Aptasensors Based on Non-Enzymatic Peroxidase Mimics: Current Progress and Challenges
Abstract
:1. Introduction
2. Functional Mimics of Horseradish Peroxidase
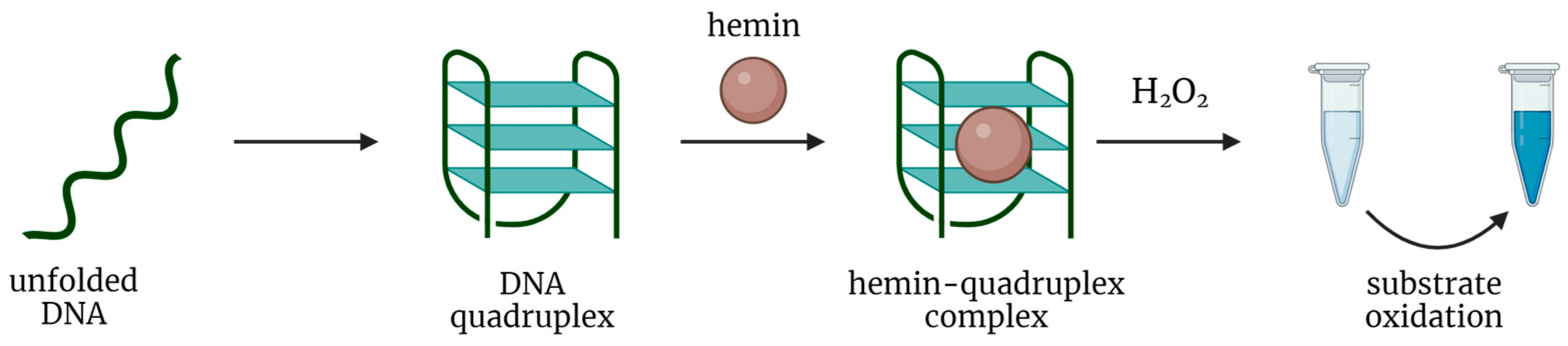
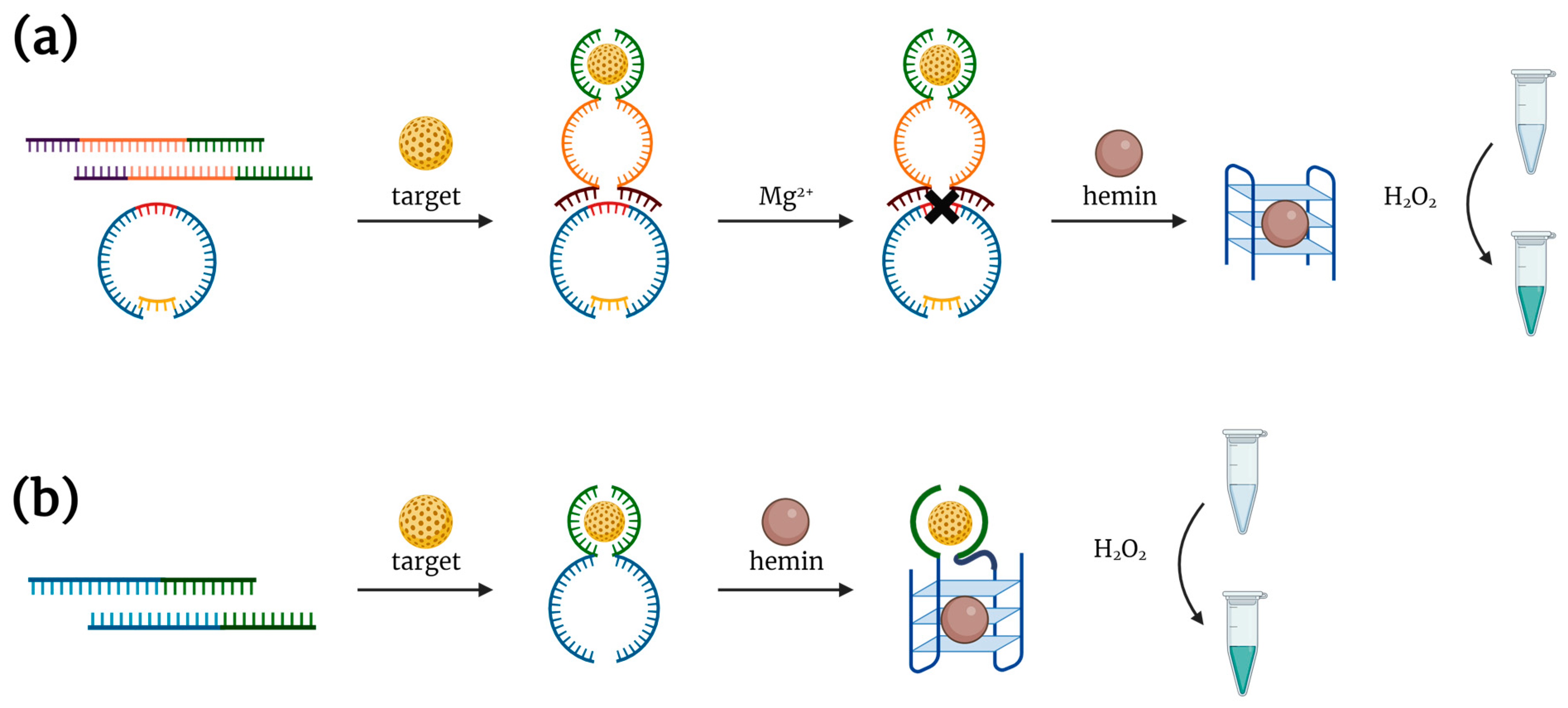
2.1. Complex of Hemin with DNA G-Quadruplex
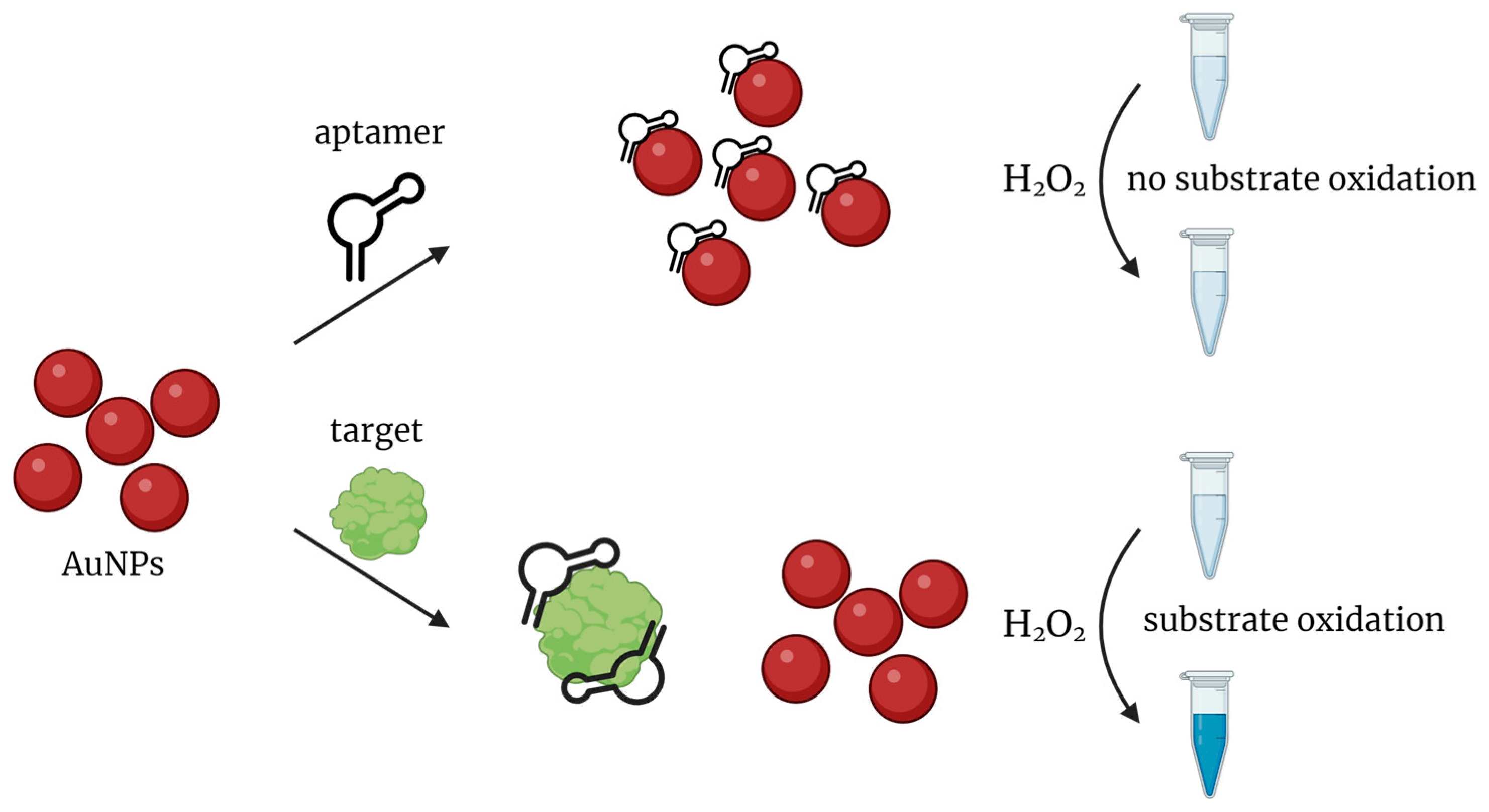
2.2. Nanozymes
2.3. Combination of Different HRP Mimics
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Type of Peroxidase Mimic | Analyte | Substrate | LOD | Selectivity | Ref. |
|---|---|---|---|---|---|
| hemin-DNAzyme complex | ochratoxin A | TMB | 2.5 nM | ochratoxin B warfarin | [19] |
| hemin-DNAzyme complex | AMP lysozyme | ABTS | 50 μM for AMP 5 pM for lysozyme | [20] | |
| hemin-DNAzyme complex | AMP lysozyme | ABTS | 4 μM for AMP 0.1 pM for lysozyme | [21] | |
| hemin-DNAzyme complex | ATP | ABTS | 1 μM | TTP, CTP, GTP | [22] |
| hemin-DNAzyme complex | ATP cocaine | ABTS | 5 μM for ATP 1 μM for cocaine | [23] | |
| hemin-GQ aptamer | VEGF | luminol | 18 nM | [24] | |
| hemin-GQ aptamer | 8-OHdG | ABTS | 0.14 nM | 8-OHdG analogs (dC, dA, dU, T, dG) and metal ions (Zn2+, Hg2+, Cu2+, Ca2+, Mn2+, Al3+, Na+) | [25] |
| hemin-DNAzyme complex | methamphetamine | ABTS | 0.5 nM | 15 general illicit drugs and metabolites: ketamine, norketamine, morphine, methadone, cocaine, mephedrone, cathinone, methcathinone, 3-trifluoromethylphenylpiperazine, 1-(3-tri-fluoromethylphenyl) piperazine, 3,4-methylenedioxy pyrovalerone, MDA, MDMA, EDDP, and mCPP | [26] |
| hemin-DNAzyme complex | СЕА | ABTS | 5.5 pM | BSA, lysozyme, insulin, prostate-specific antigen | [27] |
| hemin-DNAzyme complex | СЕА | 1,1′-oxalyldiimidazole | 0.58 ng/mL | alpha-fetoprotein, prostate-specific antigen | [28] |
| hemin-DNAzyme complex | VEGF | ABTS | 1.7 pM | [29] | |
| hemin-DNAzyme complex | S. aureus | luminol | 5 CFU/mL | S. epidermidis, S. warneri, P. aeruginosa; E. coli; C. Perfringens, and inactivated S. aureus | [30] |
| hemin-DNAzyme complex | ATP | ABTS | [32] | ||
| hemin-DNAzyme complex | cocaine | ABTS | 1 μM | chlorpromazine, diphenhydramine, promazine, scopolamine, caffeine, levamisole, lidocaine, and sucrose. minimal cross-reactivity to caffeine, chlorpromazine, promazine, and levamisole | [33] |
| hemin-DNAzyme complex | MUC-1 (exosomes) | ABTS | 3.94 × 105 particles/mL | exosomes generated by normal liver cell line L-02 | [34] |
| hemin-DNAzyme complex | quinclorac | TMB | 7.1 ng/mL | other quinolines and commonly used pesticides | [35] |
| hemin-DNAzyme complex | MUC1 | ABTS | 5 nM | BSA, thrombin, lysozyme, IgG | [36] |
| hemin-DNAzyme complex | ATP | TMB | 2.4 nM | CTP, GTP and UTP | [31] |
| hemin-DNAzyme complex | Hg2+ ions thrombin sulfadimethoxine cocaine 17β- estradiol | ABTS | 4 μM for thrombine 0.5 μM for sulfadimethoxine | [37] | |
| hemin-DNAzyme complex | ochratoxin A | TMB | 10 nM | other mycotoxins: aflatoxin B1, zearalenone, and ochratoxin B | [38] |
| nanozyme: PtNPs | thrombin | TMB | 0.4 μM | [39] | |
| nanozyme: Ag/Pt nanoclusters | thrombin | TMB | 2.6 nM | HSA, lysozyme, IgG, bovine thrombin | [40] |
| nanozyme: AuNPs | kanamycin | TMB | 1.49 nM | penicillin, ampicillin, and streptomycin | [41] |
| nanozyme: AuNPs | acetamiprid | TMB | 1.8 ppm | agritone, imidacloprid, and endothal | [42] |
| nanozyme: AuNPs | sulfadimethoxine | TMB | 10 ng/mL | kanamycin, chloramphenicol, oxytetracycline hydrochloride | [43] |
| nanozyme: AuNPs | murine norovirus | TMB | 30 particles/mL | S. aureus, E. coli, E. coli bacteriophage MS2 | [44] |
| nanozyme: AuNPs | P. aeruginosa | TMB | V. cholerae, L. monocytogens, S. aureus | [45] | |
| nanozyme: AuNPs | zearalenone | TMB | 10 ng/mL | aflatoxin B1, ochratoxin B, and metal ions (Ca2+, Na+, Mg2+, Zn2+) | [46] |
| nanozyme: AuNPs | streptomycin | ABTS | 86 nM | tetracycline, oxytetracycline, carbamazepine, penicillin, amoxicillin, and diclofenac | [47] |
| nanozyme: AuNPs | CD30+ small extracellular vesicles | TMB | 102–109 vesicles/mL | [48] | |
| nanozyme: AuNPs | malachite green | TMB | 1.8 nM | sulfaguanidine, sulfanilamide, oxytetracycline hydrochloride, chloramphenicol | [49] |
| nanozyme: AuNPs | sIL-2Ra | oPD | 1 pM | BSA, IL-5Ra, IL-13Ra2, IL-17Ra, CD166 | [50] |
| nanozyme: AuNPs | C-reactive protein | TMB | 0.07 pM | [51] | |
| nanozyme: AuNPs | retinol-binding protein 4 | luminol | 50 fM | [52] | |
| nanozyme: AgNPs (tyrosine-capped) | chlorpyrifos | TMB | 11.3 ppm | diazinon, dichlorvos, phorate, monocrotophos, methamidophos, azamethiphos, aldicarb, clothianidin, captan, thiamethoxam, mancozeb | [53] |
| nanozyme: Fe3O4 | S. typhimurium | TMB | 7.5 × 105 CFU/mL | [54] | |
| nanozyme: Au/Pd | C. jejuni | TMB | 10 cells | H. pylori, E. coli | [55] |
| nanozyme: core-shell gold nanorods | glucose insulin | TMB | 7.5 μM for glucose 0.2 pM for insulin | [56] | |
| nanozyme: Au nanoclusters conjugated to BSA | S. typhimurium | TMB | 1 CFU/mL | E. coli O157:H7, S. aureus, P. aeruginosa | [57] |
| nanozyme: Fe-MOF | thrombin | TMB | 0.8 nM | BSA, lysozyme, IgG, amino acids | [58] |
| nanozyme: Cu-MOF | C-reactive protein | TMB | 240 pg/mL | glucose, glutathione, ascorbic acid, iron, creatinine, albumin, calcium | [59] |
| nanozyme: Cu-MOF | thrombin | TMB | 360 pM | glucose, glutathione, ascorbic acid, iron, creatinine, albumin, C-reactive protein, calcium | [60] |
| nanozyme: MOF | chlorpyrifos | TMB | 4.4 ng/mL | atrazine, carbaryl, diazinon, malathion, bisphenol A, imidacloprid | [61] |
| nanozyme: MOG + PtNPs | fumonisin B1 | TMB | 2.7 pg/mL | ochratoxin A, ochratoxin B, aflatoxin B1, aflatoxin B2, zearalenone | [62] |
| nanozyme: MOF-on-MOF | chlorpyrifos | TMB luminol | 5.3 ng/mL | Na+, K+, Mg2+, Zn2+, Cl−, NO3−, SO42−, Ac−, glucose, urea, citric acid, glyphosate, trichlorfon | [63] |
| nanozyme: g-C3N4 NSs | CD63+ exosomes | TMB | 13.52 × 105 particles/μL | exosomes generated by nontumorigenic cell line | [64] |
| nanozyme: SWCNTs | CD63+ exosomes | TMB | 5.2 × 105 particles/μL | exosomes generated by nontumorigenic cell line | [65] |
| nanozyme: ZnFe2O4/rGO | S. typhimurium | TMB | 11 CFU/mL | S. aureus, E. coli, L. monocytogenes, V. parahaemolyticus | [66] |
| hybrid: DNA nanotube/magnetic beads/ hemin-DNAzyme | insulin | TMB | 0.39 µIU/mL | [71] | |
| hybrid: hemin-DNAzyme/PtNPs | thrombin | TMB | 15 pM | hemoglobin, lysozyme, BSA | [69] |
| hybrid: DNAzyme + MOF + PtNPs | chloramphenicol | TMB | 0.03 pM | kanamycin, streptomycin, oxytetracycline, gentamicin sulfate, chlortetracycline, Zn2+, Ca2+, Mg2+, casein, globulin, albumin | [70] |
References
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. General overviews on applications of ELISA. In SpringerBriefs in Applied Sciences and Technology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 19–29. [Google Scholar] [CrossRef]
- Hosseini, S.; Marco, P.V.-V.; Sergio, R.-P.; Martinez-Chapa, O. Enzyme-Linked Immunosorbent Assay (ELISA): From A to Z; Springer Briefs in Applied Sciences and Technology Forensic and Medical Bioinformatics: Singapore, 2018; ISBN 978-981-10-6765-5. [Google Scholar]
- Bradbury, A.; Lyon, O.M.F. Used in Research. Nature 2015, 518, 27–29. [Google Scholar] [CrossRef] [PubMed]
- Edfors, F.; Hober, A.; Linderbäck, K.; Maddalo, G.; Azimi, A.; Sivertsson, Å.; Tegel, H.; Hober, S.; Szigyarto, C.A.K.; Fagerberg, L.; et al. Enhanced validation of antibodies for research applications. Nat. Commun. 2018, 9, 4130. [Google Scholar] [CrossRef] [PubMed]
- In vitro selection of aptamers and their applications. Nat. Rev. Methods Prim. 2023, 3, 55. [CrossRef] [PubMed]
- Odeh, F.; Nsairat, H.; Alshaer, W.; Ismail, M.A.; Esawi, E.; Qaqish, B.; Bawab, A.A.; Ismail, S.I. Aptamers chemistry: Chemical modifications and conjugation strategies. Molecules 2020, 25, 3. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Cheng, N.; Ruan, X.; Du, D.; Lin, Y. Review—Nanozyme-Based Immunosensors and Immunoassays: Recent Developments and Future Trends. J. Electrochem. Soc. 2020, 167, 037508. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Lu, Y.; Zhou, P.; Lu, L.; Lv, H.; Hai, X. Recent Advances in the Immunoassays Based on Nanozymes. Biosensors 2022, 12, 1119. [Google Scholar] [CrossRef]
- Vashist, S.K.; Luong, J.H.T. (Eds.) Handbook of Immunoassay Technologies Approaches, Performances, and Applications; Academic Press (Elsevier): Cambridge, MA, USA, 2018. [Google Scholar]
- Davydova, A.; Vorobyeva, M. Aptamer-Based Biosensors for the Colorimetric Detection of Blood Biomarkers: Paving the Way to Clinical Laboratory Testing. Biomedicines 2022, 10, 1606. [Google Scholar] [CrossRef]
- Aslan, Y.; Atabay, M.; Chowdhury, H.K.; Göktürk, I.; Saylan, Y.; Inci, F. Aptamer-Based Point-of-Care Devices: Emerging Technologies and Integration of Computational Methods. Biosensors 2023, 13, 569. [Google Scholar] [CrossRef]
- Chang, D.; Zakaria, S.; Deng, M.; Allen, N.; Tram, K.; Li, Y. Integrating deoxyribozymes into colorimetric sensing platforms. Sensors 2016, 16, 2061. [Google Scholar] [CrossRef]
- Cozma, I.; McConnell, E.M.; Brennan, J.D.; Li, Y. DNAzymes as key components of biosensing systems for the detection of biological targets. Biosens. Bioelectron. 2021, 177, 112972. [Google Scholar] [CrossRef]
- Liu, B.; Wang, Y.; Chen, Y.; Guo, L.; Wei, G. Biomimetic two-dimensional nanozymes: Synthesis, hybridization, functional tailoring, and biosensor applications. J. Mater. Chem. B 2020, 8, 10065–10086. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Jana, D.; Zhao, Y. Metal-Organic Framework Derived Nanozymes in Biomedicine. Acc. Chem. Res. 2020, 53, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Homogeneous assays using aptamers. Analyst 2011, 136, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Krainer, F.W.; Glieder, A. An updated view on horseradish peroxidases: Recombinant production and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 1611–1625. [Google Scholar] [CrossRef] [PubMed]
- Walter, J.-G.; Stahl, F. Aptazymes: Expanding the specificity of natural catalytic nucleic acids by application of in vitro selected oligonucleotides. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2020; pp. 107–120. [Google Scholar]
- Yang, C.; Lates, V.; Prieto-Simón, B.; Marty, J.L.; Yang, X. Aptamer-DNAzyme hairpins for biosensing of Ochratoxin A. Biosens. Bioelectron. 2012, 32, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Teller, C.; Shimron, S.; Willner, I. Aptamer-DNAzyme hairpins for amplified biosensing. Anal. Chem. 2009, 81, 9114–9119. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Shlyahovsky, B.; Elbaz, J.; Willner, I. Amplified analysis of low-molecular-weight substrates or proteins by the self-assembly of DNAzyme-aptamer conjugates. J. Am. Chem. Soc. 2007, 129, 5804–5805. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, J.; Chen, R.; Chen, L.; Deng, L. Highly effective colorimetric and visual detection of ATP by a DNAzyme-aptamer sensor. Chem. Biodivers. 2011, 8, 311–316. [Google Scholar] [CrossRef]
- Elbaz, J.; Moshe, M.; Shlyahovsky, B.; Willner, I. Cooperative multicomponent self-assembly of nucleic acid structures for the activation of DNAzyme cascades: A paradigm for DNA sensors and aptasensors. Chem.-Eur. J. 2009, 15, 3411–3418. [Google Scholar] [CrossRef]
- Freeman, R.; Girsh, J.; Fang-Ju Jou, A.; Ho, J.A.A.; Hug, T.; Dernedde, J.; Willner, I. Optical aptasensors for the analysis of the vascular endothelial growth factor (VEGF). Anal. Chem. 2012, 84, 6192–6198. [Google Scholar] [CrossRef]
- Liu, H.; Wang, Y.S.; Wang, J.C.; Xue, J.H.; Zhou, B.; Zhao, H.; Liu, S.D.; Tang, X.; Chen, S.H.; Li, M.H.; et al. A colorimetric aptasensor for the highly sensitive detection of 8-hydroxy-2′-deoxyguanosine based on G-quadruplex-hemin DNAzyme. Anal. Biochem. 2014, 458, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Yang, Z.; Du, P.; Xu, Z.; Wang, Z.; Li, X. G-quadruplex-hemin DNAzyme molecular beacon probe for the detection of methamphetamine. RSC Adv. 2016, 6, 62754–62759. [Google Scholar] [CrossRef]
- Shahbazi, N.; Hosseinkhani, S.; Ranjbar, B. A facile and rapid aptasensor based on split peroxidase DNAzyme for visual detection of carcinoembryonic antigen in saliva. Sens. Actuators B Chem. 2017, 253, 794–803. [Google Scholar] [CrossRef]
- Khang, H.; Cho, K.; Chong, S.; Lee, J.H. All-in-one dual-aptasensor capable of rapidly quantifying carcinoembryonic antigen. Biosens. Bioelectron. 2017, 90, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Peng, L.; Li, M.; Ma, J.; Qi, S.; Chen, H.; Zhou, L.; Chen, X. A label-free colorimetric biosensor for sensitive detection of vascular endothelial growth factor-165. Analyst 2017, 142, 2419–2425. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Guo, J.; Maina, S.W.; Yang, Y.; Hu, Y.; Li, X.; Qiu, J.; Xin, Z. An aptasensor for staphylococcus aureus based on nicking enzyme amplification reaction and rolling circle amplification. Anal. Biochem. 2018, 549, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Shahsavar, K.; Hosseini, M.; Shokri, E.; Ganjali, M.R.; Ju, H. A sensitive colorimetric aptasensor with a triple-helix molecular switch based on peroxidase-like activity of a DNAzyme for ATP detection. Anal. Methods 2017, 9, 4726–4731. [Google Scholar] [CrossRef]
- Kang, B.; Park, S.V.; Soh, H.T.; Oh, S.S. A Dual-Sensing DNA Nanostructure with an Ultrabroad Detection Range. ACS Sens. 2019, 4, 2802–2808. [Google Scholar] [CrossRef]
- Luo, Y.; Yu, H.; Alkhamis, O.; Liu, Y.; Lou, X.; Yu, B.; Xiao, Y. Label-Free, Visual Detection of Small Molecules Using Highly Target-Responsive Multimodule Split Aptamer Constructs. Anal. Chem. 2019, 91, 7199–7207. [Google Scholar] [CrossRef]
- Zhou, Y.; Xu, H.; Wang, H.; Ye, B.C. Detection of breast cancer-derived exosomes using the horseradish peroxidase-mimicking DNAzyme as an aptasensor. Analyst 2020, 145, 107–114. [Google Scholar] [CrossRef]
- Yang, L.; Ye, X.; Li, X.; Huang, Z.; Chen, F.; Yang, W.; Wang, Z. Colorimetric aptasensor for sensitive detection of quinclorac based on exonuclease III-assisted cyclic release of phosphorodiamidate morpholino oligomer mimic enzyme strategy. Anal. Chim. Acta 2022, 1207, 339815. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, N.; Tan, C.; Fang, W.; Tan, Y.; Jiang, Y. A sensitive colorimetric aptasensor based on trivalent peroxidase-mimic DNAzyme and magnetic nanoparticles. Anal. Chim. Acta 2018, 1018, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Zhao, J.; Huang, Y.; Cheng, X.; Wang, S.; Han, Y.; Xiao, Y.; Lou, X. Universal Design of Structure-Switching Aptamers with Signal Reporting Functionality. Anal. Chem. 2019, 91, 14514–14521. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lin, Y.; Wang, X.; Xu, L.; Wang, Z.; Fu, F.F. Exonuclease-assisted multicolor aptasensor for visual detection of ochratoxin A based on G-quadruplex-hemin DNAzyme-mediated etching of gold nanorod. Microchim. Acta 2018, 185, 259. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, A.; Siao, Y.D.; Yang, S.T.; Hsieh, P.V.; Fukushima, H.; Chang, Y.; Ruaan, R.C.; Chen, W.Y. Preparation of a DNA aptamer-Pt complex and its use in the colorimetric sensing of thrombin and anti-thrombin antibodies. Anal. Chem. 2008, 80, 6580–6586. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Zheng, A.X.; Liu, B.; Zhang, X.L.; He, Y.; Yang, H.H.; Chen, G. One-pot synthesized DNA-templated Ag/Pt bimetallic nanoclusters as peroxidase mimics for colorimetric detection of thrombin. Chem. Commun. 2014, 50, 13103–13106. [Google Scholar] [CrossRef] [PubMed]
- Kumar Sharma, T.; Ramanathan, R.; Weerathunge, P.; Mohammadtaheri, M.; Kumar Daima, H.; Shukla, R.; Bansal, V. Aptamer-mediated “turn-off/turn-on” nanozyme activity of gold nanoparticles for kanamycin detection. Chem. Commun. 2014, 50, 15856–15859. [Google Scholar] [CrossRef] [PubMed]
- Weerathunge, P.; Ramanathan, R.; Shukla, R.; Sharma, T.K.; Bansal, V. Aptamer-controlled reversible inhibition of gold nanozyme activity for pesticide sensing. Anal. Chem. 2014, 86, 11937–11941. [Google Scholar] [CrossRef]
- Yan, J.; Huang, Y.; Zhang, C.; Fang, Z.; Bai, W.; Yan, M.; Zhu, C.; Chen, A. Aptamer based photometric assay for the antibiotic sulfadimethoxine based on the inhibition and reactivation of the peroxidase-like activity of gold nanoparticles. Microchim. Acta 2017, 184, 59–63. [Google Scholar] [CrossRef]
- Weerathunge, P.; Ramanathan, R.; Torok, V.A.; Hodgson, K.; Xu, Y.; Goodacre, R.; Behera, B.K.; Bansal, V. Ultrasensitive Colorimetric Detection of Murine Norovirus Using NanoZyme Aptasensor. Anal. Chem. 2019, 91, 3270–3276. [Google Scholar] [CrossRef]
- Das, R.; Dhiman, A.; Kapil, A.; Bansal, V.; Sharma, T.K. Aptamer-mediated colorimetric and electrochemical detection of Pseudomonas aeruginosa utilizing peroxidase-mimic activity of gold NanoZyme. Anal. Bioanal. Chem. 2019, 411, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Zhao, R.; Feng, S.; Xie, Y. Colorimetric zearalenone assay based on the use of an aptamer and of gold nanoparticles with peroxidase-like activity. Microchim. Acta 2018, 185, 535. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Wu, Y.; Tao, H.; Chen, H.; Yang, W.; Qiu, S. Colorimetric detection of streptomycin in milk based on peroxidase-mimicking catalytic activity of gold nanoparticles. RSC Adv. 2017, 7, 38471–38478. [Google Scholar] [CrossRef]
- Slyusarenko, M.; Shalaev, S.; Valitova, A.; Zabegina, L.; Nikiforova, N.; Nazarova, I.; Rudakovskaya, P.; Vorobiev, M.; Lezov, A.; Filatova, L.; et al. AuNP Aptasensor for Hodgkin Lymphoma Monitoring. Biosensors 2022, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Hong, C.Y.; Lin, Z.Z.; Chen, X.M.; Huang, Z.Y. Detection of Malachite Green using a colorimetric aptasensor based on the inhibition of the peroxidase-like activity of gold nanoparticles by cetyltrimethylammonium ions. Microchim. Acta 2019, 186, 322. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Jo, H.; Her, J.; Youn, H.; Park, J.; Jo, J.; Lee, J.; Chang, C.L.; Ban, C. A Rapid Colorimetric Sensor for Soluble Interleukin-2 Receptor α, Based on Aptamer-Adsorbed AuNP. ChemBioChem 2019, 20, 2236–2240. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Tang, M.Q.; Chen, J.; Zhu, Y.H.; Lei, C.B.; He, H.W.; Xu, X.H. A sandwich ELISA-like detection of C-reactive protein in blood by citicoline-bovine serum albumin conjugate and aptamer-functionalized gold nanoparticles nanozyme. Talanta 2020, 217, 121070. [Google Scholar] [CrossRef]
- Torabi, R.; Ghourchian, H. Ultrasensitive nano-aptasensor for monitoring retinol binding protein 4 as a biomarker for diabetes prognosis at early stages. Sci. Rep. 2020, 10, 594. [Google Scholar] [CrossRef]
- Weerathunge, P.; Behera, B.K.; Zihara, S.; Singh, M.; Prasad, S.N.; Hashmi, S.; Mariathomas, P.R.D.; Bansal, V.; Ramanathan, R. Dynamic interactions between peroxidase-mimic silver NanoZymes and chlorpyrifos-specific aptamers enable highly-specific pesticide sensing in river water. Anal. Chim. Acta 2019, 1083, 157–165. [Google Scholar] [CrossRef]
- Park, J.Y.; Jeong, H.Y.; Kim, M.I.; Park, T.J. Colorimetric Detection System for Salmonella typhimurium Based on Peroxidase-Like Activity of Magnetic Nanoparticles with DNA Aptamers. J. Nanomater. 2015, 2015, 527126. [Google Scholar] [CrossRef]
- Dehghani, Z.; Hosseini, M.; Mohammadnejad, J.; Bakhshi, B.; Rezayan, A.H. Colorimetric aptasensor for Campylobacter jejuni cells by exploiting the peroxidase like activity of Au@Pd nanoparticles. Microchim. Acta 2018, 185, 448. [Google Scholar] [CrossRef] [PubMed]
- Tan, F.; Wang, Z.; Yang, Y.; Xie, X.; Hua, X.; Yang, X.; Huang, H. Facile preparation of peroxidase-like core-shell nanorods and application as platform for colorimetric determination of glucose, insulin and glucose/insulin ratio. Talanta 2019, 204, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Gao, R.; Jia, L. Enhancement of the peroxidase-like activity of aptamers modified gold nanoclusters by bacteria for colorimetric detection of Salmonella typhimurium. Talanta 2021, 221, 121476. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhu, Y.; Binyam, A.; Liu, M.; Wu, Y.; Li, F. Discovering the enzyme mimetic activity of metal-organic framework (MOF) for label-free and colorimetric sensing of biomolecules. Biosens. Bioelectron. 2016, 86, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.K.; Omer, K.M. Ultrasensitive aptamer-functionalized Cu-MOF fluorescent nanozyme as an optical biosensor for detection of C-reactive protein. Anal. Biochem. 2022, 658, 114928. [Google Scholar] [CrossRef] [PubMed]
- Ali, G.K.; Omer, K.M. Nanozyme and Stimulated Fluorescent Cu-Based Metal-Organic Frameworks (Cu-MOFs) Functionalized with Engineered Aptamers as a Molecular Recognition Element for Thrombin Detection in the Plasma of COVID-19 Patients. ACS Omega 2022, 7, 36804–36810. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; He, Z.; Wang, H.; Feng, X.; Han, P. Magnetically controlled colorimetric aptasensor for chlorpyrifos based on copper-based metal-organic framework nanoparticles with peroxidase mimetic property. Microchim. Acta 2020, 187, 524. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, D.Y.; Li, Z.Y.; Hu, R.; Yang, Y.H.; Yang, T. A visual peroxidase mimicking aptasensor based on Pt nanoparticles-loaded on iron metal organic gel for fumonisin B1 analysis in corn meal. Biosens. Bioelectron. 2022, 209, 114241. [Google Scholar] [CrossRef]
- Chai, H.; Yu, K.; Zhao, Y.; Zhang, Z.; Wang, S.; Huang, C.; Zhang, X.; Zhang, G. MOF-On-MOF Dual Enzyme-Mimic Nanozyme with Enhanced Cascade Catalysis for Colorimetric/Chemiluminescent Dual-Mode Aptasensing. Anal. Chem. 2023, 95, 10785–10794. [Google Scholar] [CrossRef]
- Wang, Y.M.; Liu, J.W.; Adkins, G.B.; Shen, W.; Trinh, M.P.; Duan, L.Y.; Jiang, J.H.; Zhong, W. Enhancement of the Intrinsic Peroxidase-Like Activity of Graphitic Carbon Nitride Nanosheets by ssDNAs and Its Application for Detection of Exosomes. Anal. Chem. 2017, 89, 12327–12333. [Google Scholar] [CrossRef]
- Xia, Y.; Liu, M.; Wang, L.; Yan, A.; He, W.; Chen, M.; Lan, J.; Xu, J.; Guan, L.; Chen, J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017, 92, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Duan, N.; Qiu, Y.; Li, J.; Wang, Z. Colorimetric aptasensor for the detection of Salmonella enterica serovar typhimurium using ZnFe2O4-reduced graphene oxide nanostructures as an effective peroxidase mimetics. Int. J. Food Microbiol. 2017, 261, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.; Zhang, J.; Zhang, L.; Wang, L.; Chen, H. Aptamer biosensor for Salmonella typhimurium detection based on luminescence energy transfer from Mn2 +-doped NaYF4:Yb, Tm upconverting nanoparticles to gold nanorods. Spectrochim. Acta–Part A Mol. Biomol. Spectrosc. 2017, 171, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Bagheryan, Z.; Raoof, J.B.; Golabi, M.; Turner, A.P.F.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ren, W.; Luo, H.Q.; Li, N.B. Label-free cascade amplification strategy for sensitive visual detection of thrombin based on target-triggered hybridization chain reaction-mediated in situ generation of DNAzymes and Pt nanochains. Biosens. Bioelectron. 2016, 80, 463–470. [Google Scholar] [CrossRef]
- Luan, Q.; Xiong, X.; Gan, N.; Cao, Y.; Li, T.; Wu, D.; Dong, Y.; Hu, F. A multiple signal amplified colorimetric aptasensor for antibiotics measurement using DNAzyme labeled Fe-MIL-88-Pt as novel peroxidase mimic tags and CSDP target-triggered cycles. Talanta 2018, 187, 27–34. [Google Scholar] [CrossRef]
- Rafati, A.; Zarrabi, A.; Abediankenari, S.; Aarabi, M.; Gill, P. Sensitive colorimetric assay using insulin g-quadruplex aptamer arrays on DNA nanotubes coupled with magnetic nanoparticles. R. Soc. Open Sci. 2018, 5, 171835. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davydova, A.S.; Vorobyeva, M.A. Aptasensors Based on Non-Enzymatic Peroxidase Mimics: Current Progress and Challenges. Biosensors 2024, 14, 1. https://doi.org/10.3390/bios14010001
Davydova AS, Vorobyeva MA. Aptasensors Based on Non-Enzymatic Peroxidase Mimics: Current Progress and Challenges. Biosensors. 2024; 14(1):1. https://doi.org/10.3390/bios14010001
Chicago/Turabian StyleDavydova, Anna S., and Mariya A. Vorobyeva. 2024. "Aptasensors Based on Non-Enzymatic Peroxidase Mimics: Current Progress and Challenges" Biosensors 14, no. 1: 1. https://doi.org/10.3390/bios14010001
APA StyleDavydova, A. S., & Vorobyeva, M. A. (2024). Aptasensors Based on Non-Enzymatic Peroxidase Mimics: Current Progress and Challenges. Biosensors, 14(1), 1. https://doi.org/10.3390/bios14010001





