Abstract
Functional nucleic acid (FNA) probes have been widely used in environmental monitoring, food analysis, clinical diagnosis, and biological imaging because of their easy synthesis, functional modification, flexible design, and stable properties. However, most FNA probes are designed based on one-photon (OP) in the ultraviolet or visible regions, and the effectiveness of these OP-based FNA probes may be hindered by certain factors, such as their potential for photodamage and limited light tissue penetration. Two-photon (TP) is characterized by the nonlinear absorption of two relatively low-energy photons of near-infrared (NIR) light with the resulting emission of high-energy ultraviolet or visible light. TP-based FNA probes have excellent properties, including lower tissue self-absorption and autofluorescence, reduced photodamage and photobleaching, and higher spatial resolution, making them more advantageous than the conventional OP-based FNA probes in biomedical sensing. In this review, we summarize the recent advances of TP-excited and -activated FNA probes and detail their applications in biomolecular detection. In addition, we also share our views on the highlights and limitations of TP-based FNA probes. The ultimate goal is to provide design approaches for the development of high-performance TP-based FNA probes, thereby promoting their biological applications.
Keywords:
DNA nanotechnology; two-photon; bioimaging; fluorescent probe; biomolecule; light-activated; RNA 1. Introduction
Nucleic acids have gained prominence as a fundamental framework of functionally diverse and versatile molecular probes due to their unique molecular recognition capabilities, programmable base sequences, convenient synthesis and modification, and stimuli-triggered responsiveness [1,2,3,4]. Functional nucleic acids (FNA) mainly include aptamers (which selectively bind with numerous biological target molecules) [5,6,7,8], DNAzymes (which catalyze the cleavage of various substrates like enzymes) [9,10,11], and chemically modified nucleic acids (which possess all kinds of specific functions and can response to stimuli factors) [12,13,14]. They can be obtained by the systematic evolution of ligands by exponential enrichment (SELEX) or chemical reactions and exhibit much specific recognition capability for a variety of unique biological applications [15,16,17,18,19].
Based on the many advantages of FNA molecules, extensive efforts have been made in the development of FNA probes used in detecting a diverse array of analytes [20], such as RNAs [21,22,23,24,25], proteins [26,27,28], small molecules [29,30,31], and metal ions [32,33]. These functional nucleic acid probes mainly use the specific recognition between the base sequence and the target molecule and combine the techniques of in situ hybridization, fluorescence resonance energy transfer (FRET), and signal amplification strategy to investigate dynamic changes and interaction processes at the molecular level; they have shown significant superiority in unraveling the fundamental mechanisms of pathophysiological processes and facilitating disease diagnosis [34,35,36,37]. As a result, the ongoing advancement and utilization of FNA probes hold great promise for advancing our understanding of biology and improving diagnostic approaches [38,39,40].
Typically, most FNA probes were primarily designed based on one-photon (OP) that is required to be excited/activated by a short wavelength light source in the ultraviolet or visible (UV/Vis) regions [41,42]. OP-based FNA probes possess advantages such as high sensitivity and selectivity, programmability, ease of synthesis and modification, and real-time monitoring, which have led to their widespread application in biomedical research, nanotechnology, and materials science [43,44,45,46,47]. However, the inevitable disadvantages of OP light sources, including shallow tissue penetration, photobleaching, phototoxicity, and autofluorescence from biological systems, greatly restrict the development of OP-based FNA probes in various organelles and deep tissues.
In comparison with OP-based FNA probes that employ a short wavelength light source, FNA probes based on two-photon (TP) can be excited/activated by a long wavelength femtosecond laser [48,49,50]. The theory of TP absorption was initially introduced by Maria Goeppert-Mayer in the 1930s [51] and, with the advent of femtosecond laser technology, researchers began investigating this instrumentation in the 2000s. TP for biosensing use near-infrared (NIR) photons as the light source, which can achieve accurate regulation of the detection process of FNA probes. NIR light causes less damage to cells, reduces autofluorescence from biological tissue, allows for deeper tissue penetration [52,53,54,55], and gives out negligible autofluorescence in biological matrices [56,57]. The above advantages make TP-based FNA probes more promising for various biological applications when compared to conventional OP-based FNA probes [58,59].
This review (Figure 1) provides a comprehensive overview of the designs and applications of TP-based FNA probes used as biosensors. Initially, we emphasize the utilization of TP-excited FNA probes in the detection of various analytes, including nucleic acids, enzymes, biothiols, ATP, and metal ions. Subsequently, we delve into TP-activated FNA probes and their specific applications in RNA and ATP detection. Finally, we discuss the existing challenges and future outlooks regarding TP-based FNA probes used in the field of biomedical research.
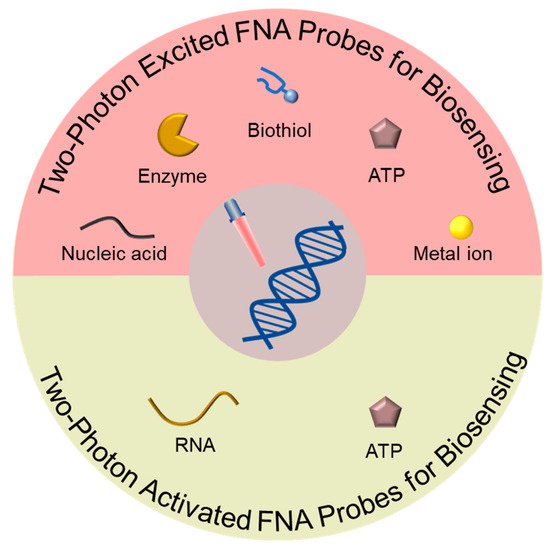
Figure 1.
Schematic overview of TP-based FNA probes used for biosensing.
2. Two-Photon Excited FNA Probes for Biosensing
FNA probes based on one-photon excitation (OPE) suffer from limitations such as limited tissue penetration depth, photodamage, and photobleaching. To overcome these challenges, two-photon excitation (TPE) fluorescence imaging has become an optimal approach for enhancing sensing capabilities in living cells and tissues. In contrast to the prevailing construction strategy of OPE-based FNA probes, TPE-based FNA probes constructed by introducing TP dye as the signal reporter can effectively eliminate the problems related to shallow imaging depth and background fluorescence interference in biological matrices while using an NIR excitation wavelength that provides increased penetration depth and reduced photodamage. Therefore, the applications of TPE-based FNA probes are extensive in the field of biosensing, allowing for the detection of biomolecules such as nucleic acids, enzymes, biothiols, ATP, and metal ions. By investigating the dynamics and interactions between molecules, the underlying mechanisms of pathophysiological processes can be better attained, thereby advancing disease diagnosis and therapeutic monitoring.
2.1. Two-Photon Excited FNA Probes for Nucleic Acid Analysis
MicroRNAs (miRNAs) are short noncoding nucleic acid fragments, typically comprising 21–23 nucleotides, that play crucial roles in gene expression regulation and cellular functions [60]. MiRNAs can bind to target mRNA, leading to its inhibition or degradation, thereby regulating the expression levels of genes [61]. This regulatory mechanism has important functions in various biological processes, including maintaining cellular homeostasis, developmental regulation, cell proliferation, and apoptosis [62,63,64]. Due to the specific expression patterns of miRNAs in different tissues and disease states, assaying miRNA levels in body fluids can contribute to early disease diagnosis, prognostic assessment, and monitoring treatment response [65,66]. Consequently, monitoring the dynamic expression of miRNAs in living cells holds significant value in gaining insights into miRNA-associated cellular processes and facilitating disease diagnosis.
In general, TPE-based FNA probes were designed by incorporating TP dye molecules that could be excited by a TP laser to produce fluorescence signals. As shown in Figure 2a, Wang et al. constructed a spherical nucleic acid (SNA)-based fluorescent probe by incorporating TPE and DNAzyme for the highly selective and sensitive analysis of target miRNAs in living cells [67]. Their approach involved functionalizing gold nanoparticles (AuNPs) with substrate strands capable of hybridizing into two split DNAzyme fragments. In the formation of the TPE-SNA nanoprobe, the substrate/split-DNAzyme duplexes were incorporated with multiple TP dye molecules, leading to a state where fluorescence emission was turned off. When target miRNA was present, the conformational rearrangement of the split DNAzyme would be induced and their catalytic ability could be activated, thus leading to the cleavage of the substrate strands and the subsequent release of the TP dye molecules. Simultaneously, the liberated target miRNA could spontaneously form a hybrid with another split DNAzyme fragment, initiating additional cleavage reactions. This sequential hybridization-activated cleavage process gradually dissociated the TP dye molecules from the surface of the AuNPs. As a result, a cascade of TPE fluorescence emission occurred, amplifying the fluorescence signals for miRNA detection. In addition, Yang et al. developed a TPE-based nanoprobe for imaging intracellular miRNA by employing metal-organic frameworks (MOFs) as the nanocarrier along with hairpins to amplify TP fluorescence signals (Figure 2b) [68]. Hairpin 2 (H2) was engineered with TP fluorophore and BHQ1 quencher modifications, where the proximity of BHQ1 in the hairpin structure led to the fluorescence quenching of TP fluorophore. After being delivered into cells by MOFs, hairpin 1 (H1) and H2 could be triggered by intracellular miRNA, leading to the formation of an H1–H2 duplex through a catalytic hairpin assembly (CHA) reaction. This contributed to the separation of the TP fluorophore from the BHQ1 quencher, resulting in the activation of TP fluorescence and the release of the miRNA. The released miRNA could further trigger the formation of additional H1–H2 duplexes, enabling the amplified target miRNA detection. The incorporation of TP fluorophore facilitated the effective imaging of miRNA in both living tumor cells and deep tumor tissues, achieving a notable penetration depth of 160 µm. This TPE-based nanoprobe demonstrated promising capabilities for sensitive and specific miRNA detection in complex biological environments.
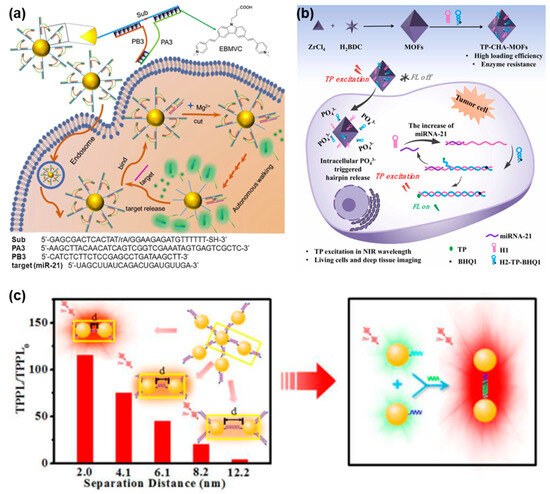
Figure 2.
FNA probes based on TPE for nucleic acid analysis. (a) Schematic illustration of a TPE sensing platform based on spherical DNAzyme for intracellular miRNA assay. Adapted with permission from [67]. (b) Schematic illustration of the synthesized TP-CHA-MOFs for miRNA imaging in tumor cells. Adapted with permission from [68]. (c) Schematic illustration of DNA-induced coupling of AuNPs and TPPL enhancement for DNA sequence detection. Adapted with permission from [69].
To accurately measure the length of DNA molecules, Yuan et al. conducted a study where they prepared assemblies of DNA-tuned AuNPs with controlled separation distances to investigate the impact of plasmon coupling strength and particle size on two-photon photoluminescence (TPPL) enhancement (Figure 2c) [69]. They observed that decreasing the separation distance between the DNA-coupled nano-assemblies resulted in a significant increase in TPPL intensity. Based on this finding, they developed a TP sensing scheme for detecting DNA sequences by utilizing the DNA-induced coupling of AuNPs and TPPL enhancement. The developed method exhibited exceptional sensitivity, achieving a limit of detection (LOD) as low as 2.9 pM, enabling the detection of DNA sequences at very low concentrations. Additionally, it demonstrated excellent selectivity against single-stranded DNA (ssDNA) with mismatched bases, allowing for the easy differentiation of even a single mismatch at room temperature. Moreover, the researchers highlighted the potential extension of this method to DNA detection in living cells or in vivo, leveraging the unique advantages of TPE. Therefore, the TPPL-based method shows promise for sensitive and selective DNA detection in biological systems.
2.2. Two-Photon Excited FNA Probes for Enzyme Analysis
Enzymes are specific and efficient biocatalysts, most of which are proteins produced by living cells that perform essential roles in living systems by accelerating biochemical reactions through their highly efficient catalytic activities [70,71,72]. Due to their specificity and efficiency, enzymes are essential for maintaining the proper functioning of biological processes. However, abnormal enzyme activity could have detrimental effects and contribute to various diseases [73,74,75,76]. By assaying the activity of specific enzymes or their levels in tissues, cells, or bodily fluids, we can assess the status and functionality of specific biological processes [77]. For example, measuring the activity of liver enzymes provides information about liver function, liver diseases, and drug metabolism. Enzyme detection can also be used to monitor changes in processes such as cell proliferation, apoptosis, signal transduction pathways, and gene expression [78,79]. Hence, there is a need to create and advance probes capable of directly detecting enzyme activity in vivo, particularly with a focus on non-invasive and real-time methods.
According to the different hydrolysis characteristics of enzymes, different nucleic acid sequences that responded to the hydrolysis of enzymes were designed on the FNA probes. As shown in Figure 3a, Ge et al. developed a FRET nanoprobe that was excited by a TP laser and successfully applied in imaging caspase-3 by assembling TP dye-labeled peptides on the surface of AuNPs [80]. This study is the first successful application of an AuNP/peptide-TP biosensor for evaluating caspase-3 activity in both live cells and liver tissues that have undergone ischemic reperfusion surgery. In a separate study, Wang et al. reported a TPE-based nanoprobe used for assessing DNase activity both in vitro and ex vivo (Figure 3b) [81]. This nanoprobe primarily consisted of a AuNP core acting as the cellular transporter and fluorescence quenching motif, along with TP dyes incorporated into the double-stranded DNA (dsDNA) to act as the reporter motif. When DNases were present, the dsDNA immobilized on the surface of AuNPs underwent hydrolysis, releasing a large amount of TP dyes. Consequently, the TP dyes exhibited fluorescence recovery, allowing for the measurement of DNase activity. This TPE-based nanoprobe has been successfully applied in the monitoring of DNase activity in living cells and in vivo. Furthermore, Wang et al. also reported a TPE-based nanoprobe for detecting RNase H activity in living cells and ex vivo tissues, as depicted in Figure 3c [82]. This nanoprobe involved binding TP dyes to spherical nucleic acids (SNA) with a DNA/RNA duplex corona and a AuNP core. The developed probe offers a convenient means for detecting RNase H activity, which is expected to pave the way for further research in the field of RNase H.
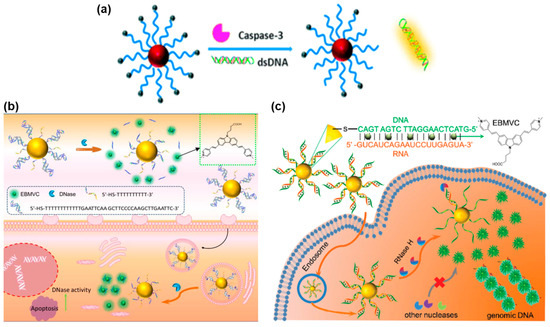
Figure 3.
FNA probes based on TPE for enzyme analysis. (a) Schematic illustration of the AuNPs/peptide-TP dye nanoprobe for assaying caspase-3. Adapted with permission from [80]. (b) Schematic illustration of the TPE-based nanoprobe for imaging DNase. Adapted with permission from [81]. (c) Schematic illustration of the SNA-based TPE sensor for detecting RNase H. Adapted with permission from [82].
2.3. Two-Photon Excited FNA Probes for Biothiol Analysis
Biothiols, which encompass thiol-containing amino acids and peptides, serve as vital structural or functional constituents in many proteins and peptides and fulfill various essential functions in biological systems [83,84]. GSH, known as the predominant intracellular non-protein thiol in the cytoplasm, acts as a crucial reducing agent involved in various cellular functions, such as maintaining intracellular redox activity, facilitating xenobiotic metabolism, enabling intracellular signal transduction, and regulating gene expression [85,86,87]. Measuring the levels of biothiols provides valuable information to assess oxidative-reductive status, antioxidant capacity, and the risks and progression of certain diseases, such as cardiovascular diseases, cancer, and neurodegenerative diseases [88]. Given their significance in physiological activities, monitoring biothiol molecules has garnered considerable interest [89,90].
As shown in Figure 4a, Tang et al. presented a TPE-based biosensor for GSH detection in living cells and tissues [91]. The sensor was prepared by forming DNA-templated silver nanoparticles (AgNPs) and binding TP dye to the DNA. The resulting conjugate, AgNPs/DNA/TP dye, exhibited favorable TP-sensitized fluorescence characteristics, excellent cell permeability and biocompatibility. Importantly, the conjugation of DNA/TP dye with DNA-templated AgNPs was achieved through a simple process that did not require surface functionalization or covalent labeling. By conducting assays in vitro and in vivo, the AgNPs/DNA/TP dye nanoprobe demonstrated the capability for quantitative GSH detection, even within complex biological environments.
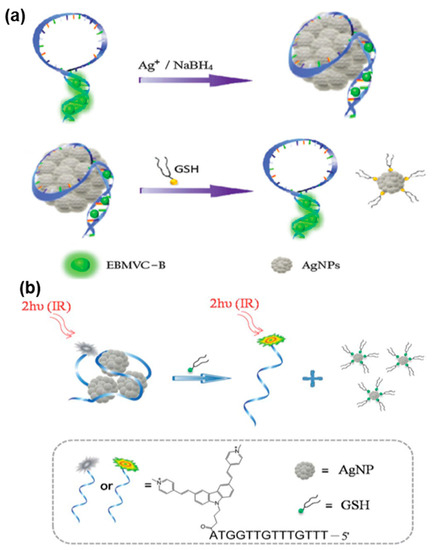
Figure 4.
FNA probes based on TPE for biothiols analysis. (a) Schematic illustration of the AgNPs/DNA/TP dye nanoprobe for GSH detection. Adapted with permission from [91]. (b) Schematic illustration of the AgNPs/DNA-TP dye conjugate used for biothiols detection. Adapted with permission from [92].
Similarly, Liu et al. fabricated AgNPs/DNA-TP dye conjugates as a TPE-based nanoprobe for imaging biothiols in living cells (Figure 4b) [92]. The DNA-templated AgNPs served as biocompatible nanoplatforms for delivering DNA into living cells, while also effectively quenching fluorescence. When biothiols were present, such as GSH, the robust thiol–silver interaction led to the detachment of TP dye-labeled ssDNA from the surface of AgNPs. This detachment subsequently induced the fluorescence emission of the TP dye, allowing for the detection of biothiols.
2.4. Two-Photon Excited FNA Probes for ATP Analysis
Adenosine 5′-triphosphate (ATP), an essential cellular component present in all living organisms, serves as an indispensable intracellular energy source and plays a role as a signaling molecule associated with energy status [93,94,95]. It fulfills a wide range of functions in numerous biological processes, including the regulation of ion channels, modulation of signaling cascades, and facilitation of protein transport activities, as well as having involvement in DNA replication and transcription [96,97]. ATP homeostasis, which refers to the maintenance of constant cellular ATP concentrations, is critical for cell, tissue, and organ system functionality [98,99]. Deficiency in the ATP level is considered to be related to many diseases such as malignant tumors [100,101]; by determining the ATP content in cells, tissues, or bodily fluids, it becomes possible to assess the state of cellular energy metabolism and gain insights into cell vitality and health [102]. Consequently, monitoring the changes in ATP concentration both in vitro and in vivo holds significant research importance for understanding real-time biological processes [103,104].
As shown in Figure 5, Yi et al. designed a TPE-based fluorescent nanoprobe, consisting of an ATP aptamer and TP dye, for molecular probing in various biological environments [105]. This approach harnessed the impressive quenching capability of graphene oxide (GO) towards nearby TP dyes. Notably, the binding affinity between ssDNA and GO surpassed that of the aptamer-target complex, facilitating the successful detection of ATP. The results revealed that the GO/aptamer-TP dye nanoprobe served as a sensitive, reliable, and selective sensor for quantitatively detecting ATP in complex biological environments. After efficient delivery into living cells or tissues, this nanoprobe acted as an in vivo sensor, enabling the specific and high-contrast TP imaging of ATP. This innovative design established a methodological foundation for the development of TPE-based fluorescent probes utilizing carbon nanomaterials to determine biologically relevant analytes in vitro or in vivo.
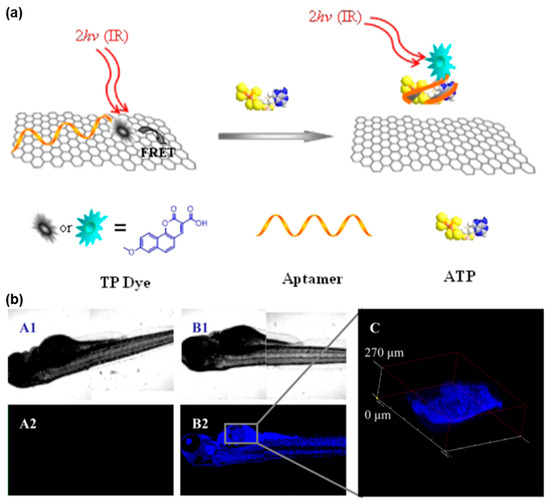
Figure 5.
FNA probes based on TPE for ATP analysis. (a) Schematic illustration of TP GO/aptamer-based nanoprobe conjugate for ATP detection. (b) TP confocal microscopy of zebrafish treated with different probes. Adapted with permission from [105].
2.5. Two-Photon Excited FNA Probes for Metal Ions Analysis
Metal ions are vital components in biological systems, where they play critical roles in numerous biological processes [106,107,108]. Disruptions in metal homeostasis have been shown to be associated with many severe diseases, including various cancers, where irregular concentrations of metal ions are implicated [109]. Both excessive and deficient levels of metal ions can negatively impact their normal functions [110]. By quantifying the content of metal ions in samples, it becomes possible to assess the steady-state and dynamic changes of metal ions in cells [111], providing information about cellular metabolism, toxicity, and disease development [112]. Thus, there is significant interest in monitoring the concentrations of metal ions in living cells and their subcellular locations to gain insights into their roles in diverse human health and disease pathological processes [113,114,115].
As shown in Figure 6, Yang et al. developed an innovative FNA probe for the live cell imaging of Zn2+ by utilizing an RNA-cleaving DNAzyme coupled with TP fluorophores [116]. To facilitate cellular uptake, AuNPs were employed as effective transporters. The modified TP fluorophores exhibited outstanding TP excitation efficiency and photostability. In the absence of Zn2+, both AuNPs and a molecular quencher contributed to the quenching of TP fluorophores. However, the presence of Zn2+ facilitated the specific cleavage of the substrate strand labeled with TP fluorophores by the DNAzyme, resulting in enhanced fluorescence and TP imaging. Notably, the FNA probe demonstrated remarkable selectivity for Zn2+ in comparison to other metal ions commonly existing in biological environments. With the aid of TP fluorophores, the TP probe could be excited by NIR light, enabling deep tissue penetration of up to 160 µm for the imaging of Zn2+. This TP-based analysis strategy holds promise for the detection of various metal ions in biological systems, offering enhanced tissue penetration capabilities and reduced phototoxicity.
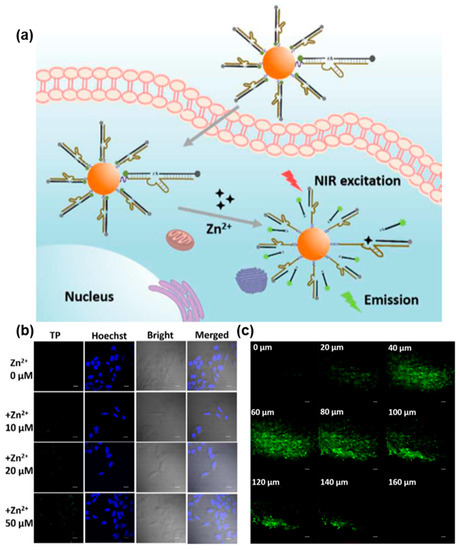
Figure 6.
FNA probes based on TPE for metal ions analysis. (a) Schematic illustration of TPE-based fluorescent probe for selectively imaging intracellular Zn2+. (b) TP confocal microscopy images of intracellular different Zn2+ concentrations. (c) The TP fluorescence images of rat liver tissue with different penetration depths. Adapted with permission from [116].
3. Two-Photon Activated FNA Probes for Biosensing
Light, as an external stimulus, can be instantly and accurately transmitted to specific locations. Additionally, it possesses different wavelengths and is thereby capable of inducing a wide range of chemical reactions [117,118]. The integration of a photocleavable (PC) linker into the DNA strand allows for the formation of light-activated structures, enabling the precise spatiotemporal activation of FNA probes through controlled light irradiation [119,120,121]. Therefore, FNA probes based on two-photon activation (TPA) exhibit exceptional properties, such as heightened temporal and spatial resolution, which enables them to be used as powerful tools for obtaining detailed information about analytes in living cells, especially in single cells [122]. Through single-cell detection using TPA-based FNA probes, we can understand the heterogeneity of different cells within the same sample and reveal the differences between cells in normal physiological or disease states. This method has important applications in biomedical research, clinical diagnostics, and personalized treatments.
3.1. Two-Photon-Activated FNA Probes for RNA Analysis
Traditional methods for RNA detection often necessitate a substantial number of cell samples, making it challenging to precisely measure RNA expression in heterogeneous cells [123,124]. However, the utilization of TPA-based FNA probes enables single-cell RNA detection, facilitating the quantitative analysis of RNA in individual cells. This approach unveils RNA differences between different cells, enhancing our comprehension of its roles in cellular functions, regulatory mechanisms, and disease progression [125,126,127]. The application of this technology extends to diverse fields including cancer research, neuroscience, immunology, and more, guiding personalized treatment and precision medicine and deepening our understanding of miRNA regulatory networks.
As shown in Figure 7a, Lin et al. designed TPA-based nanoflares that enabled high spatiotemporal control for monitoring intracellular mRNA [128]. Such nanoflares comprised light-responsive DNA hairpin probes and AuNPs. In the absence of UV irradiation, the DNA hairpin could remain awakened, unresponsive to the target analyte. Upon UV activation, the hairpin structures were disrupted, revealing sticky domains that acted as toeholds for initiating strand displacement reactions, eventually leading to the release of fluorophores and subsequent fluorescence enhancement. With the ability to adjust light irradiation, temporal control over mRNA detection in living cells was achieved using TPA-based nanoflares. Under TP laser irradiation, these nanoflares could selectively detect mRNA in the single living cell at desired time points. In contrast to the conventional nanoflares, the novel TPA-based nanoflares exhibited heightened detection sensitivity and enabled the detection of biomarkers in a single cell with precise spatiotemporal control.
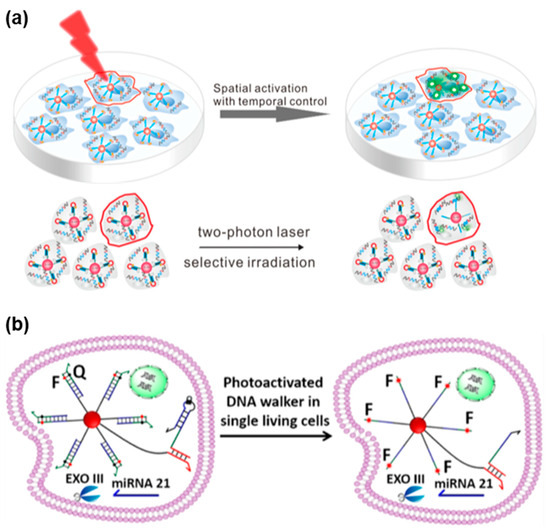
Figure 7.
FNA probes based on TPA for RNA analysis. (a) Schematic illustration of TPA-based nanoflares for mRNA detection in the single living cells via selective TP irradiation at 740 nm. Adapted with permission from [128]. (b) Schematic illustration of TPA-based DNA walker probe for signal amplification miRNA imaging via selective TP irradiation at 740 nm. Adapted with permission from [129].
Chen et al. devised a TPA-based DNA walker probe that enabled light-controllable signal amplification for imaging cancer-related miRNA in a single living cell (Figure 7b) [129]. This developed probe combined a light-activated nucleic acid strand displacement reaction with the traditional exonuclease III (EXO III)-assisted DNA walker, utilizing DNA nanoflares as the foundation. The researchers successfully demonstrated the amplification imaging of light-activated signals for miRNA-21 in individual living HeLa cells via selective TP irradiation (λ = 740 nm) using femtosecond laser-equipped confocal microscopy.
3.2. Two-Photon-Activated FNA Probes for ATP Analysis
As with the previously mentioned RNA assays, traditional ATP detection methods also face the issue of requiring a large amount of sample, which masks the heterogeneity between cells [130]. However, using TPA-based FNA probes for single-cell ATP detection allows for the precise measurement of ATP content in individual cells, revealing variations in ATP levels among different cells. This helps us understand the functional features of cells, metabolic states, and differences between cell types [131]. Through single-cell detection, we can gain insight into cellular energy metabolism, including ATP synthesis pathways, energy production, and consumption balance, among others. This contributes to our understanding of cellular energy regulation mechanisms and the changes in energy demand during different physiological or diseased states [132]. In a word, single-cell ATP detection holds great significance in elucidating cellular energy metabolism, uncovering cell heterogeneity, investigating disease mechanisms, and advancing precision medicine.
As shown in Figure 8, Duan et al. introduced a light-activated FNA probe constructed with a PC linker for ATP detection in a single living cell [133]. Two distinct methods for spatiotemporal activation of the probe were discussed. The first method involved the use of a micrometer-sized optical fiber to selectively direct UV light (λ = 365 nm) for activating the light-activated FNA probe in living cells. Alternatively, a TP laser confocal scanning microscope was employed as the second method to selectively activate the probe via TP irradiation (λ = 740 nm) in single living cells. The aptamer sequence integrated into the light-activated FNA probe selectively interacted with ATP, leading to the generation of the indicated signal. With TP irradiation, the developed light-activated biosensor enabled high spatiotemporal resolution for ATP detection in single living cells at a desired time and location. These findings highlight the potential of TPA-based FNA probes for monitoring biomarkers at different cell stages.
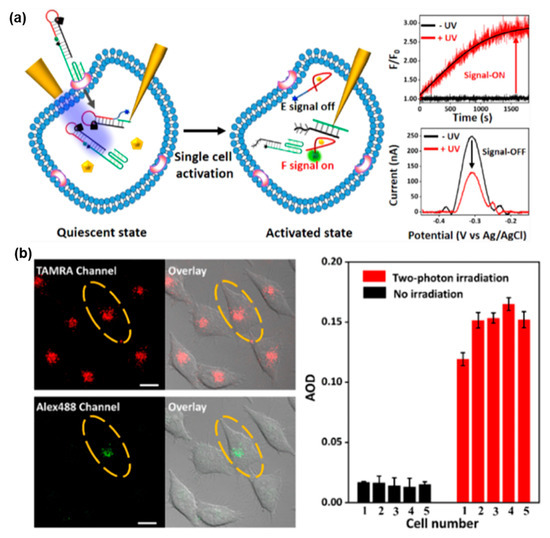
Figure 8.
FNA probe based on TPA for ATP analysis. (a) Schematic illustration of light-activated FNA probe for the fluorescence and electrochemical bimodal imaging of ATP in a single living cell. (b) The fluorescence signal of a single living cell vis TP irradiation at 740 nm. Adapted with permission from [133].
4. Conclusions and Future Prospects
Typically, TP-based FNA probes are composed of FNA sequences along with fluorophores or linkers that are sensitive to TP irradiation. The FNA sequences serve a crucial role in specific interactions with target biomolecules and can include aptamers, DNAzymes, modified nucleic acids, and other related elements. TP technology serves a dual purpose in TP-based FNA probes. Not only does it function as an excitation light source for TP dye, producing fluorescence signals, but it can also serve as an activation light source, enabling spatiotemporal control over FNA probes. As a result, TP-based FNA probes offer advantages such as high spatiotemporal resolution, deep tissue penetration, low photodamage, and high sensitivity. These probes have garnered significant attention in the field of studying molecular interactions in cells, monitoring and imaging drug delivery processes, and quantifying biomarkers. In addition, they are also expected to be used in the regulation of biomimetic materials, the synthesis of artificial tissues, and the preparation of biological hydrogels.
Despite the years of development and utilization of TP-based FNA probes in biomedicine, they are still considered to be in their early stages. For the further development of high-performance TP-based FNA probes and the promotion of their commercialization, there are still some limitations with TP-based FNA probes that should be addressed. Priority should be given to the development of TP dye molecules with higher TP excitation efficiency and environmental stability, thereby improving the imaging resolution and reliability. Additionally, there is a need to develop a user-friendly TP platform that can achieve different regulation effects by adjusting the wavelength, power, and spot size of the two-photon laser. Lastly, efforts should focus on developing additional FNA probe vectors to improve their delivery and targeting efficiency.
Author Contributions
Writing—original draft preparation, K.W.; writing—review and editing, C.M.; supervision and funding acquisition, Y.W. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 61988102), Key Research and Development Program of Guangdong Province (grant no. 2019B090917007), Science and Technology Planning Project of Guangdong Province (grant no. 2019B090909011), Chinese Academy of Sciences Pioneer Hundred Talents Program (grant no. E1Z1D10900), and Chinese Academy of Sciences funding (grant no. E3Z2D10300).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Seeman, N.C. DNA in a material world. Nature 2003, 421, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Leung, H.M.; Lo, P.K. Stimuli-Responsive Self-Assembled DNA Nanomaterials for Biomedical Applications. Small 2017, 13, 1602881. [Google Scholar] [CrossRef] [PubMed]
- Tam, D.Y.; Zhuang, X.; Wong, S.W.; Lo, P.K. Photoresponsive Self-Assembled DNA Nanomaterials: Design, Working Principles, and Applications. Small 2019, 15, 1805481. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zuo, X.; Li, Q.; Chen, F.; Chen, Y.R.; Deng, J.; Han, D.; Hao, C.; Huang, F.; Huang, Y.; et al. Nucleic Acids Analysis. Sci. China Chem. 2020, 64, 171–203. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, Y.; Xu, X.; Liu, Y.; Lin, B.; Zhang, M.; Zhang, J.; Wan, S.; Yang, C.; Tan, W. Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chem. Rev. 2021, 121, 12035–12105. [Google Scholar] [CrossRef]
- Meng, H.-M.; Liu, H.; Kuai, H.; Peng, R.; Mo, L.; Zhang, X.-B. Aptamer-integrated DNA nanostructures for biosensing, bioimaging and cancer therapy. Chem. Soc. Rev. 2016, 45, 2583–2602. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, J.; Sun, Y.; Mo, L.; Liu, B.; Pan, X.; Liu, Z.; Tan, W. Aptamer-Based Logic Computing Reaction on Living Cells to Enable Non-Antibody Immune Checkpoint Blockade Therapy. J. Am. Chem. Soc. 2021, 143, 8391–8401. [Google Scholar] [CrossRef]
- DeRosa, M.C.; Lin, A.; Mallikaratchy, P.; McConnell, E.M.; McKeague, M.; Patel, R.; Shigdar, S. In Vitro selection of aptamers and their applications. Nat. Rev. Methods Primers 2023, 3, 54. [Google Scholar] [CrossRef]
- Morrison, D.; Rothenbroker, M.; Li, Y.F. DNAzymes: Selected for Applications. Small Methods 2018, 2, 1700319. [Google Scholar] [CrossRef]
- Wang, X.; Kim, G.; Chu, J.L.; Song, T.; Yang, Z.; Guo, W.; Shao, X.; Oelze, M.L.; Li, K.C.; Lu, Y. Noninvasive and Spatiotemporal Control of DNAzyme-Based Imaging of Metal Ions In Vivo Using High-Intensity Focused Ultrasound. J. Am. Chem. Soc. 2022, 144, 5812–5819. [Google Scholar] [CrossRef]
- Zhao, D.; Chang, D.; Zhang, Q.; Chang, Y.; Liu, B.; Sun, C.; Li, Z.; Dong, C.; Liu, M.; Li, Y. Rapid and Specific Imaging of Extracellular Signaling Molecule Adenosine Triphosphate with a Self-Phosphorylating DNAzyme. J. Am. Chem. Soc. 2021, 143, 15084–15090. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, M.P.; Duan, Z.; Huang, F.; Laps, S.; Dong, J.; Xia, F.; Willner, I. Photocleavable Ortho-Nitrobenzyl-Protected DNA Architectures and Their Applications. Chem. Rev. 2023, 123, 6839–6887. [Google Scholar] [CrossRef] [PubMed]
- Vyborna, Y.; Galas, J.-C.; Estevez-Torres, A. DNA-Controlled Spatiotemporal Patterning of a Cytoskeletal Active Gel. J. Am. Chem. Soc. 2021, 143, 20022–20026. [Google Scholar] [CrossRef] [PubMed]
- Shi, P.; Wang, Y. Synthetic DNA for Cell-Surface Engineering. Angew. Chem. Int. Ed. 2021, 60, 11580–11591. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Xing, H.; Zhang, J.; Lu, Y. Functional DNA Molecules Enable Selective and Stimuli-Responsive Nanoparticles for Biomedical Applications. Acc. Chem. Res. 2019, 52, 2415–2426. [Google Scholar] [CrossRef]
- Liu, J.; Cao, Z.; Lu, Y. Functional Nucleic Acid Sensors. Chem. Rev. 2009, 109, 1948–1998. [Google Scholar] [CrossRef]
- Pei, H.; Zuo, X.; Zhu, D.; Huang, Q.; Fan, C. Functional DNA Nanostructures for Theranostic Applications. Acc. Chem. Res. 2014, 47, 550–559. [Google Scholar] [CrossRef]
- Liang, H.; Zhang, X.-B.; Lv, Y.; Gong, L.; Wang, R.; Zhu, X.; Yang, R.; Tan, W. Functional DNA-Containing Nanomaterials: Cellular Applications in Biosensing, Imaging, and Targeted Therapy. Acc. Chem. Res. 2014, 47, 1891–1901. [Google Scholar] [CrossRef]
- Melnychuk, N.; Klymchenko, A.S. DNA-Functionalized Dye-Loaded Polymeric Nanoparticles: Ultrabright FRET Platform for Amplified Detection of Nucleic Acids. J. Am. Chem. Soc. 2018, 140, 10856–10865. [Google Scholar] [CrossRef]
- Reid, M.S.; Le, X.C.; Zhang, H. Exponential Isothermal Amplification of Nucleic Acids and Assays for Proteins, Cells, Small Molecules, and Enzyme Activities: An EXPAR Example. Angew. Chem. Int. Ed. Engl. 2018, 57, 11856–11866. [Google Scholar] [CrossRef]
- Sheng, C.; Zhao, J.; Di, Z.; Huang, Y.; Zhao, Y.; Li, L. Spatially resolved In Vivo imaging of inflammation-associated mRNA via enzymatic fluorescence amplification in a molecular beacon. Nat. Biomed. Eng. 2022, 6, 1074–1084. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, Z.; Shao, Y.; Hu, W.; Li, L. Spatially Selective Imaging of Mitochondrial MicroRNAs via Optically Programmable Strand Displacement Reactions. Angew. Chem. Int. Ed. 2021, 60, 17937–17941. [Google Scholar] [CrossRef] [PubMed]
- Qing, Z.; Luo, G.; Xing, S.; Zou, Z.; Lei, Y.; Liu, J.; Yang, R. Pt-S Bond-Mediated Nanoflares for High-Fidelity Intracellular Applications by Avoiding Thiol Cleavage. Angew. Chem. Int. Ed. 2020, 59, 14044–14048. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Liu, C.; Li, Y.; Ma, Y.; Deng, J.; Li, L.; Sun, J. Thermophoretic Detection of Exosomal microRNAs by Nanoflares. J. Am. Chem. Soc. 2020, 142, 4996–5001. [Google Scholar] [CrossRef] [PubMed]
- Qing, Z.; Xu, J.; Hu, J.; Zheng, J.; He, L.; Zou, Z.; Yang, S.; Tan, W.; Yang, R. In Situ Amplification-Based Imaging of RNA in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 11574–11585. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Zhao, J.; Yi, D.; Di, Z.; Li, L. Peptide Nucleic Acid (PNA)-Guided Peptide Engineering of an Aptamer Sensor for Protease-Triggered Molecular Imaging. Angew. Chem. Int. Ed. 2021, 60, 22659–22663. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Zhao, J.; Yuan, J.; Zhao, Y.; Li, L. Organelle-Specific Photoactivation of DNA Nanosensors for Precise Profiling of Subcellular Enzymatic Activity. Angew. Chem. Int. Ed. 2021, 60, 8923–8931. [Google Scholar] [CrossRef] [PubMed]
- Albright, S.; Cacace, M.; Tivon, Y.; Deiters, A. Cell Surface Labeling and Detection of Protein Tyrosine Kinase 7 via Covalent Aptamers. J. Am. Chem. Soc. 2023, 145, 16458–16463. [Google Scholar] [CrossRef]
- Wu, K.; Yao, C.; Yang, D.; Liu, D. A functional DNA nanosensor for highly sensitive and selective imaging of ClO− in atherosclerotic plaques. Biosens. Bioelectron. 2022, 209, 114273. [Google Scholar] [CrossRef]
- Di, Z.; Lu, X.; Zhao, J.; Jaklenec, A.; Zhao, Y.; Langer, R.; Li, L. Mild Acidosis-Directed Signal Amplification in Tumor Microenvironment via Spatioselective Recruitment of DNA Amplifiers. Angew. Chem. Int. Ed. 2022, 61, e202205436. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, Q.; Shi, L.; Peng, P.; Shi, L.; Li, T. Logic-Gated Proximity Aptasensing for Cell-Surface Real-Time Monitoring of Apoptosis. Angew. Chem. Int. Ed. 2021, 60, 20858–20864. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.; Zhao, J.; Li, L. An Enzyme-Activatable Engineered DNAzyme Sensor for Cell-Selective Imaging of Metal Ions. Angew. Chem. Int. Ed. 2021, 60, 6300–6304. [Google Scholar] [CrossRef]
- Xiong, M.; Yang, Z.; Lake, R.J.; Li, J.; Hong, S.; Fan, H.; Zhang, X.B.; Lu, Y. DNAzyme-Mediated Genetically Encoded Sensors for Ratiometric Imaging of Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2020, 59, 1891–1896. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Deng, J.; Han, Z.; Liu, C.; Tian, F.; Xu, R.; Han, D.; Zhang, S.; Sun, J. Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation. J. Am. Chem. Soc. 2021, 143, 1290–1295. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, J.; Yang, Z.; Mou, Q.; Ma, Y.; Xiong, Y.; Lu, Y. Functional DNA Regulated CRISPR-Cas12a Sensors for Point-of-Care Diagnostics of Non-Nucleic-Acid Targets. J. Am. Chem. Soc. 2020, 142, 207–213. [Google Scholar] [CrossRef]
- Fan, Z.; Xiao, K.; Lin, J.; Liao, Y.; Huang, X. Functionalized DNA Enables Programming Exosomes/Vesicles for Tumor Imaging and Therapy. Small 2019, 15, e1903761. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yao, C.; Zhu, Y.; Yang, L.; Luo, D.; Yang, D. DNA Functional Materials Assembled from Branched DNA: Design, Synthesis, and Applications. Chem. Rev. 2020, 120, 9420–9481. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Wu, P.; Kim, T.; Lei, L.; Tian, S.; Wang, Y.; Lu, Y. Photocaged DNAzymes as a General Method for Sensing Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2014, 53, 13798–13802. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, X.; Wang, J.; Zhang, Y.; Zeng, W.; Liu, Y.; Zhang, X. Photoactivatable CRISPR/Cas12a Strategy for One-Pot DETECTR Molecular Diagnosis. Anal. Chem. 2022, 94, 9724–9731. [Google Scholar] [CrossRef]
- Li, J.X.; Khan, S.; Gu, J.; Filipe, C.D.M.; Didar, T.F.; Li, Y.F. A Simple Colorimetric Au-on-Au Tip Sensor with a New Functional Nucleic Acid Probe for Food-borne Pathogen Salmonella typhimurium. Angew. Chem. Int. Ed. 2023, 62, e202300828. [Google Scholar] [CrossRef]
- Li, M.; Zhao, J.; Chu, H.; Mi, Y.; Zhou, Z.; Di, Z.; Zhao, M.; Li, L. Light-Activated Nanoprobes for Biosensing and Imaging. Adv. Mater. 2019, 31, e1804745. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.L.; Shi, Y.A.; Zhang, Y.Q.; He, J.Y.; Li, M.D.; Huang, W.X.; Yuan, R.; Xu, W.J. Modular DNA Tetrahedron Nanomachine-Guided Dual-Responsive Hybridization Chain Reactions for Discernible Bivariate Assay and Cell Imaging. Anal. Chem. 2023, 95, 10337–10345. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Gao, J.; Cao, L.; Xie, X.; Fan, J.; Wang, H.; Wang, H.-H.; Nie, Z. A DNA Molecular Robot that Autonomously Walks on the Cell Membrane to Drive Cell Motility. Angew. Chem. Int. Ed. 2021, 60, 26087–26095. [Google Scholar] [CrossRef]
- He, S.; Gao, F.; Ma, J.; Ma, H.; Dong, G.; Sheng, C. Aptamer-PROTAC Conjugates (APCs) for Tumor-Specific Targeting in Breast Cancer. Angew. Chem. Int. Ed. 2021, 60, 23299–23305. [Google Scholar] [CrossRef]
- Zhou, Z.X.; Lin, N.N.; Ouyang, Y.; Liu, S.Q.; Zhang, Y.J.; Willner, I. Cascaded, Feedback-Driven, and Spatially Localized Emergence of Constitutional Dynamic Networks Driven by Enzyme-Free Catalytic DNA Circuits. J. Am. Chem. Soc. 2023, 145, 12617–12629. [Google Scholar] [CrossRef]
- Shen, X.T.; Ouyang, Q.W.; Tan, H.W.; Ouyang, J.; Na, N. Computation-Assisted Design of ssDNA Framework Nanorobots for Cancer Logical Recognition, Toehold Disintegration, Visual Dual- Diagnosis, and Synergistic Therapy. Anal. Chem. 2023, 95, 5903–5910. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Li, W.; Du, W.; Yue, A.; Zhao, J.; Liu, D. In-situ monitoring of nitrile-bearing pesticide residues by background-free surface-enhanced Raman spectroscopy. Chin. Chem. Lett. 2022, 33, 519–522. [Google Scholar] [CrossRef]
- Croissant, J.G.; Zink, J.I.; Raehm, L.; Durand, J.-O. Two-Photon-Excited Silica and Organosilica Nanoparticles for Spatiotemporal Cancer Treatment. Adv. Healthc. Mater. 2018, 7, 1701248. [Google Scholar] [CrossRef]
- van Kesteren, S.; Shen, X.; Aldeghi, M.; Isa, L. Printing on Particles: Combining Two-Photon Nanolithography and Capillary Assembly to Fabricate Multimaterial Microstructures. Adv. Mater. 2023, 35, e2207101. [Google Scholar] [CrossRef]
- Abele, T.; Messer, T.; Jahnke, K.; Hippler, M.; Bastmeyer, M.; Wegener, M.; Gopfrich, K. Two-Photon 3D Laser Printing Inside Synthetic Cells. Adv. Mater. 2022, 34, 2106709. [Google Scholar] [CrossRef]
- Grzybowski, A.; Pietrzak, K. Maria Goeppert-Mayer (1906–1972): Two-photon effect on dermatology. Clin. Dermatol. 2013, 31, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Shuhendler, A.J.; Ye, D.; Xu, J.J.; Chen, H.Y. Two-photon excitation nanoparticles for photodynamic therapy. Chem. Soc. Rev. 2016, 45, 6725–6741. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, J.; Yu, Z.; Zhu, X.; Yu, J.; Wu, Z.; Wang, S.; Zhou, H. Molecular Tailoring Based on Forster Resonance Energy Transfer for Initiating Two-Photon Theranostics with Amplified Reactive Oxygen Species. Anal. Chem. 2022, 94, 14029–14037. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Wu, Y.-X.; Yu, S.; Wang, X.; He, K.; Li, D.; Cao, Y.; Gan, N. Two-Photon CQDs-Based Dual-Mode Nanoprobe for Fluorescence Imaging and Magnetic Resonance Imaging of Intracellular Wide pH. Anal. Chem. 2021, 93, 5691–5699. [Google Scholar] [CrossRef]
- Wu, X.; Wang, R.; Qi, S.; Kwon, N.; Han, J.; Kim, H.; Li, H.; Yu, F.; Yoon, J. Rational Design of a Highly Selective Near-Infrared Two-Photon Fluorogenic Probe for Imaging Orthotopic Hepatocellular Carcinoma Chemotherapy. Angew. Chem. Int. Ed. 2021, 60, 15418–15425. [Google Scholar] [CrossRef]
- Tang, F.; Liu, J.-Y.; Wu, C.-Y.; Liang, Y.-X.; Lu, Z.-L.; Ding, A.-X.; Xu, M.-D. Two-Photon Near-Infrared AIE Luminogens as Multifunctional Gene Carriers for Cancer Theranostics. ACS Appl. Mater. Interfaces 2021, 13, 23384–23395. [Google Scholar] [CrossRef] [PubMed]
- Juvekar, V.; Lee, H.W.; Lee, D.J.; Kim, H.M. Two-photon fluorescent probes for quantitative bio-imaging analysis in live tissues. TrAC Trends Anal. Chem. 2022, 157, 116787. [Google Scholar] [CrossRef]
- Wu, Y.-X.; Zhang, D.; Hu, X.; Peng, R.; Li, J.; Zhang, X.; Tan, W. Multicolor Two-Photon Nanosystem for Multiplexed Intracellular Imaging and Targeted Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 12569–12576. [Google Scholar] [CrossRef]
- Jin, H.; Yang, M.; Sun, Z.; Gui, R. Ratiometric two-photon fluorescence probes for sensing, imaging and biomedicine applications at living cell and small animal levels. Coord. Chem. Rev. 2021, 446, 214114. [Google Scholar] [CrossRef]
- Gao, X.; Li, S.; Ding, F.; Fan, H.; Shi, L.; Zhu, L.; Li, J.; Feng, J.; Zhu, X.; Zhang, C. Rapid Detection of Exosomal MicroRNAs Using Virus-Mimicking Fusogenic Vesicles. Angew. Chem. Int. Ed. 2019, 58, 8719–8723. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, T.; He, Y.; Deng, Z.; Li, J.; Liu, H.; Nie, J.; Wang, D.; Huang, J.; Zhong, S. Nanoflare Couple: Multiplexed mRNA Imaging and Logic-Controlled Combinational Therapy. Anal. Chem. 2022, 94, 12204–12212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Ouyang, Y.; Sohn, Y.S.; Fadeev, M.; Karmi, O.; Nechushtai, R.; Stein, I.; Pikarsky, E.; Willner, I. miRNA-Guided Imaging and Photodynamic Therapy Treatment of Cancer Cells Using Zn(II)-Protoporphyrin IX-Loaded Metal–Organic Framework Nanoparticles. ACS Nano 2022, 16, 1791–1801. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, K.; Padmanabhan, G. MiRNAs as potential biomarker of kidney diseases: A review. Cell Biochem. Funct. 2020, 38, 990–1005. [Google Scholar] [CrossRef] [PubMed]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhou, Y.; Sun, Y.; Wu, S.; Xu, T.; Chang, Y.-C.; Bi, S.; Jiang, L.-P.; Zhu, J.-J. Endogenous mRNA Triggered DNA-Au Nanomachine for In Situ Imaging and Targeted Multimodal Synergistic Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 5948–5958. [Google Scholar] [CrossRef]
- Zhu, R.; Feng, H.; Li, Q.; Su, L.; Fu, Q.; Li, J.; Song, J.; Yang, H. Asymmetric Core–Shell Gold Nanoparticles and Controllable Assemblies for SERS Ratiometric Detection of MicroRNA. Angew. Chem. Int. Ed. 2021, 60, 12560–12568. [Google Scholar] [CrossRef]
- Wang, N.; Song, L.; Qiu, Y.; Xing, H.; Li, J. Hybridization-activated spherical DNAzyme for cascading two-photon fluorescence emission: Applied for intracellular miRNA measurement by two-photon microscopy. Sens. Actuators B-Chem. 2019, 286, 250–257. [Google Scholar] [CrossRef]
- Yang, C.; Wang, K.; Li, Z.; Mo, L.; Lin, W. A two-photon metal-organic framework nanoprobe with catalytic hairpin assembly for amplified MicroRNA imaging in living cells and tissues. Sens. Actuators B-Chem. 2022, 359, 131593. [Google Scholar] [CrossRef]
- Yuan, P.; Ma, R.; Guan, Z.; Gao, N.; Xu, Q.-H. Tuning Two-Photon Photoluminescence of Gold Nanoparticle Aggregates with DNA and Its Application as Turn-on Photoluminescence Probe for DNA Sequence Detection. ACS Appl. Mater. Interfaces 2014, 6, 13149–13156. [Google Scholar] [CrossRef]
- Yi, M.; Wang, F.; Tan, W.; Hsieh, J.-T.; Egelman, E.H.; Xu, B. Enzyme Responsive Rigid-Rod Aromatics Target “Undruggable” Phosphatases to Kill Cancer Cells in a Mimetic Bone Microenvironment. J. Am. Chem. Soc. 2022, 144, 13055–13059. [Google Scholar] [CrossRef]
- Beasley, S.; Vandewalle, A.; Singha, M.; Nguyen, K.; England, W.; Tarapore, E.; Dai, N.; Corrêa, I.R.; Atwood, S.X.; Spitale, R.C. Exploiting Endogenous Enzymes for Cancer-Cell Selective Metabolic Labeling of RNA In Vivo. J. Am. Chem. Soc. 2022, 144, 7085–7088. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Zhan, J.; Xu, G.; Chen, Y.; Qin, Q.; Liao, X.; Ma, S.; Yang, Z.; Cai, Y. Enzyme-Instructed Self-Assembly Enabled Monomer–Excimer Transition to Construct Higher Ordered Luminescent Supramolecular Assembly for Activity-based Bioimaging. Angew. Chem. Int. Ed. 2021, 60, 8121–8129. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Ling, S.; Huang, H.; Zhang, Y.; Chen, G.; Huang, S.; Li, C.; Guo, W.; Wang, Q. Rapid Unperturbed-Tissue Analysis for Intraoperative Cancer Diagnosis Using an Enzyme-Activated NIR-II Nanoprobe. Angew. Chem. Int. Ed. 2021, 60, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Li, Z.; Fu, F.; Chen, X.; Song, J.; Yang, H. Stimuli-Responsive Plasmonic Assemblies and Their Biomedical Applications. Nano Today 2021, 36, 101014. [Google Scholar] [CrossRef]
- Zhao, X.; Ji, X.; Qu, J.; Xie, K.; Wang, Z.; Fang, P.; Wang, Y.; Wan, Y.; Yang, Y.; Zhang, W.; et al. Sequencing-free Analysis of Multiple Methylations on Gene-Specific mRNAs. J. Am. Chem. Soc. 2022, 144, 6010–6018. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Zhang, L.; Fu, S.; Su, X. Ultra-specific fluorescence detection of DNA modifying enzymes by dissipation system. Biosens. Bioelectron. 2022, 215, 114561. [Google Scholar] [CrossRef]
- Farag, N.; Mattossovich, R.; Merlo, R.; Nierzwicki, L.; Palermo, G.; Porchetta, A.; Perugino, G.; Ricci, F. Folding-upon-Repair DNA Nanoswitches for Monitoring the Activity of DNA Repair Enzymes. Angew. Chem. Int. Ed. Engl. 2021, 60, 7283–7289. [Google Scholar] [CrossRef]
- Hong, M.; Xu, L.; Xue, Q.; Li, L.; Tang, B. Fluorescence Imaging of Intracellular Telomerase Activity Using Enzyme-Free Signal Amplification. Anal. Chem. 2016, 88, 12177–12182. [Google Scholar] [CrossRef]
- Li, T.; Liu, L.; Wei, N.; Yang, J.Y.; Chapla, D.G.; Moremen, K.W.; Boons, G.J. An automated platform for the enzyme-mediated assembly of complex oligosaccharides. Nat. Chem. 2019, 11, 229–236. [Google Scholar] [CrossRef]
- Ge, Q.; Wang, N.; Li, J.; Yang, R. Peptide-fluorophore/AuNP conjugate-based two-photon excited fluorescent nanosensor for caspase-3 activity imaging assay in living cells and tissue. MedChemComm 2017, 8, 1435–1439. [Google Scholar] [CrossRef]
- Wang, N.; Li, J.; He, B.; Deng, T.; Yang, J.; Li, J. Two-photon excitation nanoprobe for DNases activity imaging assay in hepatic ischemia reperfusion injury. Sens. Actuators B-Chem. 2019, 298, 126853. [Google Scholar] [CrossRef]
- Wang, N.; Song, L.; Xing, H.; Zhang, K.; Yang, R.; Li, J. A spherical nucleic acid-based two-photon nanoprobe for RNase H activity assay in living cells and tissues. Nanoscale 2019, 11, 8133–8137. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhou, T.; Jin, M.; Zhou, K.; Liu, D.; Li, X.; Huo, F.; Li, W.; Yin, C. Thiol-Chromene “Click” Reaction Triggered Self-Immolative for NIR Visualization of Thiol Flux in Physiology and Pathology of Living Cells and Mice. J. Am. Chem. Soc. 2020, 142, 1614–1620. [Google Scholar] [CrossRef] [PubMed]
- Yin, G.; Niu, T.; Yu, T.; Gan, Y.; Sun, X.; Yin, P.; Chen, H.; Zhang, Y.; Li, H.; Yao, S. Simultaneous Visualization of Endogenous Homocysteine, Cysteine, Glutathione, and their Transformation through Different Fluorescence Channels. Angew. Chem. Int. Ed. Engl. 2019, 58, 4557–4561. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Cai, R.; Chen, X.; Wu, Q.; Zhang, L.; Jiang, Y.; Cui, C.; Wan, S.; Tan, W. Facile approach to prepare HSA-templated MnO2 nanosheets as oxidase mimic for colorimetric detection of glutathione. Talanta 2019, 195, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Xiao, C.; Li, Z.; Yang, X. Engineering nanomedicine for glutathione depletion-augmented cancer therapy. Chem. Soc. Rev. 2021, 50, 6013–6041. [Google Scholar] [CrossRef]
- Chai, X.; Fan, Z.; Yu, M.M.; Zhao, J.; Li, L. A Redox-Activatable DNA Nanodevice for Spatially-Selective, AND-Gated Imaging of ATP and Glutathione in Mitochondria. Nano Lett. 2021, 21, 10047–10053. [Google Scholar] [CrossRef]
- Yin, C.-X.; Xiong, K.-M.; Huo, F.-J.; Salamanca, J.C.; Strongin, R.M. Fluorescent Probes with Multiple Binding Sites for the Discrimination of Cys, Hcy, and GSH. Angew. Chem. Int. Ed. 2017, 56, 13188–13198. [Google Scholar] [CrossRef]
- Zhang, L.; Cai, Q.Y.; Li, J.; Ge, J.; Wang, J.Y.; Dong, Z.Z.; Li, Z.H. A label-free method for detecting biothiols based on poly(thymine)-templated copper nanoparticles. Biosens. Bioelectron. 2015, 69, 77–82. [Google Scholar] [CrossRef]
- Niu, L.-Y.; Chen, Y.-Z.; Zheng, H.-R.; Wu, L.-Z.; Tung, C.-H.; Yang, Q.-Z. Design strategies of fluorescent probes for selective detection among biothiols. Chem. Soc. Rev. 2015, 44, 6143–6160. [Google Scholar] [CrossRef]
- Tang, Q.; Wang, N.; Zhou, F.; Deng, T.; Zhang, S.; Li, J.; Yang, R.; Zhong, W.; Tan, W. A novel AgNP/DNA/TPdye conjugate-based two-photon nanoprobe for GSH imaging in cell apoptosis of cancer tissue. Chem. Commun. 2015, 51, 16810–16812. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, M.; Tang, Q.; Deng, T.; Yan, H.; Li, J.; Li, Y.; Yang, R. Two-photon AgNP/DNA-TP dye nanosensing conjugate for biothiol probing in live cells. Analyst 2014, 139, 6185–6191. [Google Scholar] [CrossRef] [PubMed]
- Lobas, M.A.; Tao, R.; Nagai, J.; Kronschlager, M.T.; Borden, P.M.; Marvin, J.S.; Looger, L.L.; Khakh, B.S. A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat. Commun. 2019, 10, 711. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Gao, J.; Xue, W.; Di, Z.; Xing, H.; Lu, Y.; Li, L. Upconversion Luminescence-Activated DNA Nanodevice for ATP Sensing in Living Cells. J. Am. Chem. Soc. 2018, 140, 578–581. [Google Scholar] [CrossRef]
- Patel, A.; Malinovska, L.; Saha, S.; Wang, J.; Alberti, S.; Krishnan, Y.; Hyman, A.A. ATP as a biological hydrotrope. Science 2017, 356, 753–756. [Google Scholar] [CrossRef]
- Di, Z.; Zhao, J.; Chu, H.; Xue, W.; Zhao, Y.; Li, L. An Acidic-Microenvironment-Driven DNA Nanomachine Enables Specific ATP Imaging in the Extracellular Milieu of Tumor. Adv. Mater. 2019, 31, 1901885. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, R.; Gaffney, K.; Kim, M.; Muhammednazaar, S.; Tian, W.; Wang, B.; Liang, J.; Hong, H. Folding-Degradation Relationship of a Membrane Protein Mediated by the Universally Conserved ATP-Dependent Protease FtsH. J. Am. Chem. Soc. 2018, 140, 4656–4665. [Google Scholar] [CrossRef]
- Li, F.; Liu, Y.; Dong, Y.; Chu, Y.; Song, N.; Yang, D. Dynamic Assembly of DNA Nanostructures in Living Cells for Mitochondrial Interference. J. Am. Chem. Soc. 2022, 144, 4667–4677. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, L.; Wei, J.; Ma, K.; Gong, X.; Shang, J.; Yu, S.; Wang, F. An Autonomous Nonenzymatic Concatenated DNA Circuit for Amplified Imaging of Intracellular ATP. Anal. Chem. 2019, 91, 15229–15234. [Google Scholar] [CrossRef]
- Cai, S.J.; Wang, J.L.; Li, J.; Zhou, B.; He, C.M.; Meng, X.X.; Huang, J.; Wang, K.M. A self-assembled DNA nanostructure as a FRET nanoflare for intracellular ATP imaging. Chem. Commun. 2021, 57, 6257–6260. [Google Scholar] [CrossRef]
- Tan, K.Y.; Li, C.Y.; Li, Y.F.; Fei, J.; Yang, B.; Fu, Y.J.; Li, F. Real-Time Monitoring ATP in Mitochondrion of Living Cells: A Specific Fluorescent Probe for ATP by Dual Recognition Sites. Anal. Chem. 2017, 89, 1749–1756. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, D.; Yuan, R.; Xiang, Y. A catalytic and dual recycling amplification ATP sensor based on target-driven allosteric structure switching of aptamer beacons. Biosens. Bioelectron. 2018, 105, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.; Zhang, X.; Lake, R.J.; Pawel, G.T.; Guo, Z.; Pei, R.; Lu, Y. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of ATP in the mitochondria of living cells. Chem. Sci. 2019, 11, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Shen, X.; Li, B.; Li, X.; Zhou, X. Signal amplification by strand displacement in a carbon dot based fluorometric assay for ATP. Mikrochim. Acta 2018, 185, 392. [Google Scholar] [CrossRef]
- Yi, M.; Yang, S.; Peng, Z.; Liu, C.; Li, J.; Zhong, W.; Yang, R.; Tan, W. Two-Photon Graphene Oxide/Aptamer Nanosensing Conjugate for In Vitro or In Vivo Molecular Probing. Anal. Chem. 2014, 86, 3548–3554. [Google Scholar] [CrossRef]
- Qian, R.-C.; Zhou, Z.-R.; Guo, W.; Wu, Y.; Yang, Z.; Lu, Y. Cell Surface Engineering Using DNAzymes: Metal Ion Mediated Control of Cell–Cell Interactions. J. Am. Chem. Soc. 2021, 143, 5737–5744. [Google Scholar] [CrossRef]
- Zhou, W.; Saran, R.; Liu, J. Metal Sensing by DNA. Chem. Rev. 2017, 117, 8272–8325. [Google Scholar] [CrossRef]
- Wu, Z.; Fan, H.; Satyavolu, N.S.R.; Wang, W.; Lake, R.; Jiang, J.H.; Lu, Y. Imaging Endogenous Metal Ions in Living Cells Using a DNAzyme-Catalytic Hairpin Assembly Probe. Angew. Chem. Int. Ed. 2017, 56, 8721–8725. [Google Scholar] [CrossRef]
- Wang, W.; Satyavolu, N.S.R.; Wu, Z.; Zhang, J.R.; Zhu, J.J.; Lu, Y. Near-Infrared Photothermally Activated DNAzyme-Gold Nanoshells for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2017, 56, 6798–6802. [Google Scholar] [CrossRef]
- Wang, W.-Z.; Huang, L.-B.; Zheng, S.-P.; Moulin, E.; Gavat, O.; Barboiu, M.; Giuseppone, N. Light-Driven Molecular Motors Boost the Selective Transport of Alkali Metal Ions through Phospholipid Bilayers. J. Am. Chem. Soc. 2021, 143, 15653–15660. [Google Scholar] [CrossRef]
- Saidur, M.R.; Aziz, A.R.A.; Basirun, W.J. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017, 90, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Yang, Z.; Lake, R.J.; Zheng, C.; Lu, Y. Enzyme-Mediated Endogenous and Bioorthogonal Control of a DNAzyme Fluorescent Sensor for Imaging Metal Ions in Living Cells. Angew. Chem. Int. Ed. 2019, 58, 17061–17067. [Google Scholar] [CrossRef] [PubMed]
- Lake, R.J.; Yang, Z.; Zhang, J.; Lu, Y. DNAzymes as Activity-Based Sensors for Metal Ions: Recent Applications, Demonstrated Advantages, Current Challenges, and Future Directions. Acc. Chem. Res. 2019, 52, 3275–3286. [Google Scholar] [CrossRef]
- Dai, D.; Li, Z.; Yang, J.; Wang, C.; Wu, J.R.; Wang, Y.; Zhang, D.; Yang, Y.W. Supramolecular Assembly-Induced Emission Enhancement for Efficient Mercury(II) Detection and Removal. J. Am. Chem. Soc. 2019, 141, 4756–4763. [Google Scholar] [CrossRef]
- Yang, Z.; Loh, K.Y.; Chu, Y.T.; Feng, R.; Satyavolu, N.S.R.; Xiong, M.; Nakamata Huynh, S.M.; Hwang, K.; Li, L.; Xing, H.; et al. Optical Control of Metal Ion Probes in Cells and Zebrafish Using Highly Selective DNAzymes Conjugated to Upconversion Nanoparticles. J. Am. Chem. Soc. 2018, 140, 17656–17665. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Yin, X.; Huan, S.-Y.; Chen, L.; Hu, X.-X.; Xiong, M.-Y.; Chen, K.; Zhang, X.-B. Two-Photon DNAzyme-Gold Nanoparticle Probe for Imaging Intracellular Metal Ions. Anal. Chem. 2018, 90, 3118–3123. [Google Scholar] [CrossRef]
- Yamahira, S.; Misawa, R.; Kosaka, T.; Tan, M.; Izuta, S.; Yamashita, H.; Heike, Y.; Okamoto, A.; Nagamune, T.; Yamaguchi, S. Photoactivatable Materials for Versatile Single-Cell Patterning Based on the Photocaging of Cell-Anchoring Moieties through Lipid Self-Assembly. J. Am. Chem. Soc. 2022, 144, 13154–13162. [Google Scholar] [CrossRef]
- Weinstain, R.; Slanina, T.; Kand, D.; Klan, P. Visible-to-NIR-Light Activated Release: From Small Molecules to Nanomaterials. Chem. Rev. 2020, 120, 13135–13272. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, W.; Fang, Y.; Zhang, X.; Liu, Y.; Ju, H. Activating a DNA Nanomachine via Computation across Cancer Cell Membranes for Precise Therapy of Solid Tumors. J. Am. Chem. Soc. 2021, 143, 15233–15242. [Google Scholar] [CrossRef]
- Ye, M.; Kong, Y.; Zhang, C.; Lv, Y.; Cheng, S.; Hou, D.; Xian, Y. Near-Infrared Light Controllable DNA Walker Driven by Endogenous Adenosine Triphosphate for in Situ Spatiotemporal Imaging of Intracellular MicroRNA. ACS Nano 2021, 15, 14253–14262. [Google Scholar] [CrossRef]
- Huang, F.; Chen, M.; Zhou, Z.; Duan, R.; Xia, F.; Willner, I. Spatiotemporal patterning of photoresponsive DNA-based hydrogels to tune local cell responses. Nat. Commun. 2021, 12, 2364. [Google Scholar] [CrossRef] [PubMed]
- Jun, Y.W.; Wang, T.; Hwang, S.; Kim, D.; Ma, D.; Kim, K.H.; Kim, S.; Jung, J.; Ahn, K.H. A Ratiometric Two-Photon Fluorescent Probe for Tracking Lysosomal ATP: Direct In Cellulo Observation of Lysosomal Membrane Fusion Processes. Angew. Chem. Int. Ed. 2018, 57, 10142–10147. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Deng, R.; Zhang, K.; Sun, Y.; Li, Y.; Li, J. Single-Cell Imaging of m6A Modified RNA Using m6A-Specific In Situ Hybridization Mediated Proximity Ligation Assay (m6AISH-PLA). Angew. Chem. Int. Ed. 2021, 60, 22646–22651. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Ruan, Y.-F.; Zhu, L.-B.; Shi, X.-M.; Zhao, W.-W.; Chen, H.-Y.; Xu, J.-J. An Integrated Electrochemical Nanodevice for Intracellular RNA Collection and Detection in Single Living Cell. Angew. Chem. Int. Ed. 2021, 60, 13244–13250. [Google Scholar] [CrossRef]
- Qian, R.C.; Cao, Y.; Zhao, L.J.; Gu, Z.; Long, Y.T. A Two-Stage Dissociation System for Multilayer Imaging of Cancer Biomarker-Synergic Networks in Single Cells. Angew. Chem. Int. Ed. 2017, 56, 4802–4805. [Google Scholar] [CrossRef]
- Deng, R.; Tang, L.; Tian, Q.; Wang, Y.; Lin, L.; Li, J. Toehold-initiated rolling circle amplification for visualizing individual microRNAs In Situ in single cells. Angew. Chem. Int. Ed. 2014, 53, 2389–2393. [Google Scholar] [CrossRef]
- Porichis, F.; Hart, M.G.; Griesbeck, M.; Everett, H.L.; Hassan, M.; Baxter, A.E.; Lindqvist, M.; Miller, S.M.; Soghoian, D.Z.; Kavanagh, D.G.; et al. High-throughput detection of miRNAs and gene-specific mRNA at the single-cell level by flow cytometry. Nat. Commun. 2014, 5, 5641. [Google Scholar] [CrossRef]
- Lin, M.; Yi, X.; Huang, F.; Ma, X.; Zuo, X.; Xia, F. Photoactivated Nanoflares for mRNA Detection in Single Living Cells. Anal. Chem. 2019, 91, 2021–2027. [Google Scholar] [CrossRef]
- Chen, M.X.; Duan, R.L.; Xu, S.J.; Duan, Z.J.; Yuan, Q.; Xia, F.; Huang, F.J. Photoactivated DNA Walker Based on DNA Nanoflares for Signal-Amplified MicroRNA Imaging in Single Living Cells. Anal. Chem. 2021, 93, 16264–16272. [Google Scholar] [CrossRef]
- Deng, J.; Wang, K.; Wang, M.; Yu, P.; Mao, L. Mitochondria Targeted Nanoscale Zeolitic Imidazole Framework-90 for ATP Imaging in Live Cells. J. Am. Chem. Soc. 2017, 139, 5877–5882. [Google Scholar] [CrossRef]
- Zhang, Z.; Oni, O.; Liu, J. New insights into a classic aptamer: Binding sites, cooperativity and more sensitive adenosine detection. Nucleic Acids Res. 2017, 45, 7593–7601. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tozzi, F.; Chen, J.; Fan, F.; Xia, L.; Wang, J.; Gao, G.; Zhang, A.; Xia, X.; Brasher, H.; et al. Intracellular ATP Levels Are a Pivotal Determinant of Chemoresistance in Colon Cancer Cells. Cancer Res. 2012, 72, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Tan, L.; Duan, R.; Chen, M.; Xia, F.; Huang, F. Photoactivated Biosensing Process for Dictated ATP Detection in Single Living Cells. Anal. Chem. 2021, 93, 11547–11556. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).