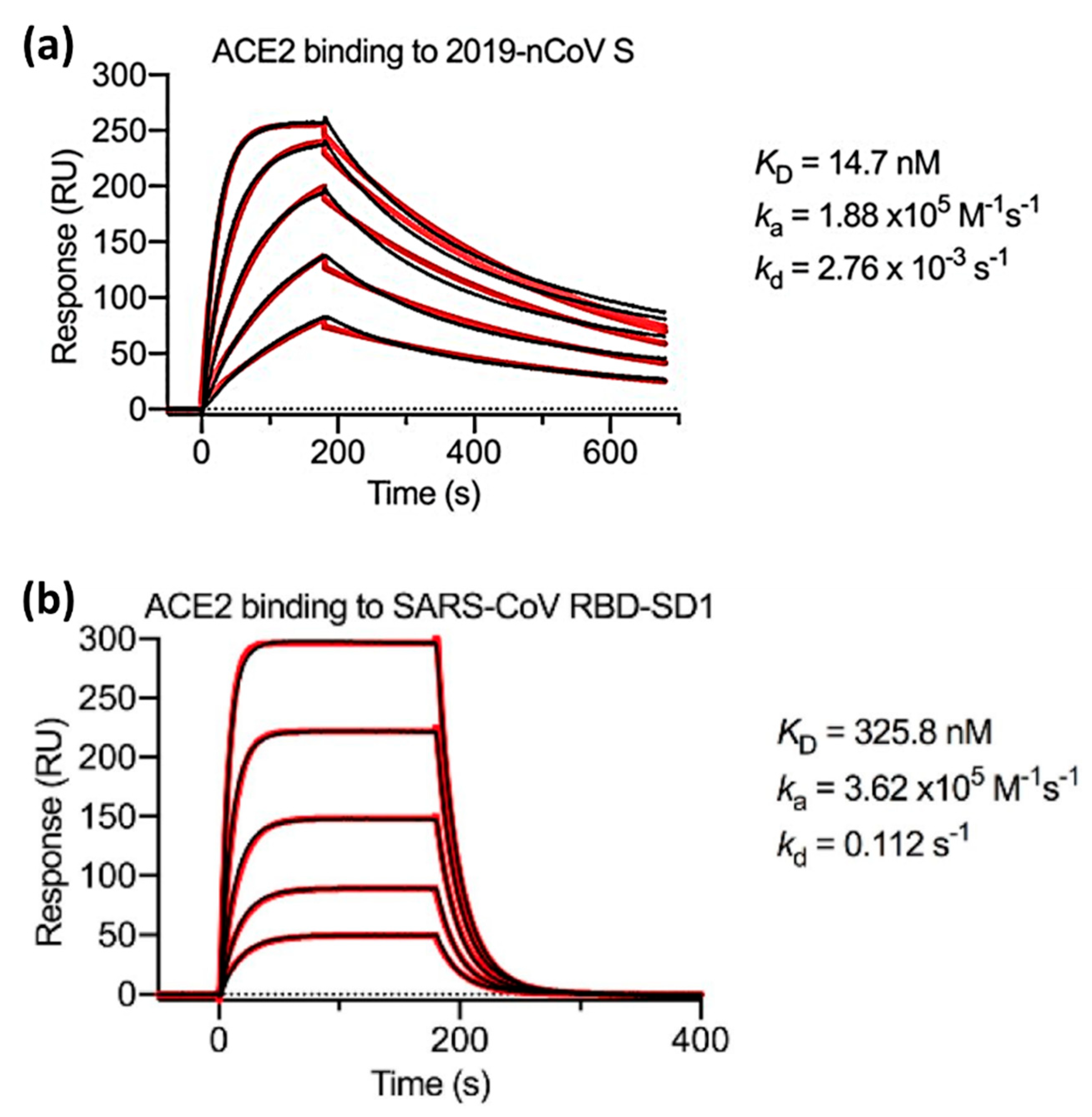
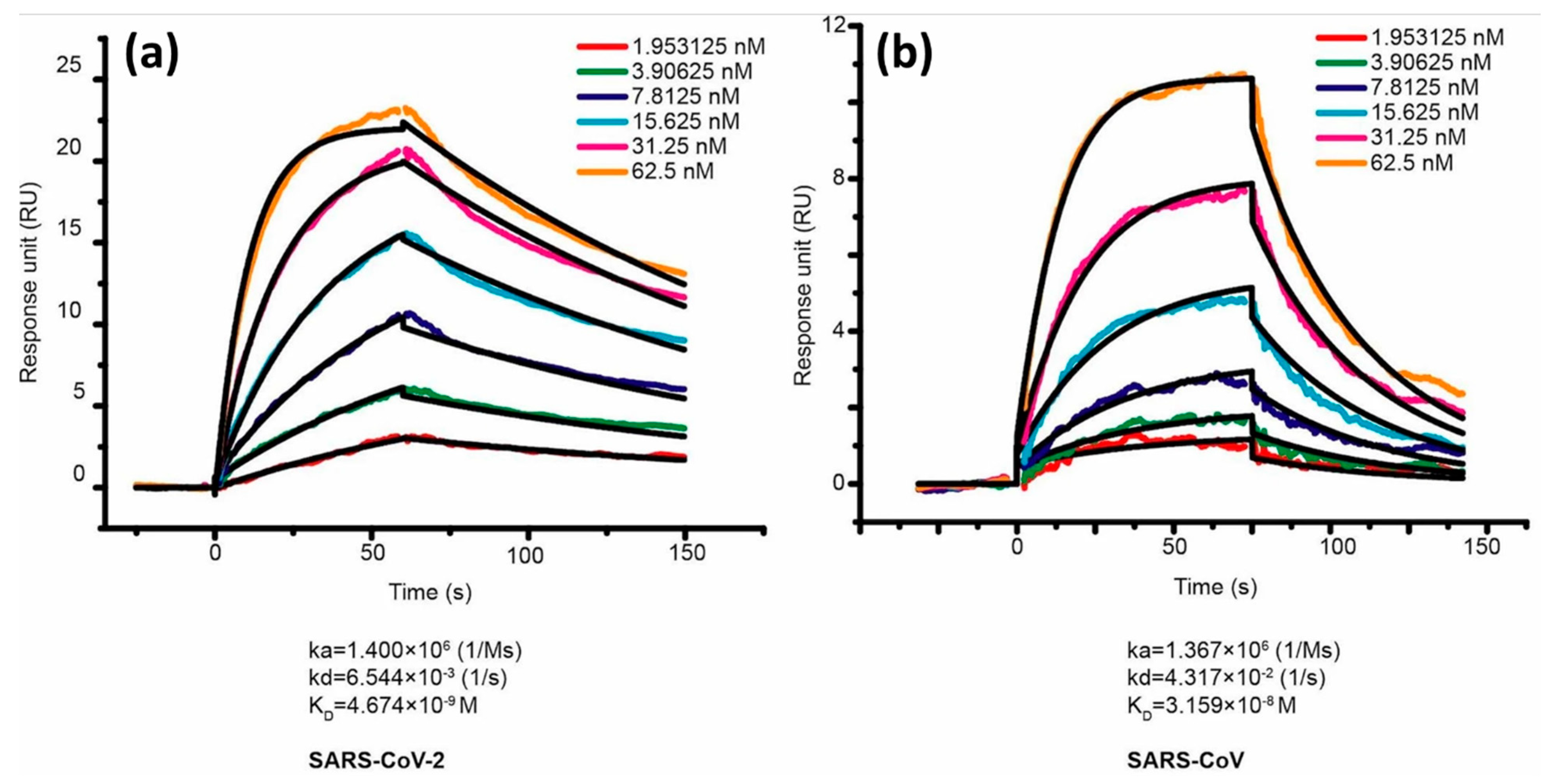
| Definition | ||||
| Unit | L/mol | mol/L | ||
| Typical range | 103 − 107 | 10−1 − 5 × 10−6 | 105 − 1012 | 10−5 − 10−12 |
| Variants | Lineage | Mutations on the RBD |
|---|---|---|
| Alpha | B.1.1.7 | N501Y |
| Beta | B.1.351 | K417N, E484K, N501Y |
| Gamma | P.1 | K417T/N, E484K, N501Y |
| Delta | B.1.617.2 | L452R, T478K |
| Omicron | B.1.1.529 | G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H |
| Optical Effect | Ligands | Analytes | Ref. | |
|---|---|---|---|---|
| SERS | ACE2 | SARS-CoV-2 S Protein | 0.1 fg/mL | [57] |
| LSPR | Aptamer | SRBD and SARS-CoV-2 pseudovirus | 21.9 pM | [58] |
| SPR | anti-SARS-CoV-2 spike S1 protein | S1 protein | 12 fg/mL | [59] |
| SPR | anti-SARS-CoV-2 spike MAbs | SARS-CoV-2 spike antigen | 0.08 pg/mL | [60] |
| Fiber optics | ACE2 | SARS-CoV-2 spike protein | ∼3.05 ng/mL | [61] |
| SERS | SARS-CoV-2 spike antibody | Spike protein | 0.77 fg/mL | [62] |
| SERRS | SARS-CoV-2 antibodies | SARS-CoV-2 spike glycoprotein (S1) | 0.60 pM | [63] |
| BLI | ACE2 | S protein | 500 pg/mL | [52] |
| Fiber optics | single-stranded DNA (ssDNA) aptamer | SARS-CoV-2 Nucleocapsid Protein | 2.8 nM | [64] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nurrohman, D.T.; Chiu, N.-F. Unraveling the Dynamics of SARS-CoV-2 Mutations: Insights from Surface Plasmon Resonance Biosensor Kinetics. Biosensors 2024, 14, 99. https://doi.org/10.3390/bios14020099
Nurrohman DT, Chiu N-F. Unraveling the Dynamics of SARS-CoV-2 Mutations: Insights from Surface Plasmon Resonance Biosensor Kinetics. Biosensors. 2024; 14(2):99. https://doi.org/10.3390/bios14020099
Chicago/Turabian StyleNurrohman, Devi Taufiq, and Nan-Fu Chiu. 2024. "Unraveling the Dynamics of SARS-CoV-2 Mutations: Insights from Surface Plasmon Resonance Biosensor Kinetics" Biosensors 14, no. 2: 99. https://doi.org/10.3390/bios14020099
APA StyleNurrohman, D. T., & Chiu, N.-F. (2024). Unraveling the Dynamics of SARS-CoV-2 Mutations: Insights from Surface Plasmon Resonance Biosensor Kinetics. Biosensors, 14(2), 99. https://doi.org/10.3390/bios14020099






