Identification of Podoplanin Aptamers by SELEX for Protein Detection and Inhibition of Platelet Aggregation Stimulated by C-Type Lectin-like Receptor 2
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Systematic Evolution of Ligands by Exponential Enrichment (SELEX)
2.3. Next-Generation Sequencing (NGS) and Data Processing
2.4. Dot Blot Analysis
2.5. Bio-Layer Interferometry Assay
2.6. Plasmid Construction
2.7. Transient Transfection
2.8. Western Blot Analysis
2.9. Flow Cytometry Analysis
2.10. Preparation of Washed Platelets
2.11. Platelet Aggregation Assay
2.12. Measurement of Circular Dichroism (CD)
2.13. Statistical Analysis
3. Results
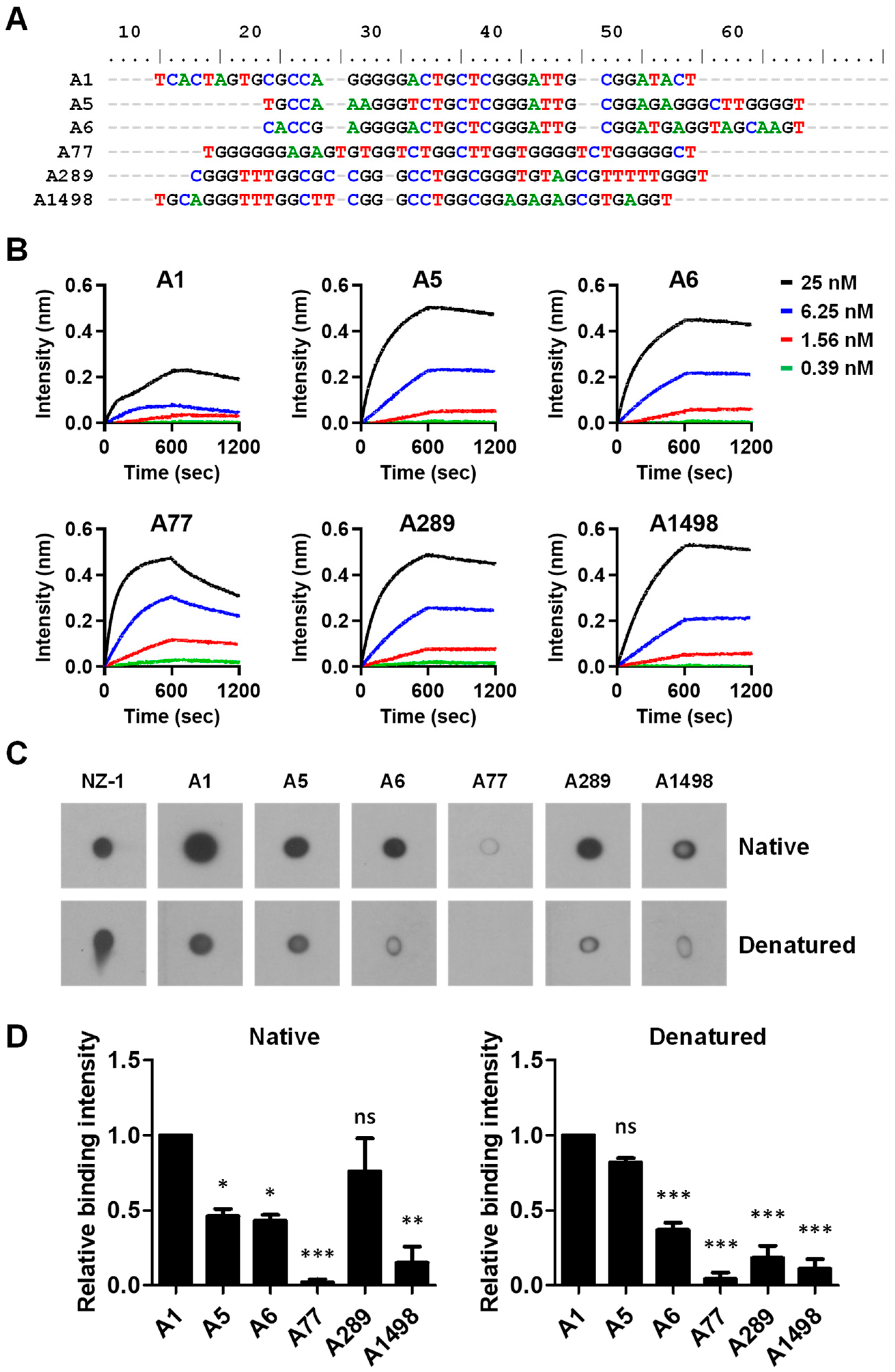
3.1. Screening for Aptamers Recognizing PDPN
3.2. Analysis of the Putative PDPN Aptamers Structures
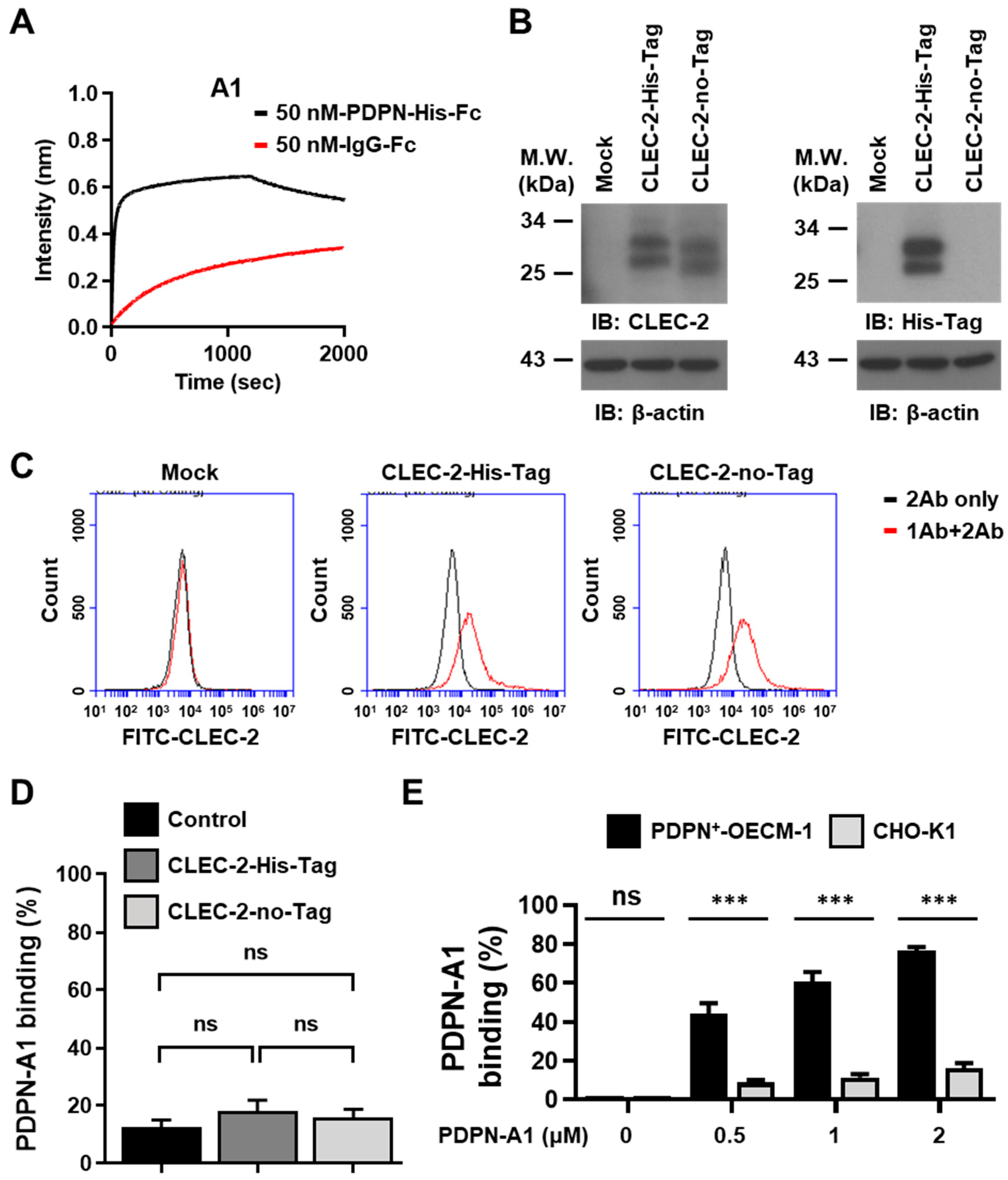
3.3. Characterization of the PDPN Aptamer A1
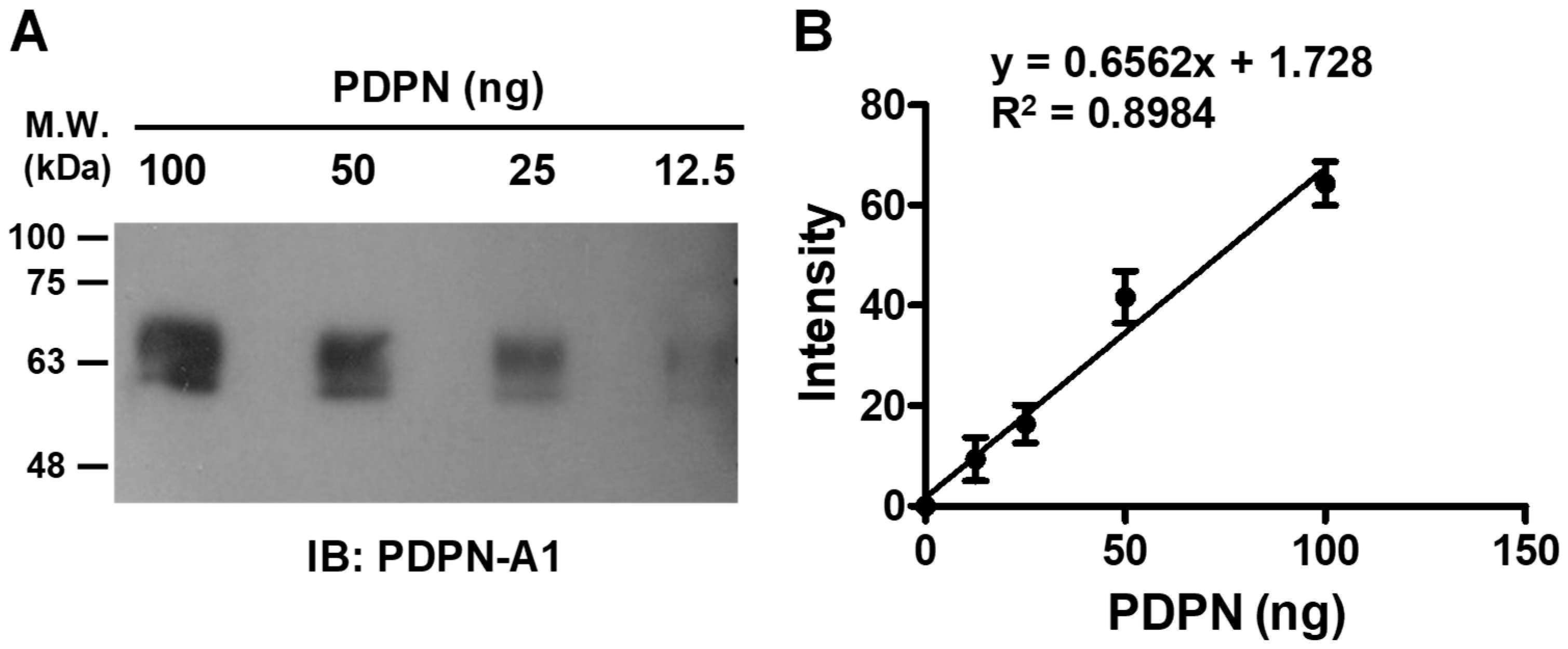
3.4. Quantitative Detection of PDPN by Aptamer A1
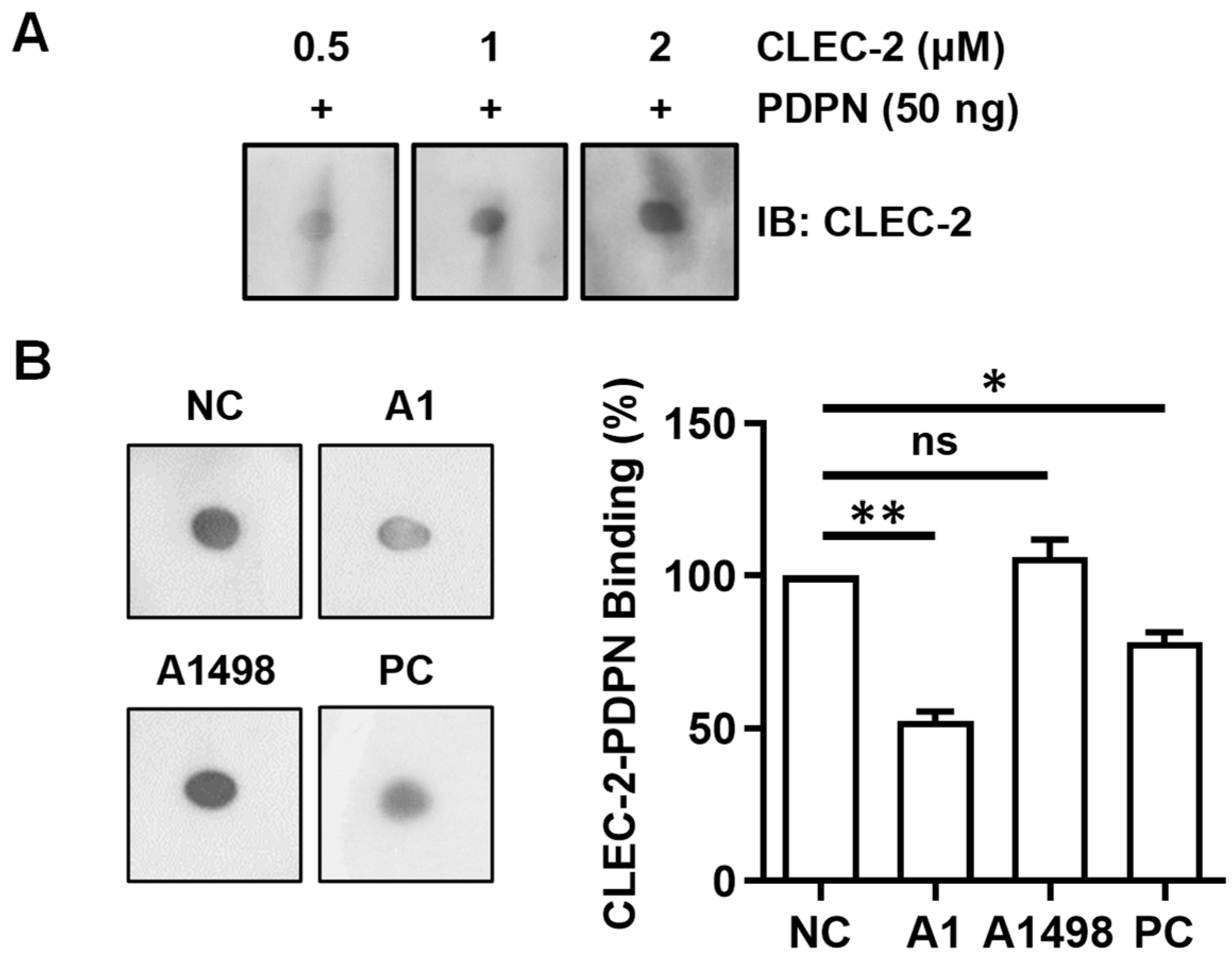
3.5. Aptamer A1 Interfering with the Interaction between PDPN and CLEC-2
3.6. Aptamer A1 Inhibiting PDPN-Induced Platelet Aggregation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, G.P.; Massague, J. Platelets, and metastasis revisited: A novel fatty link. J. Clin. Investig. 2004, 114, 1691–1693. [Google Scholar] [CrossRef] [PubMed]
- Stegner, D.; Dutting, S.; Nieswandt, B. Mechanistic explanation for platelet contribution to cancer metastasis. Thromb. Res. 2014, 133 (Suppl. S2), S149–S157. [Google Scholar] [CrossRef]
- Medina, C.; Harmon, S.; Inkielewicz, I.; Santos-Martinez, M.J.; Jones, M.; Cantwell, P.; Bazou, D.; Ledwidge, M.; Radomski, M.W.; Gilmer, J.F. Differential inhibition of tumour cell-induced platelet aggregation by the nicotinate aspirin prodrug (ST0702) and aspirin. Br. J. Pharmacol. 2012, 166, 938–949. [Google Scholar] [CrossRef]
- Uluckan, O.; Eagleton, M.C.; Floyd, D.H.; Morgan, E.A.; Hirbe, A.C.; Kramer, M.; Dowland, N.; Prior, J.L.; Piwnica-Worms, D.; Jeong, S.S.; et al. APT102, a novel adpase, cooperates with aspirin to disrupt bone metastasis in mice. J. Cell. Biochem. 2008, 104, 1311–1323. [Google Scholar] [CrossRef]
- Zara, M.; Canobbio, I.; Visconte, C.; Canino, J.; Torti, M.; Guidetti, G.F. Molecular mechanisms of platelet activation and aggregation induced by breast cancer cells. Cell. Signal. 2018, 48, 45–53. [Google Scholar] [CrossRef]
- Cohen, S.A.; Trikha, M.; Mascelli, M.A. Potential future clinical applications for the GPIIb/IIIa antagonist, abciximab in thrombosis, vascular and oncological indications. Pathol. Oncol. Res. 2000, 6, 163–174. [Google Scholar] [CrossRef] [PubMed]
- Erpenbeck, L.; Schon, M.P. Deadly allies: The fatal interplay between platelets and metastasizing cancer cells. Blood 2010, 115, 3427–3436. [Google Scholar] [CrossRef]
- Jurasz, P.; Alonso-Escolano, D.; Radomski, M.W. Platelet-cancer interactions: Mechanisms and pharmacology of tumour cell-induced platelet aggregation. Br. J. Pharmacol. 2004, 143, 819–826. [Google Scholar] [CrossRef] [PubMed]
- Klerk, C.P.; Smorenburg, S.M.; Otten, H.M.; Lensing, A.W.; Prins, M.H.; Piovella, F.; Prandoni, P.; Bos, M.M.; Richel, D.J.; van Tienhoven, G.; et al. The effect of low molecular weight heparin on survival in patients with advanced malignancy. J. Clin. Oncol. 2005, 23, 2130–2135. [Google Scholar] [CrossRef] [PubMed]
- Lian, L.; Li, W.; Li, Z.Y.; Mao, Y.X.; Zhang, Y.T.; Zhao, Y.M.; Chen, K.; Duan, W.M.; Tao, M. Inhibition of MCF-7 breast cancer cell-induced platelet aggregation using a combination of antiplatelet drugs. Oncol. Lett. 2013, 5, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ogose, A.; Kawashima, H.; Hotta, T.; Ariizumi, T.; Li, G.; Umezu, H.; Endo, N. High-level expression of podoplanin in benign and malignant soft tissue tumors: Immunohistochemical and quantitative real-time RT-PCR analysis. Oncol. Rep. 2011, 25, 599–607. [Google Scholar] [CrossRef][Green Version]
- Astarita, J.L.; Acton, S.E.; Turley, S.J. Podoplanin: Emerging functions in development, the immune system, and cancer. Front. Immunol. 2012, 3, 283. [Google Scholar] [CrossRef] [PubMed]
- Kunita, A.; Kashima, T.G.; Morishita, Y.; Fukayama, M.; Kato, Y.; Tsuruo, T.; Fujita, N. The platelet aggregation-inducing factor aggrus/podoplanin promotes pulmonary metastasis. Am. J. Pathol. 2007, 170, 1337–1347. [Google Scholar] [CrossRef]
- Suzuki-Inoue, K.; Kato, Y.; Inoue, O.; Kaneko, M.K.; Mishima, K.; Yatomi, Y.; Yamazaki, Y.; Narimatsu, H.; Ozaki, Y. Involvement of the snake toxin receptor CLEC-2, in podoplanin-mediated platelet activation, by cancer cells. J. Biol. Chem. 2007, 282, 25993–26001. [Google Scholar] [CrossRef] [PubMed]
- Christou, C.M.; Pearce, A.C.; Watson, A.A.; Mistry, A.R.; Pollitt, A.Y.; Fenton-May, A.E.; Johnson, L.A.; Jackson, D.G.; Watson, S.P.; O’Callaghan, C.A. Renal cells activate the platelet receptor CLEC-2 through podoplanin. Biochem. J. 2008, 411, 133–140. [Google Scholar] [CrossRef]
- Ozaki, Y.; Suzuki-Inoue, K.; Inoue, O. Novel interactions in platelet biology: CLEC-2/podoplanin and laminin/GPVI. J. Thromb. Haemost. 2009, 7 (Suppl. S1), 191–194. [Google Scholar] [CrossRef]
- Watson, S.P.; Herbert, J.M.; Pollitt, A.Y. GPVI and CLEC-2 in hemostasis and vascular integrity. J. Thromb. Haemost. 2010, 8, 1456–1467. [Google Scholar] [CrossRef]
- Severin, S.; Pollitt, A.Y.; Navarro-Nunez, L.; Nash, C.A.; Mourao-Sa, D.; Eble, J.A.; Senis, Y.A.; Watson, S.P. Syk-dependent phosphorylation of CLEC-2: A novel mechanism of hem-immunoreceptor tyrosine-based activation motif signaling. J. Biol. Chem. 2011, 286, 4107–4116. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Inoue, K.; Inoue, O.; Ozaki, Y. Novel platelet activation receptor CLEC-2: From discovery to prospects. J. Thromb. Haemost. 2011, 9 (Suppl. S1), 44–55. [Google Scholar] [CrossRef]
- Abe, S.; Morita, Y.; Kaneko, M.K.; Hanibuchi, M.; Tsujimoto, Y.; Goto, H.; Kakiuchi, S.; Aono, Y.; Huang, J.; Sato, S.; et al. A novel targeting therapy of malignant mesothelioma using anti-podoplanin antibody. J. Immunol. 2013, 190, 6239–6249. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Takagi, S.; Sato, S.; Oh-hara, T.; Koike, S.; Takami, M.; Arai, H.; Fujita, N. Prevention of hematogenous metastasis by neutralizing mice and its chimeric anti-Aggrus/podoplanin antibodies. Cancer Sci. 2011, 102, 2051–2057. [Google Scholar] [CrossRef]
- Chang, Y.W.; Hsieh, P.W.; Chang, Y.T.; Lu, M.H.; Huang, T.F.; Chong, K.Y.; Liao, H.R.; Cheng, J.C.; Tseng, C.P. Identification of a novel platelet antagonist that binds to CLEC-2 and suppresses podoplanin-induced platelet aggregation and cancer metastasis. Oncotarget 2015, 6, 42733–42748. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Yu, N.Y.; Lee, S.H.; Tsai, H.J.; Wu, C.C.; Cheng, J.C.; Chen, D.P.; Wang, Y.R.; Tseng, C.P. Podoplanin promotes cancer-associated thrombosis and contributes to the unfavorable overall survival in an ectopic xenograft mouse model of oral cancer. Biomed. J. 2020, 43, 146–162. [Google Scholar] [CrossRef] [PubMed]
- Tsukiji, N.; Osada, M.; Sasaki, T.; Shirai, T.; Satoh, K.; Inoue, O.; Umetani, N.; Mochizuki, C.; Saito, T.; Kojima, S.; et al. Cobalt hematoporphyrin inhibits CLEC-2-podoplanin interaction, tumor metastasis, and arterial/venous thrombosis in mice. Blood Adv. 2018, 2, 2214–2225. [Google Scholar] [CrossRef]
- Meng, D.; Ma, X.; Li, H.; Wu, X.; Cao, Y.; Miao, Z.; Zhang, X. A Role of the podoplanin-CLEC-2 axis in promoting inflammatory response after ischemic stroke in mice. Neurotox. Res. 2021, 39, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Damaskinaki, F.N.; Moran, L.A.; Garcia, A.; Kellam, B.; Watson, S.P. Overcoming challenges in developing small molecule inhibitors for GPVI and CLEC-2. Platelets 2021, 32, 744–752. [Google Scholar] [CrossRef]
- Tseng, C.P.; Huang, Y.L.; Chang, Y.W.; Liao, H.R.; Chen, Y.L.; Hsieh, P.W. Polysaccharide-containing fraction from Artemisia Argyi inhibits tumor cell-induced platelet aggregation by blocking interaction of podoplanin with C-type lectin-like receptor 2. J Food Drug Anal. 2020, 28, 115–123. [Google Scholar] [CrossRef]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the therapeutics and diagnostics pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.X.; Kwon, Y.J. Aptamers: The “evolution” of SELEX. Methods 2016, 106, 21–28. [Google Scholar] [CrossRef]
- Li, Z.; Fu, X.; Huang, J.; Zeng, P.; Huang, Y.; Chen, X.; Liang, C. Advances in screening and development of therapeutic aptamers against cancer cells. Front. Cell Dev. Biol. 2021, 9, 662791. [Google Scholar] [CrossRef]
- Avino, A.; Fabrega, C.; Tintore, M.; Eritja, R. Thrombin binding aptamer, more than a simple aptamer: Chemically modified derivatives and biomedical applications. Curr. Pharm. Des. 2012, 18, 2036–2047. [Google Scholar] [CrossRef]
- Ng, E.W.; Shima, D.T.; Calias, P.; Cunningham, E.T., Jr.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Wang, P.; Li, S.; Geng, X.; Zou, L.; Jin, M.; Zou, Q.; Wang, Q.; Yang, X.; Wang, K. Development of DNA aptamer as a beta-amyloid aggregation inhibitor. ACS Appl. Bio Mater. 2020, 3, 8611–8618. [Google Scholar] [CrossRef] [PubMed]
- Cha, B.S.; Jang, Y.J.; Lee, E.S.; Kim, D.Y.; Woo, J.S.; Son, J.; Kim, S.; Shin, J.; Han, J.; Kim, S.; et al. Development of a novel DNA aptamer targeting colorectal cancer cell-derived small extracellular vesicles as a potential diagnostic and therapeutic agent. Adv. Healthc. Mater. 2023, 12, e2300854. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Long, S.; Zhao, M.; Zhao, J.; Zhang, Y.; He, W.; Xiang, L.; Tan, J.; Ye, M.; Tan, W.; et al. Regulation of cellular signaling with an aptamer inhibitor to impede cancer metastasis. J. Am. Chem. Soc. 2024, 146, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Alshaer, W.; Ababneh, N.; Hatmal, M.; Izmirli, H.; Choukeife, M.; Shraim, A.; Sharar, N.; Abu-Shiekah, A.; Odeh, F.; Al Bawab, A.; et al. Selection and targeting of EpCAM protein by ssDNA aptamer. PLoS ONE 2017, 12, e0189558. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Zadeh, J.N.; Steenberg, C.D.; Bois, J.S.; Wolfe, B.R.; Pierce, M.B.; Khan, A.R.; Dirks, R.M.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid systems. J. Comput. Chem. 2011, 32, 170–173. [Google Scholar] [CrossRef] [PubMed]
- Fornace, M.E.; Huang, J.; Newman, C.T.; Porubsky, N.J.; Pierce, M.B.; Pierce, N.A. NUPACK: Analysis and design of nucleic acid structures, devices, and systems. ChemRxiv 2022. [Google Scholar] [CrossRef]
- Huang, C.W.; Chuang, C.P.; Chen, Y.J.; Wang, H.Y.; Lin, J.J.; Huang, C.Y.; Wei, K.C.; Huang, F.T. Integrin alpha(2)beta(1)-targeting ferritin nanocarrier traverses the blood-brain barrier for effective glioma chemotherapy. J. Nanobiotechnol. 2021, 19, 180. [Google Scholar] [CrossRef]
- Hung, W.S.; Ling, P.; Cheng, J.C.; Chang, S.S.; Tseng, C.P. Disabled-2 is a negative immune regulator of lipopolysaccharide-stimulated Toll-like receptor 4 internalization and signaling. Sci. Rep. 2016, 6, 35343. [Google Scholar] [CrossRef]
- Tsai, H.J.; Chien, K.Y.; Liao, H.R.; Shih, M.S.; Lin, Y.C.; Chang, Y.W.; Cheng, J.C.; Tseng, C.P. Functional links between Disabled-2 Ser723 phosphorylation and thrombin signaling in human platelets. J. Thromb. Haemost. 2017, 15, 2029–2044. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.V.; Armstrong, P.C.; Warner, T.D. 96-well plate-based aggregometry. Platelets 2018, 29, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Bishop, G.R.; Chaires, J.B. Characterization of DNA structures by circular dichroism. Curr. Protoc. Nucleic Acid Chem. 2003, 11, 7.11.1–7.11.8. [Google Scholar] [CrossRef] [PubMed]
- Masiero, S.; Trotta, R.; Pieraccini, S.; De Tito, S.; Perone, R.; Randazzo, A.; Spada, G.P. A non-empirical chromophoric interpretation of CD spectra of DNA G-quadruplex structures. Org. Biomol. Chem. 2010, 8, 2683–2692. [Google Scholar] [CrossRef]
- Tothova, P.; Krafcikova, P.; Viglasky, V. Formation of highly ordered multimers in G-quadruplexes. Biochemistry 2014, 53, 7013–7027. [Google Scholar] [CrossRef]
- Paramasivan, S.; Rujan, I.; Bolton, P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods 2007, 43, 324–331. [Google Scholar] [CrossRef]
- Ivanov, V.I.; Minchenkova, L.E.; Schyolkina, A.K.; Poletayev, A.I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers 1973, 12, 89–110. [Google Scholar] [CrossRef]
- Dapic, V.; Abdomerovic, V.; Marrington, R.; Peberdy, J.; Rodger, A.; Trent, J.O.; Bates, P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31, 2097–2107. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, Z.; An, Y.; Zhang, W.; Zhang, H.; Liu, D.; Yu, C.; Duan, W.; Yang, C.J. Selection of DNA aptamers against epithelial cell adhesion molecule for cancer cell imaging and circulating tumor cell capture. Anal. Chem. 2013, 85, 4141–4149. [Google Scholar] [CrossRef]
- Sharma, S.; Byrne, H.; O’Kennedy, R.J. Antibodies and antibody-derived analytical biosensors. Essays Biochem. 2016, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A Highlight of Recent Advances in Aptamer Technology and Its Application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed]
- Domsicova, M.; Korcekova, J.; Poturnayova, A.; Breier, A. New Insights into Aptamers: An Alternative to Antibodies in the Detection of Molecular Biomarkers. Int. J. Mol. Sci. 2024, 25, 6833. [Google Scholar] [CrossRef]
- Di Mauro, V.; Lauta, F.C.; Modica, J.; Appleton, S.L.; De Franciscis, V.; Catalucci, D. Diagnostic and Therapeutic Aptamers: A Promising Pathway to Improved Cardiovascular Disease Management. JACC Basic Transl. Sci. 2024, 9, 260–277. [Google Scholar] [CrossRef]
- Park, K.S.; Park, T.I.; Lee, J.E.; Hwang, S.Y.; Choi, A.; Pack, S.P. Aptamers and Nanobodies as New Bioprobes for SARS-CoV-2 Diagnostic and Therapeutic System Applications. Biosensors 2024, 14, 146. [Google Scholar] [CrossRef] [PubMed]
- Vavalle, J.P.; Cohen, M.G. The REG1 anticoagulation system: A novel actively controlled factor IX inhibitor using RNA aptamer technology for treatment of acute coronary syndrome. Future Cardiol. 2012, 8, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Russo Krauss, I.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, 46, 12177–12185. [Google Scholar] [CrossRef]
- Liu, M.; Zaman, K.; Fortenberry, Y.M. Overview of the Therapeutic Potential of Aptamers Targeting Coagulation Factors. Int. J. Mol. Sci. 2021, 22, 3897. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Mao, Z.; Li, W.; Pei, R. Anti-PD-L1 DNA aptamer antagonizes the interaction of PD-1/PD-L1 with antitumor effect. J. Mater. Chem. B 2021, 9, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Z.; Yu, H. Selection of threose nucleic acid aptamers to block PD-1/PD-L1 interaction for cancer immunotherapy. Chem. Commun. 2020, 56, 14653–14656. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Liu, Y.; Shi, M.; Chen, J.; Li, K.; Tan, Y.; Chen, L.; Wu, Y.; Wang, T.; Liu, X.; et al. Aptamer-based biosensors for virus protein detection. TrAC Trends Anal. Chem. 2022, 157, 116738. [Google Scholar] [CrossRef]
- Strehlitz, B.; Nikolaus, N.; Stoltenburg, R. Protein Detection with Aptamer Biosensors. Sensors 2008, 8, 4296–4307. [Google Scholar] [CrossRef] [PubMed]
- Sequeira-Antunes, B.; Ferreira, H.A. Nucleic Acid Aptamer-Based Biosensors: A Review. Biomedicines 2023, 11, 3201. [Google Scholar] [CrossRef] [PubMed]
- Shraim, A.S.; Abdel Majeed, B.A.; Al-Binni, M.A.; Hunaiti, A. Therapeutic Potential of Aptamer-Protein Interactions. ACS Pharmacol. Transl. Sci. 2022, 5, 1211–1227. [Google Scholar] [CrossRef] [PubMed]
- Civit, L.; Taghdisi, S.M.; Jonczyk, A.; Hassel, S.K.; Grober, C.; Blank, M.; Stunden, H.J.; Beyer, M.; Schultze, J.; Latz, E.; et al. Systematic evaluation of cell-SELEX enriched aptamers binding to breast cancer cells. Biochimie 2018, 145, 53–62. [Google Scholar] [CrossRef]
- Civit, L.; Moradzadeh, N.; Jonczyk, A.; Neckermann, P.; Asbach, B.; Peterhoff, D.; Wagner, R.; Famulok, M.; Mayer, G.; Kjems, J.; et al. A Multi-Faceted Binding Assessment of Aptamers Targeting the SARS-CoV-2 Spike Protein. Int. J. Mol. Sci. 2024, 25, 4642. [Google Scholar] [CrossRef]
- Kelly, L.; Maier, K.E.; Yan, A.; Levy, M. A comparative analysis of cell surface targeting aptamers. Nat. Commun. 2021, 12, 6275. [Google Scholar] [CrossRef]
- Chen, C.H.; Chernis, G.A.; Hoang, V.Q.; Landgraf, R. Inhibition of heregulin signaling by an aptamer that preferentially binds to the oligomeric form of human epidermal growth factor receptor-3. Proc. Natl. Acad. Sci. USA 2003, 100, 9226–9231. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ebright, J.N.; Stovall, G.M.; Chen, X.; Nguyen, H.H.; Singh, A.; Syrett, A.; Ellington, A.D. Technical and biological issues relevant to cell typing with aptamers. J. Proteome Res. 2009, 8, 2438–2448. [Google Scholar] [CrossRef]
- Suzuki-Inoue, K.; Inoue, O.; Ding, G.; Nishimura, S.; Hokamura, K.; Eto, K.; Kashiwagi, H.; Tomiyama, Y.; Yatomi, Y.; Umemura, K.; et al. Essential in vivo roles of the C-type lectin receptor CLEC-2: Embryonic/neonatal lethality of CLEC-2-deficient mice by blood/lymphatic misconnections and impaired thrombus formation of CLEC-2-deficient platelets. J. Biol. Chem. 2010, 285, 24494–24507. [Google Scholar] [CrossRef] [PubMed]
- Sheng, M.; Sun, R.; Fu, J.; Lu, G. The podoplanin-CLEC-2 interaction promotes platelet-mediated melanoma pulmonary metastasis. BMC Cancer 2024, 24, 399. [Google Scholar] [CrossRef] [PubMed]
- Meng, D.; Luo, M.; Liu, B. The Role of CLEC-2 and Its Ligands in Thromboinflammation. Front. Immunol. 2021, 12, 688643. [Google Scholar] [CrossRef] [PubMed]
- Osada, M.; Inoue, O.; Ding, G.; Shirai, T.; Ichise, H.; Hirayama, K.; Takano, K.; Yatomi, Y.; Hirashima, M.; Fujii, H.; et al. Platelet activation receptor CLEC-2 regulates blood/lymphatic vessel separation by inhibiting proliferation, migration, and tube formation of lymphatic endothelial cells. J. Biol. Chem. 2012, 287, 22241–22252. [Google Scholar] [CrossRef] [PubMed]
- Honma, M.; Minami-Hori, M.; Takahashi, H.; Iizuka, H. Podoplanin expression in wound and hyperproliferative psoriatic epidermis: Regulation by TGF-beta and STAT-3 activating cytokines, IFN-gamma, IL-6, and IL-22. J. Dermatol. Sci. 2012, 65, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Peters, A.; Burkett, P.R.; Sobel, R.A.; Buckley, C.D.; Watson, S.P.; Bettelli, E.; Kuchroo, V.K. Podoplanin negatively regulates CD4+ effector T cell responses. J. Clin. Investig. 2015, 125, 129–140. [Google Scholar] [CrossRef]
- Kumar, V.; Dasoveanu, D.C.; Chyou, S.; Tzeng, T.C.; Rozo, C.; Liang, Y.; Stohl, W.; Fu, Y.X.; Ruddle, N.H.; Lu, T.T. A dendritic-cell-stromal axis maintains immune responses in lymph nodes. Immunity 2015, 42, 719–730. [Google Scholar] [CrossRef]
- Herzog, B.H.; Fu, J.; Wilson, S.J.; Hess, P.R.; Sen, A.; McDaniel, J.M.; Pan, Y.; Sheng, M.; Yago, T.; Silasi-Mansat, R.; et al. Podoplanin maintains high endothelial venule integrity by interacting with platelet CLEC-2. Nature 2013, 502, 105–109. [Google Scholar] [CrossRef]
- Lowe, K.L.; Finney, B.A.; Deppermann, C.; Hagerling, R.; Gazit, S.L.; Frampton, J.; Buckley, C.; Camerer, E.; Nieswandt, B.; Kiefer, F.; et al. Podoplanin and CLEC-2 drive cerebrovascular patterning and integrity during development. Blood 2015, 125, 3769–3777. [Google Scholar] [CrossRef]
- Tamura, S.; Suzuki-Inoue, K.; Tsukiji, N.; Shirai, T.; Sasaki, T.; Osada, M.; Satoh, K.; Ozaki, Y. Podoplanin-positive periarteriolar stromal cells promote megakaryocyte growth and proplatelet formation in mice by CLEC-2. Blood 2016, 127, 1701–1710. [Google Scholar] [CrossRef]
- Harami, G.M.; Kovacs, Z.J.; Pancsa, R.; Palinkas, J.; Barath, V.; Tarnok, K.; Malnasi-Csizmadia, A.; Kovacs, M. Phase separation by ssDNA binding protein controlled via protein-protein and protein-DNA interactions. Proc. Natl. Acad. Sci. USA 2020, 117, 26206–26217. [Google Scholar] [CrossRef] [PubMed]
- Morin, J.A.; Cerron, F.; Jarillo, J.; Beltran-Heredia, E.; Ciesielski, G.L.; Arias-Gonzalez, J.R.; Kaguni, L.S.; Cao, F.J.; Ibarra, B. DNA synthesis determines the binding mode of the human mitochondrial single-stranded DNA-binding protein. Nucleic Acids Res. 2017, 45, 7237–7248. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, R.; Horita, M.; Yamaguchi, D.; Nakai, H.; Nakano, S.I. Evaluation of weak interactions of proteins and organic cations with DNA duplex structures. Biophys. J. 2022, 121, 2873–2881. [Google Scholar] [CrossRef] [PubMed]




| Aptamer | Sequences | Frequency (%) | Kd (nM) a |
|---|---|---|---|
| A1 | 5′-ATCCAGAGTGACGCAGCATCACTAGTGCGCCAGGGGGA CTGCTCGGGATTGCGGATACTGGACACGGTGGCTTAGT-3′ | 68.35 | 1.3 ± 1.2 |
| A5 | 5′-ATCCAGAGTGACGCAGCATGCCAAAGGGTCTGCTCGGG ATTGCGGAGAGGGCTTGGGGTGGACACGGTGGCTTAGT-3′ | 9.40 | 0.2 ± 0.1 |
| A6 | 5′-ATCCAGAGTGACGCAGCACACCGAGGGGACTGCTCGGG ATTGCGGATGAGGTAGCAAGTGGACACGGTGGCTTAGT-3′ | 4.76 | 0.2 ± 0.0 |
| A77 | 5′-ATCCAGAGTGACGCAGCATGGGGGGAGAGTGTGGTCTG GCTTGGTGGGGTCTGGGGGCTGGACACGGTGGCTTAGT-3′ | 0.09 | 1.7 ± 0.1 |
| A289 | 5′-ATCCAGAGTGACGCAGCACGGGTTTGGCGCCGGGCCTG GCGGGTGTAGCGTTTTTGGGTGGACACGGTGGCTTAGT-3′ | 0.03 | 0.5 ± 0.0 |
| A1498 | 5′-ATCCAGAGTGACGCAGCATGCAGGGTTTGGCTTCGGGC CTGGCGGAGAGAGCGTGAGGTGGACACGGTGGCTTAGT-3′ | 0.01 | 0.4 ± 0.0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsai, H.-J.; Cheng, K.-W.; Li, J.-C.; Ruan, T.-X.; Chang, T.-H.; Wang, J.-R.; Tseng, C.-P. Identification of Podoplanin Aptamers by SELEX for Protein Detection and Inhibition of Platelet Aggregation Stimulated by C-Type Lectin-like Receptor 2. Biosensors 2024, 14, 464. https://doi.org/10.3390/bios14100464
Tsai H-J, Cheng K-W, Li J-C, Ruan T-X, Chang T-H, Wang J-R, Tseng C-P. Identification of Podoplanin Aptamers by SELEX for Protein Detection and Inhibition of Platelet Aggregation Stimulated by C-Type Lectin-like Receptor 2. Biosensors. 2024; 14(10):464. https://doi.org/10.3390/bios14100464
Chicago/Turabian StyleTsai, Hui-Ju, Kai-Wen Cheng, Jou-Chen Li, Tsai-Xiang Ruan, Ting-Hsin Chang, Jin-Ru Wang, and Ching-Ping Tseng. 2024. "Identification of Podoplanin Aptamers by SELEX for Protein Detection and Inhibition of Platelet Aggregation Stimulated by C-Type Lectin-like Receptor 2" Biosensors 14, no. 10: 464. https://doi.org/10.3390/bios14100464
APA StyleTsai, H.-J., Cheng, K.-W., Li, J.-C., Ruan, T.-X., Chang, T.-H., Wang, J.-R., & Tseng, C.-P. (2024). Identification of Podoplanin Aptamers by SELEX for Protein Detection and Inhibition of Platelet Aggregation Stimulated by C-Type Lectin-like Receptor 2. Biosensors, 14(10), 464. https://doi.org/10.3390/bios14100464





