Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Photonic Crystal Aminosilanization and Surface Activation
2.3. Deposition of Protein Recognition Units
2.4. Photonic Crystal Surface Mode Imaging
2.5. Oligonucleotide–RAD51 Association
2.6. Data Analysis and Statistical Treatment
3. Results and Discussion
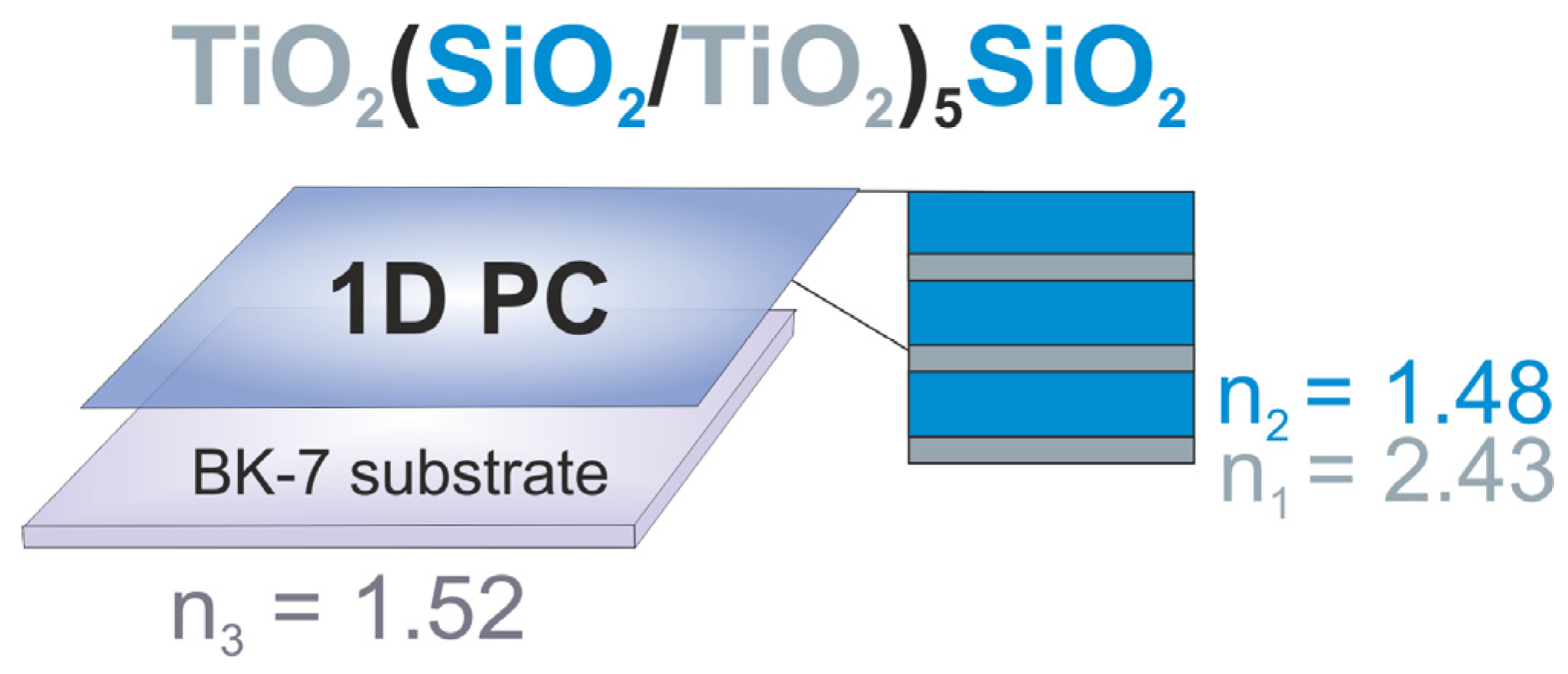
3.1. Modification of the Surface of a Photonic Crystal–Based Sensor with Protein Recognition Units
3.2. Surface Blocking and Immobilization of the Model Oligonucleotide
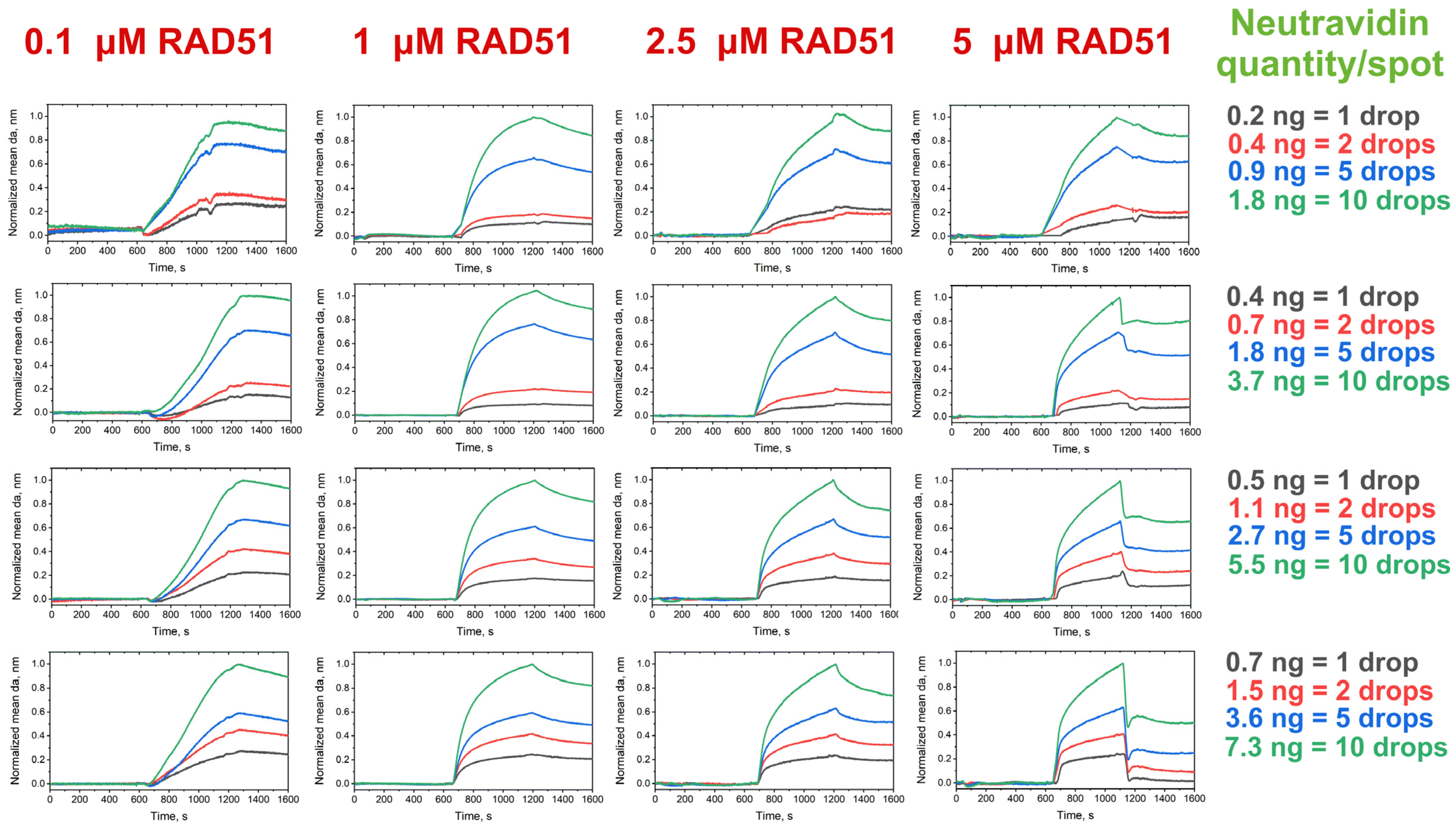
3.3. Analysis of RAD51–Oligonucleotide Association
3.4. Limit of Detection and Linearity of the Sensor Response
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kim, H.-M.; Jeong, D.H.; Lee, H.-Y.; Park, J.-H.; Lee, S.-K. Design and validation of fiber optic localized surface plasmon resonance sensor for thyroglobulin immunoassay with high sensitivity and rapid detection. Sci. Rep. 2021, 11, 15985. [Google Scholar] [CrossRef] [PubMed]
- Soltermann, F.; Struwe, W.B.; Kukura, P. Label-free methods for optical in vitro characterization of protein–protein interactions. Phys. Chem. Chem. Phys. 2021, 23, 16488–16500. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, J.E.; Jaramillo, S.A.; Settles, E.; Salazar, J.J.V.; Lehr, A.; Gonzalez, J.; Aranda, C.R.; Navarro-Contreras, H.R.; Raniere, M.O.; Harvey, M.; et al. Detection of SARS-CoV-2 and its S and N proteins using surface enhanced Raman spectroscopy. RSC Adv. 2021, 11, 25788–25794. [Google Scholar] [CrossRef] [PubMed]
- Yano, T.; Kajisa, T.; Ono, M.; Miyasaka, Y.; Hasegawa, Y.; Saito, A.; Otsuka, K.; Sakane, A.; Sasaki, T.; Yasutomo, K.; et al. Ultrasensitive detection of SARS-CoV-2 nucleocapsid protein using large gold nanoparticle-enhanced surface plasmon resonance. Sci. Rep. 2022, 12, 1060. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Geng, Z.; Su, Y.; Fan, Z.; Wang, S.; Fang, W.; Chen, H. Label-free exosome detection based on a low-cost plasmonic biosensor array integrated with microfluidics. Langmuir 2019, 35, 9816–9824. [Google Scholar] [CrossRef]
- Soares, M.S.; Silva, L.C.B.; Vidal, M.; Loyez, M.; Facão, M.; Caucheteur, C.; Segatto, M.E.V.; Costa, F.M.; Leitão, C.; Pereira, S.O.; et al. Label-free plasmonic immunosensor for cortisol detection in a D-shaped optical fiber. Biomed. Opt. Express 2022, 13, 3259–3274. [Google Scholar] [CrossRef] [PubMed]
- Bekmurzayeva, A.; Ashikbayeva, Z.; Myrkhiyeva, Z.; Nugmanova, A.; Shaimerdenova, M.; Ayupova, T.; Tosi, D. Label-free fiber-optic spherical tip biosensor to enable picomolar-level detection of CD44 protein. Sci. Rep. 2021, 11, 19583. [Google Scholar] [CrossRef]
- Premaratne, G.; Mubarak, Z.H.A.; Senavirathna, L.; Liu, L.; Krishnan, S. Measuring ultra-low levels of nucleotide biomarkers using quartz crystal microbalance and SPR microarray imaging methods: A comparative analysis. Sens. Actuators B 2017, 253, 368–375. [Google Scholar] [CrossRef]
- Nawattanapaiboon, K.; Kiatpathomchai, W.; Santanirand, P.; Vongsakulyanon, A.; Amarit, R.; Somboonkaew, A.; Sutapun, B.; Srikhirin, T. SPR-DNA array for detection of methicillin-resistant Staphylococcus Aureus (MRSA) in combination with loop-mediated isothermal amplification. Biosens. Bioelectron. 2015, 74, 335–340. [Google Scholar] [CrossRef]
- Nifontova, G.; Petrova, I.; Gerasimovich, E.; Konopsky, V.N.; Ayadi, N.; Charlier, C.; Fleury, F.; Karaulov, A.; Sukhanova, A.; Nabiev, I. Label-free multiplexed microfluidic analysis of protein interactions based on photonic crystal surface mode imaging. Int. J. Mol. Sci. 2023, 24, 4347. [Google Scholar] [CrossRef]
- Sizova, S.; Shakurov, R.; Mitko, T.; Shirshikov, F.; Solovyeva, D.; Konopsky, V.; Alieva, E.; Klinov, D.; Bespyatykh, J.; Basmanov, D. The elaboration of effective coatings for photonic crystal chips in optical biosensors. Polymers 2022, 14, 152. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, D.; Yamada, H.; Sueyoshi, K.; Hisamoto, H.; Endo, T. Imprinted photonic crystal-film-based smartphone-compatible label-free optical sensor for SARS-CoV-2 testing. Biosensors 2022, 12, 200. [Google Scholar] [CrossRef] [PubMed]
- Sinibaldi, A.; Sampaoli, C.; Danz, N.; Munzert, P.; Sibilio, L.; Sonntag, F.; Occhicone, A.; Falvo, E.; Tremante, E.; Giacomini, P.; et al. Detection of soluble ERBB2 in breast cancer cell lysates using a combined label-free/fluorescence platform based on Bloch surface waves. Biosens. Bioelectron. 2017, 92, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Resende, S.; Frasco, M.F.; Sales, M.G.F. A biomimetic photonic crystal sensor for label-free detection of urinary venous thromboembolism biomarker. Sens. Actuators B 2020, 312, 127947. [Google Scholar] [CrossRef]
- Cetin, A.E.; Topkaya, S.N. Photonic crystal and plasmonic nanohole based label-free biodetection. Biosens. Bioelectron. 2019, 132, 196–202. [Google Scholar] [CrossRef]
- Li, T.; Liu, G.; Kong, H.; Yang, G.; Wei, G.; Zhou, X. Recent advances in photonic crystal-based sensors. Coord. Chem. Rev. 2023, 475, 214909. [Google Scholar] [CrossRef]
- Parandin, F.; Heidari, F.; Aslinezhad, M.; Mehdi, M.; Parandin, M.M.; Roshani, S.; Roshani, S. Design of 2D photonic crystal biosensor to detect blood components. Opt. Quant. Electron. 2022, 54, 618. [Google Scholar] [CrossRef]
- Kaňok, R.; Hlubina, P.; Gembalová, L.; Ciprian, D. Efficient optical sensing based on phase shift of waves supported by a one-dimensional photonic crystal. Sensors 2021, 21, 6535. [Google Scholar] [CrossRef]
- Sinibaldi, A.; Danz, N.; Anopchenko, A.; Munzert, P.; Schmieder, S.; Chandrawati, R.; Rizzo, R.; Rana, S.; Sonntag, F.; Occhicone, A.; et al. Label-free detection of tumor angiogenesis biomarker angiopoietin 2 using Bloch surface waves on one dimensional photonic crystals. J. Light. Technol. 2015, 33, 3385–3393. [Google Scholar] [CrossRef]
- Occhicone, A.; Pea, M.; Polito, R.; Giliberti, V.; Sinibaldi, A.; Mattioli, F.; Cibella, S.; Notargiacomo, A.; Nucara, A.; Biagioni, P.; et al. Spectral characterization of mid-infrared Bloch surface waves excited on a truncated 1D photonic crystal. ACS Photonics 2021, 8, 350–359. [Google Scholar] [CrossRef]
- Haryanto, A.; Eo, Y.J.; Do, Y.R.; Lee, C.W. Layer-by-layer assembly of 1-D photonic crystal for wavelength-selective optical filter. Appl. Surf. Sci. 2023, 611, 155762. [Google Scholar] [CrossRef]
- Konopsky, V.N.; Alieva, E.V. Photonic crystal surface mode imaging biosensor based on wavelength interrogation of resonance peak. Sens. Actuators B 2018, 276, 271–278. [Google Scholar] [CrossRef]
- Konopsky, V.; Mitko, T.; Aldarov, K.; Alieva, E.; Basmanov, D.; Moskalets, A.; Matveeva, A.; Morozova, O.; Klinov, D. Photonic crystal surface mode imaging for multiplexed and high-throughput label-free biosensing. Biosens. Bioelectron. 2020, 168, 112575. [Google Scholar] [CrossRef] [PubMed]
- Konopsky, V.N.; Alieva, E.V. Imaging biosensor based on planar optical waveguide. Opt. Laser Technol. 2019, 115, 171–175. [Google Scholar] [CrossRef]
- Gaur, G.; Hu, S.; Mernaugh, R.L.; Kravchenko, I.I.; Retterer, S.T.; Weiss, S.M. Label-free detection of Herceptin® using suspended silicon microring resonators. Sens. Actuators B 2018, 275, 394–401. [Google Scholar] [CrossRef]
- Chi, J.; Wu, Y.; Qin, F.; Su, M.; Cheng, N.; Zhang, J.; Li, C.; Lian, Z.; Yang, X.; Cheng, L.; et al. All-printed point-of-care immunosensing biochip for one drop blood diagnostics. Lab Chip 2022, 22, 3008–3014. [Google Scholar] [CrossRef]
- Lehmann, S.; Kraft, F.A.; Gerken, M. Spatially resolved protein binding kinetics analysis in microfluidic photonic crystal sensors. Sensors 2023, 23, 5637. [Google Scholar] [CrossRef]
- Laurini, E.; Marson, D.; Fermeglia, A.; Aulic, S.; Fermeglia, M.; Pricl, S. Role of Rad51 and DNA repair in cancer: A molecular perspective. Pharmacol. Ther. 2020, 208, 107492. [Google Scholar] [CrossRef]
- Tsai, Y.-F.; Chan, L.-P.; Chen, Y.-K.; Su, C.-W.; Hsu, C.-W.; Wang, Y.-Y.; Yuan, S.-S.F. RAD51 is a poor prognostic marker and a potential therapeutic target for oral squamous cell carcinoma. Cancer Cell Int. 2023, 23, 231. [Google Scholar] [CrossRef]
- Budke, B.; Tueckmantel, W.; Miles, K.; Kozikowski, A.P.; Connell, P.P. Optimization of drug candidates that inhibit the D-Loop activity of RAD51. ChemMedChem 2019, 14, 1031–1040. [Google Scholar] [CrossRef]
- McPherson, K.S.; Korzhnev, D.M. Targeting protein–protein interactions in the DNA damage response pathways for cancer chemotherapy. RSC Chem. Biol. 2021, 2, 1167–1195. [Google Scholar] [CrossRef] [PubMed]
- Subramanyam, S.; Spies, M. RAD51 discrimination between single- and double-strand DNA: A matter of flexibility and enthalpy. EMBO J. 2020, 39, e104547. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, F.; El-Sagheer, A.H.; Allard, J.; Brown, T.; Dushek, O.; Esashi, F. Molecular flexibility of dna as a key determinant of RAD51 recruitment. EMBO J. 2020, 39, e103002. [Google Scholar] [CrossRef] [PubMed]
- Velic, D.; Demeyer, A.; Peterlini, T.; Benhelli-Mokrani, H.; Mathé-Allainmat, M.; Masson, J.-Y.; Fleury, F. Molecular determinant of DIDS analogs targeting RAD51 activity. Molecules 2021, 26, 5460. [Google Scholar] [CrossRef] [PubMed]
- Song, D.; Graham, T.G.W.; Loparo, J.J. A general approach to visualize protein binding and DNA conformation without protein labelling. Nat. Commun. 2016, 7, 10976. [Google Scholar] [CrossRef]
- Altmannova, V.; Spirek, M.; Orlic, L.; Jēkabsons, A.; Clarence, T.; Henggeler, A.; Mlcouskova, J.; Chaleil, R.A.G.; Matos, J.; Krejci, L. The role of bivalent ions in the regulation of D-loop extension mediated by DMC1 during meiotic recombination. iScience 2022, 25, 105439. [Google Scholar] [CrossRef]
- Shkundina, I.S.; Gall, A.A.; Dick, A.; Cocklin, S.; Mazin, A.V. New RAD51 inhibitors to target homologous recombination in human cells. Genes 2021, 12, 920. [Google Scholar] [CrossRef]
- Rawal, Y.; Jia, L.; Meir, A.; Zhou, S.; Kaur, H.; Ruben, E.A.; Kwon, Y.; Bernstein, K.A.; Jasin, M.; Taylor, A.B.; et al. Structural insights into BCDX2 complex function in homologous recombination. Nature 2023, 619, 640–649. [Google Scholar] [CrossRef]
- Sut, T.N.; Park, H.; Koo, D.J.; Yoon, B.K.; Jackman, J.A. Distinct binding properties of neutravidin and streptavidin proteins to biotinylated supported lipid bilayers: Implications for sensor functionalization. Sensors 2022, 22, 5185. [Google Scholar] [CrossRef]
- Khan, S.; Burciu, B.; Filipe, C.D.M.; Li, Y.; Dellinger, K.; Didar, T.F. DNAzyme-based biosensors: Immobilization strategies, applications, and future prospective. ACS Nano 2021, 15, 13943–13969. [Google Scholar] [CrossRef]
- Schmidt, C.; Schierack, P.; Gerber, U.; Schröder, C.; Choi, Y.; Bald, I.; Lehmann, W.; Rödiger, S. Streptavidin homologues for applications on solid surfaces at high temperatures. Langmuir 2020, 36, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Barve, A.; Zhao, Z.; Jin, W.; Cheng, K. Comparison of avidin, neutravidin, and streptavidin as nanocarriers for efficient siRNA delivery. Mol. Pharm. 2017, 14, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Chi, J.; Zhang, D.; Ma, B.; Dong, X.; Yang, J.; Zhao, C.; Liu, H. Bio-inspired photonic crystals for naked eye quantification of nucleic acids. Analyst 2019, 144, 5413–5419. [Google Scholar] [CrossRef] [PubMed]
- Ionescu, R.E. Use of cysteamine and glutaraldehyde chemicals for robust functionalization of substrates with protein biomarkers—An overview on the construction of biosensors with different transductions. Biosensors 2022, 12, 581. [Google Scholar] [CrossRef] [PubMed]
- Juste-Dolz, A.; Fernández, E.; Puchades, R.; Avella-Oliver, M.; Maquieira, Á. Patterned biolayers of protein antigens for label-free biosensing in cow milk allergy. Biosensors 2023, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Arnfinnsdottir, N.B.; Chapman, C.A.; Bailey, R.C.; Aksnes, A.; Stokke, B.T. Impact of silanization parameters and antibody immobilization strategy on binding capacity of photonic ring resonators. Sensors 2020, 20, 3163. [Google Scholar] [CrossRef] [PubMed]
- Kotlarek, D.; Liu, K.; Quilis, N.G.; Bernhagen, D.; Timmerman, P.; Kouwer, P.; Dostalek, J. Thin-film polyisocyanide-based hydrogels for affinity biosensors. J. Phys. Chem. C 2021, 125, 12960–12967. [Google Scholar] [CrossRef]
- Perçin, I.; Idil, N.; Denizli, A. Molecularly imprinted poly(n-isopropylacrylamide) thermosensitive based cryogel for immunoglobulin G purification. Process Biochem. 2019, 80, 181–189. [Google Scholar] [CrossRef]
- Fabri-Faja, N.; Calvo-Lozano, O.; Dey, P.; Terborg, R.A.; Estevez, M.-C.; Belushkin, A.; Yesilköy, F.; Duempelmann, L.; Altug, H.; Pruneri, V.; et al. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta 2019, 1077, 232–242. [Google Scholar] [CrossRef]
- Ma, G.J.; Ferhan, A.R.; Jackman, J.A.; Cho, N.-J. Conformational flexibility of fatty acid-free bovine serum albumin proteins enables superior antifouling coatings. Commun. Mater. 2020, 1, 45. [Google Scholar] [CrossRef]
- Doose, S.; Barsch, H.; Sauer, M. Polymer properties of polythymine as revealed by translational diffusion. Biophys. J. 2007, 93, 1224–1234. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Frykholm, K.; Fornander, L.H.; Svedhem, S.; Westerlund, F.; Åkerman, B. Sensing conformational changes in DNA upon ligand binding using QCM-D. Polyamine condensation and Rad51 extension of DNA layers. J. Phys. Chem. B 2014, 118, 11895–11904. [Google Scholar] [CrossRef] [PubMed]
- Špírek, M.; Taylor, M.R.G.; Belan, O.; Boulton, S.J.; Krejci, L. Nucleotide proofreading functions by nematode RAD51 paralogs facilitate optimal RAD51 filament function. Nat. Commun. 2021, 12, 5545. [Google Scholar] [CrossRef] [PubMed]
- Normand, A.; Rivière, E.; Renodon-Cornière, A. Identification and characterization of human Rad51 inhibitors by screening of an existing drug library. Biochem. Pharmacol. 2014, 91, 293–300. [Google Scholar] [CrossRef]
- RAD51—DNA Repair Protein RAD51 Homolog 1—Homo Sapiens (Human). UniProt. Available online: https://www.uniprot.org/uniprotkb/Q06609/entry (accessed on 4 January 2024).
- Špírek, M.; Mlčoušková, J.; Beláň, O.; Gyimesi, M.; Harami, G.M.; Molnár, E.; Novacek, J.; Kovács, M.; Krejci, L. Human RAD51 rapidly forms intrinsically dynamic nucleoprotein filaments modulated by nucleotide binding state. Nucleic Acids Res. 2018, 46, 3967–3980. [Google Scholar] [CrossRef]

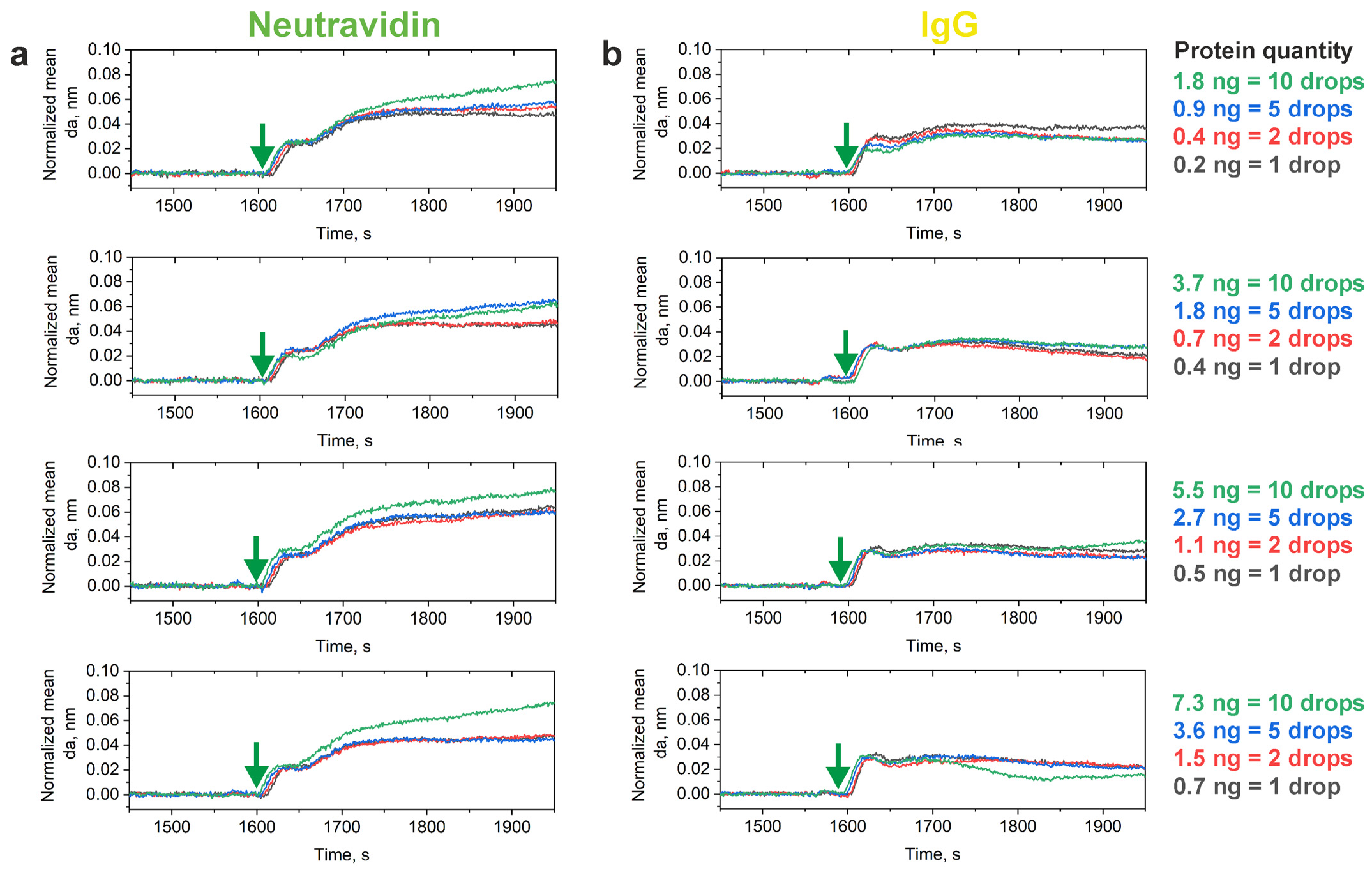
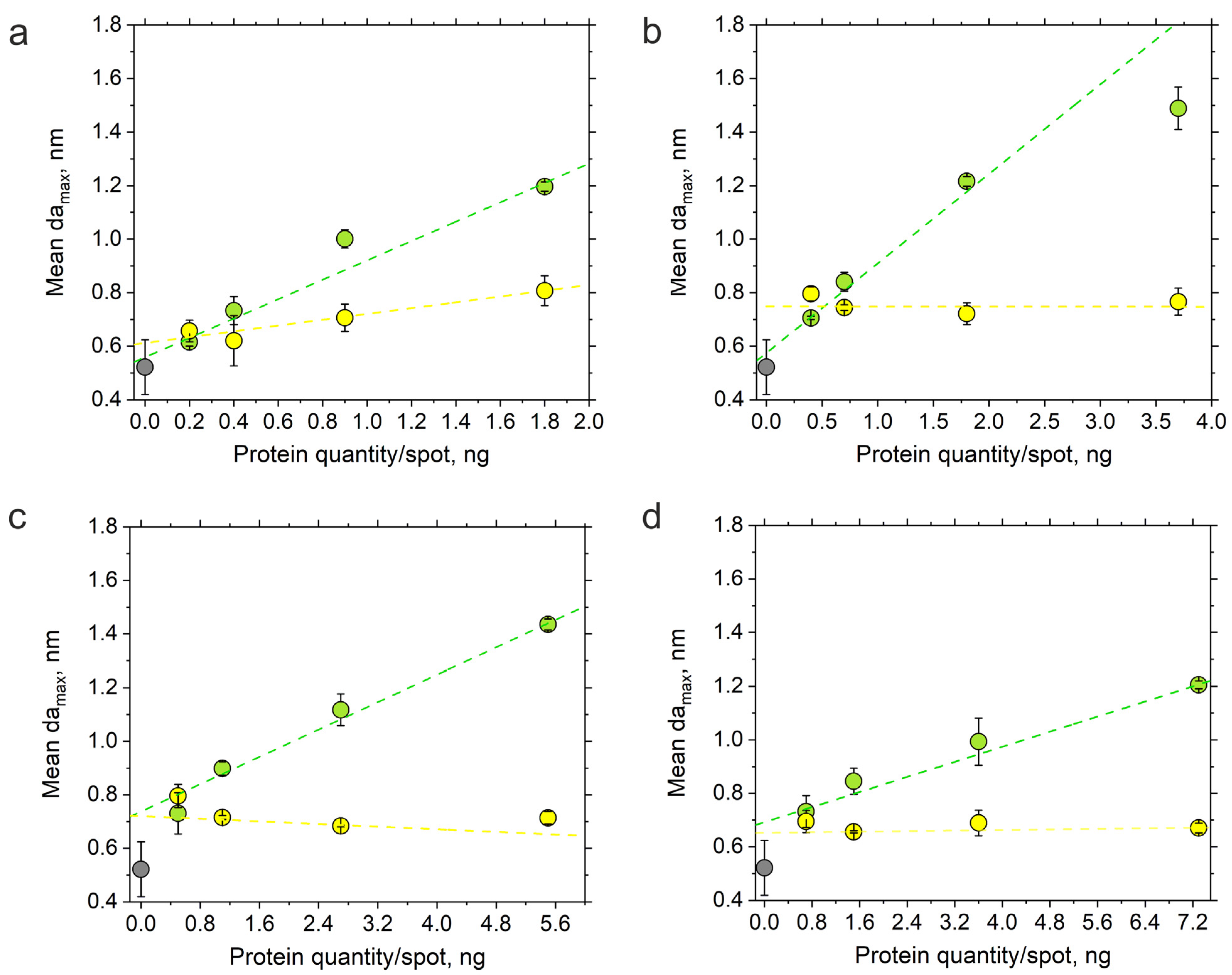
| Protein Quantity/Spot, ng | damax in Neutravidin Spots *, nm | damax in IgG Spots *, nm |
|---|---|---|
| 0.2 (1 drop) | 0.0503 | 0.0405 |
| 0.4 (2 drops) | 0.0553 | 0.0363 |
| 0.9 (5 drops) | 0.0586 | 0.0341 |
| 1.8 (10 drops) | 0.0754 | 0.0317 |
| 0.4 (1 drop) | 0.0485 | 0.0332 |
| 0.7 (2 drops) | 0.0497 | 0.0319 |
| 1.8 (5 drops) | 0.0663 | 0.0342 |
| 3.7 (10 drops) | 0.0637 | 0.0355 |
| 0.5 (1 drop) | 0.0655 | 0.0347 |
| 1.1 (2 drops) | 0.0635 | 0.0297 |
| 2.7 (5 drops) | 0.0626 | 0.0306 |
| 5.5 (10 drops) | 0.0796 | 0.0370 |
| 0.7 (1 drop) | 0.0487 | 0.0335 |
| 1.5 (2 drops) | 0.0491 | 0.0290 |
| 3.6 (5 drops) | 0.0467 | 0.0320 |
| 7.3 (10 drops) | 0.0749 | 0.0317 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nifontova, G.; Charlier, C.; Ayadi, N.; Fleury, F.; Karaulov, A.; Sukhanova, A.; Nabiev, I. Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate. Biosensors 2024, 14, 43. https://doi.org/10.3390/bios14010043
Nifontova G, Charlier C, Ayadi N, Fleury F, Karaulov A, Sukhanova A, Nabiev I. Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate. Biosensors. 2024; 14(1):43. https://doi.org/10.3390/bios14010043
Chicago/Turabian StyleNifontova, Galina, Cathy Charlier, Nizar Ayadi, Fabrice Fleury, Alexander Karaulov, Alyona Sukhanova, and Igor Nabiev. 2024. "Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate" Biosensors 14, no. 1: 43. https://doi.org/10.3390/bios14010043
APA StyleNifontova, G., Charlier, C., Ayadi, N., Fleury, F., Karaulov, A., Sukhanova, A., & Nabiev, I. (2024). Photonic Crystal Surface Mode Real-Time Imaging of RAD51 DNA Repair Protein Interaction with the ssDNA Substrate. Biosensors, 14(1), 43. https://doi.org/10.3390/bios14010043






