Rapid and Quantitative Detection of Lung Cancer Biomarker ENOX2 Using a Novel Aptamer in an Electrochemical DNA-Based (E-DNA) Biosensor
Abstract
1. Introduction
2. Materials and Methods
2.1. Bead-Based Aptamer Selection
2.2. ENOX2 Binding of Fluorescein-Labeled Putative Aptamer
2.3. Aptamer Design and Preparation
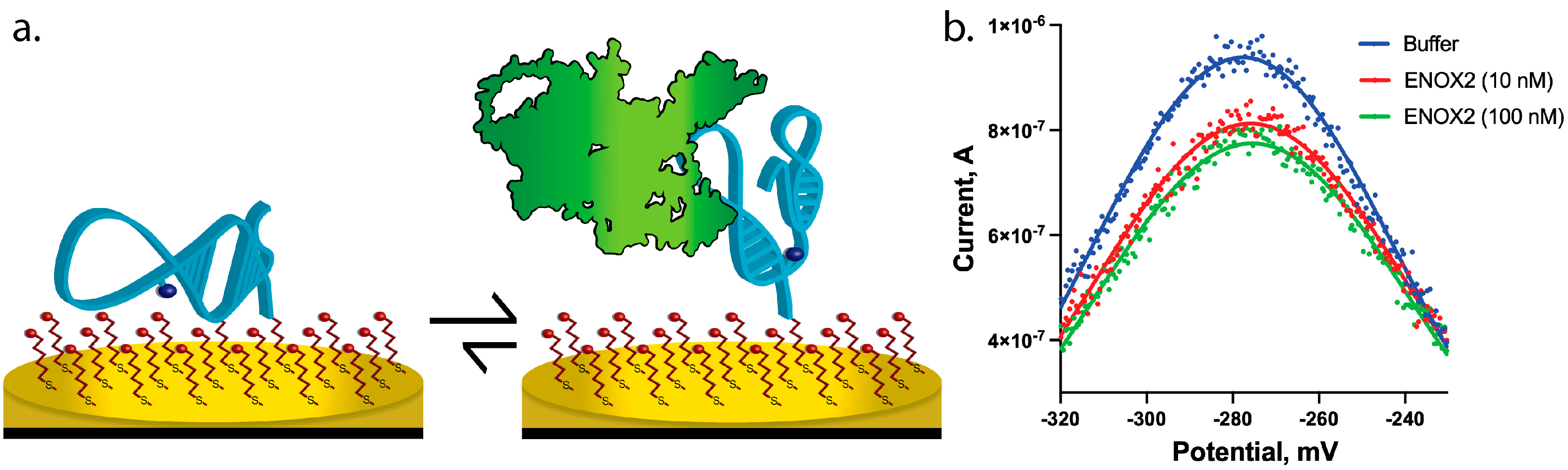
2.4. Biosensor Design and Preparation
2.5. Electrochemical and Control Parameters
3. Results
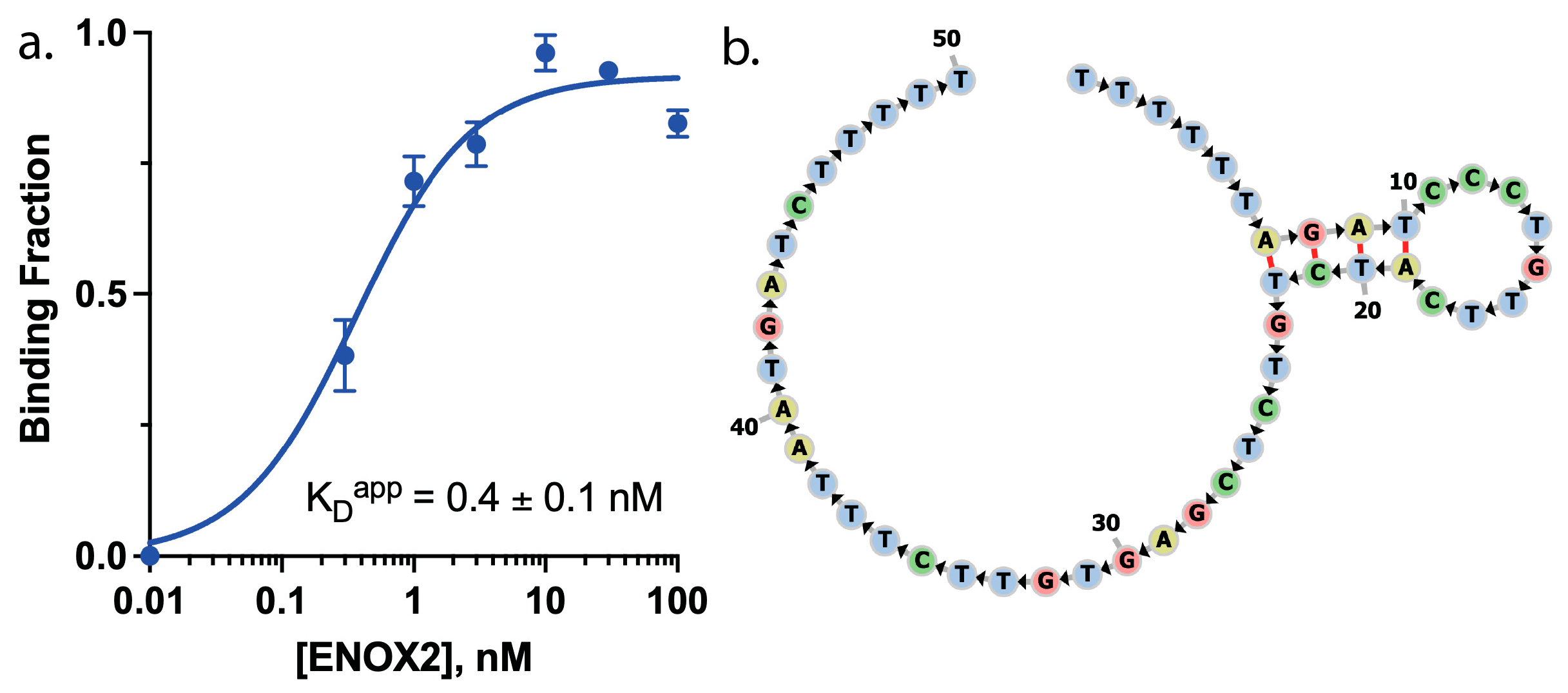
3.1. ENOX2-Binding Aptamer Screening
3.2. ENOX2-Binding Aptamer Optimization to Enable Two Conformations
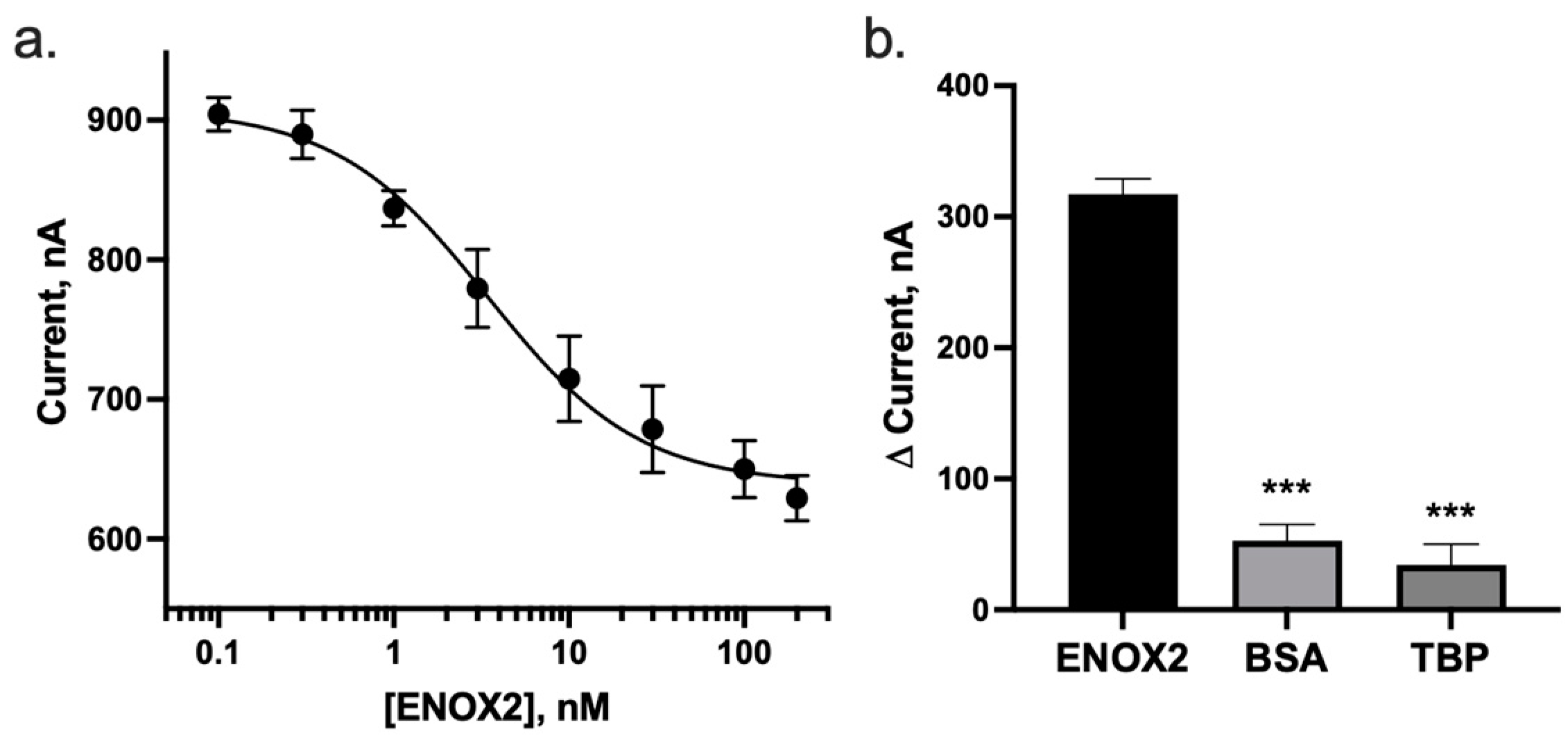
3.3. ENOX2 Binding Results in Detectable and Specific Peak Current Decrease
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Crosby, D.; Bhatia, S.; Brindle, K.M.; Coussens, L.M.; Dive, C.; Emberton, M.; Esener, S.; Fitzgerald, R.C.; Gambhir, S.S.; Kuhn, P.; et al. Early Detection of Cancer. Science 2022, 375, eaay9040. [Google Scholar] [CrossRef]
- Srinivas, P.R.; Kramer, B.S.; Srivastava, S. Trends in Biomarker Research for Cancer Detection. Lancet Oncol. 2001, 2, 698–704. [Google Scholar] [CrossRef]
- Kelker, M.; Kim, C.; Chueh, P.-J.; Guimont, R.; Morré, D.M.; Morré, D.J. Cancer Isoform of a Tumor-Associated Cell Surface NADH Oxidase (TNOX) Has Properties of a Prion. Biochemistry 2001, 40, 7351–7354. [Google Scholar] [CrossRef]
- Morré, D.J.; Morré, D.M. ENOX2 (TNOX) and Cancer. In ECTO-NOX Proteins: Growth, Cancer, and Aging; Morré, D.J., Morré, D.M., Eds.; Springer: New York, NY, USA, 2013; pp. 211–259. ISBN 978-1-4614-3958-5. [Google Scholar]
- Morré, D.J.; Morré, D.M. Cancer Diagnostic Applications of ENOX2 Proteins. In ECTO-NOX Proteins: Growth, Cancer, and Aging; Morré, D.J., Morré, D.M., Eds.; Springer: New York, NY, USA, 2013; pp. 419–434. ISBN 978-1-4614-3958-5. [Google Scholar]
- Morre, D.J.; Hignite, H.; Hostetler, B.; McClain, N.; Juang, J.; Gilmartin, D.; Miner, M.P.; Iannuzzi, M.; Morre, D.M. A TNOX-Based Protocol for Early Detection of Lung Cancer in Smokers and Non-Smokers. Clin. Cancer Res. 2014, 12, A26. [Google Scholar]
- Ronconi, G.; Lessiani, G.; Spinas, E.; Kritas, S.K.; Caraffa, A.; Saggini, A.; Antinolfi, P.; Pizzicannella, J.; Toniato, E.; Conti, P. ENOX2 (or TNOX): A New and Old Molecule with Cancer Activity Involved in Tumor Prevention and Therapy. J. Biol. Regul. Homeost. Agents 2016, 30, 649–653. [Google Scholar] [PubMed]
- Morré, D.J.; Hostetler, B.; Taggart, D.J.; Morré, D.M.; Musk, A.W.; Robinson, B.W.S.; Creaney, J. ENOX2-Based Early Detection (ONCOblot) of Asbestos-Induced Malignant Mesothelioma 4–10 Years in Advance of Clinical Symptoms. Clin. Proteom. 2016, 13, 2. [Google Scholar] [CrossRef]
- Yoo, H.; Jo, H.; Soo Oh, S. Detection and beyond: Challenges and Advances in Aptamer-Based Biosensors. Mater. Adv. 2020, 1, 2663–2687. [Google Scholar] [CrossRef]
- Baykal-Köse, S.; Voldoire, M.; Desterke, C.; Sorel, N.; Cayssials, E.; Johnson-Ansah, H.; Guerci-Bresler, A.; Bennaceur-Griscelli, A.; Chomel, J.-C.; Turhan, A. ENOX2 NADH Oxidase: A BCR-ABL1-Dependent Cell Surface and Secreted Redox Protein in Chronic Myeloid Leukemia (CML). bioRxiv 2021. [Google Scholar] [CrossRef]
- Lymperopoulos, K.; Crawford, R.; Torella, J.P.; Heilemann, M.; Hwang, L.C.; Holden, S.J.; Kapanidis, A.N. Single-Molecule DNA Biosensors for Protein and Ligand Detection. Angew. Chem. Int. Ed. 2010, 49, 1316–1320. [Google Scholar] [CrossRef]
- Yousefi, M.; Dehghani, S.; Nosrati, R.; Zare, H.; Evazalipour, M.; Mosafer, J.; Tehrani, B.S.; Pasdar, A.; Mokhtarzadeh, A.; Ramezani, M. Aptasensors as a New Sensing Technology Developed for the Detection of MUC1 Mucin: A Review. Biosens. Bioelectron. 2019, 130, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Justino, C.I.L.; Freitas, A.C.; Pereira, R.; Duarte, A.C.; Rocha Santos, T.A.P. Recent Developments in Recognition Elements for Chemical Sensors and Biosensors. TrAC Trends Anal. Chem. 2015, 68, 2–17. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Hoggarth, D.A.; Maliniak, D.; Ploense, K.; White, R.J.; Woodward, N.; Hsieh, K.; Bonham, A.J.; Eisenstein, M.; Kippin, T.E.; et al. Real-Time, Aptamer-Based Tracking of Circulating Therapeutic Agents in Living Animals. Sci. Transl. Med. 2013, 5, 213ra165. [Google Scholar] [CrossRef] [PubMed]
- Arroyo-Currás, N.; Somerson, J.; Vieira, P.A.; Ploense, K.L.; Kippin, T.E.; Plaxco, K.W. Real-Time Measurement of Small Molecules Directly in Awake, Ambulatory Animals. Proc. Natl. Acad. Sci. USA 2017, 114, 645–650. [Google Scholar] [CrossRef]
- Fetter, L.; Richards, J.; Daniel, J.; Roon, L.; Rowland, T.J.; Bonham, A.J. Electrochemical Aptamer Scaffold Biosensors for Detection of Botulism and Ricin Toxins. Chem. Commun. 2015, 51, 15137–15140. [Google Scholar] [CrossRef] [PubMed]
- Lubin, A.A.; Lai, R.Y.; Baker, B.R.; Heeger, A.J.; Plaxco, K.W. Sequence-Specific, Electronic Detection of Oligonucleotides in Blood, Soil, and Foodstuffs with the Reagentless, Reusable E-DNA Sensor. Anal. Chem. 2006, 78, 5671–5677. [Google Scholar] [CrossRef]
- White, R.J.; Rowe, A.A.; Plaxco, K.W. Re-Engineering Aptamers to Support Reagentless, Self-Reporting Electrochemical Sensors. Analyst 2010, 135, 589–594. [Google Scholar] [CrossRef]
- Lubin, A.A.; Hunt, B.V.; White, R.J.; Plaxco, K.W. Effects of Probe Length, Probe Geometry, and Redox-Tag Placement on the Performance of the Electrochemical E-DNA Sensor. Anal. Chem. 2009, 81, 2150–2158. [Google Scholar] [CrossRef]
- White, R.J.; Plaxco, K.W. Exploiting Binding-Induced Changes in Probe Flexibility for the Optimization of Electrochemical Biosensors. Anal. Chem. 2009, 82, 73–76. [Google Scholar] [CrossRef]
- Lokesh, G.L.; Wang, H.; Lam, C.H.; Thiviyanathan, V.; Ward, N.; Gorenstein, D.G.; Volk, D.E. X-Aptamer Selection and Validation. In RNA Nanostructures: Methods and Protocols; Bindewald, E., Shapiro, B.A., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2017; pp. 151–174. ISBN 978-1-4939-7138-1. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Xu, Z.Z.; Mathews, D.H. Secondary Structure Prediction of Single Sequences Using RNA structure. Methods Mol. Biol. 2016, 1490, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Bonham, A.J.; Hsieh, K.; Ferguson, B.S.; Valle-Belisle, A.; Ricci, F.; Soh, H.T.; Plaxco, K.W. Quantification of Transcription Factor Binding in Cell Extracts Using an Electrochemical, Structure-Switching Biosensor. J. Am. Chem. Soc. 2012, 134, 3346–3348. [Google Scholar] [CrossRef] [PubMed]
- Vallée-Bélisle, A.; Plaxco, K.W. Structure-Switching Biosensors: Inspired by Nature. Curr. Opin. Struct. Biol. 2010, 20, 518–526. [Google Scholar] [CrossRef]
- Markham, N.R.; Zuker, M. UNAFold: Software for Nucleic Acid Folding and Hybridization. Methods Mol. Biol. 2008, 453, 3–31. [Google Scholar] [CrossRef] [PubMed]
- Dauphin-Ducharme, P.; Plaxco, K.W. Maximizing the Signal Gain of Electrochemical-DNA Sensors. Anal. Chem. 2016, 88, 11654–11662. [Google Scholar] [CrossRef]
- Rowe, A.A.; White, R.J.; Bonham, A.J.; Plaxco, K.W. Fabrication of Electrochemical-DNA Biosensors for the Reagentless Detection of Nucleic Acids, Proteins and Small Molecules. J. Vis. Exp. 2011, 52, e2922. [Google Scholar] [CrossRef]
- Butterworth, A.; Blues, E.; Williamson, P.; Cardona, M.; Gray, L.; Corrigan, D.K. SAM Composition and Electrode Roughness Affect Performance of a DNA Biosensor for Antibiotic Resistance. Biosensors 2019, 9, 22. [Google Scholar] [CrossRef]
- White, R.J.; Phares, N.; Lubin, A.A.; Xiao, Y.; Plaxco, K.W. Optimization of Electrochemical Aptamer-Based Sensors via Optimization of Probe Packing Density and Surface Chemistry. Langmuir 2008, 24, 10513–10518. [Google Scholar] [CrossRef]
- Armbruster, D.A.; Pry, T. Limit of Blank, Limit of Detection and Limit of Quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar]
- Markham, N.R.; Zuker, M. DINAMelt Web Server for Nucleic Acid Melting Prediction. Nucleic Acids Res. 2005, 33, W577–W581. [Google Scholar] [CrossRef]
- Taggart, D.J.; Morré, D.J. Estimation of Lower Limit of Detection of ENOX2 in Serum. ONCOblot Rep. 2015, 1. Available online: https://prevent-cancer.ca/wp-content/uploads/2015/12/ONCOblot-Reports-Vol-1-No-5-September-2015.pdf (accessed on 12 January 2022).
- Rowe, A.A.; Bonham, A.J.; White, R.J.; Zimmer, M.P.; Yadgar, R.J.; Hobza, T.M.; Honea, J.W.; Ben-Yaacov, I.; Plaxco, K.W. CheapStat: An Open-Source, “Do-It-Yourself” Potentiostat for Analytical and Educational Applications. PLoS ONE 2011, 6, e23783. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quansah, M.; Fetter, L.; Fineran, A.; Colling, H.V.; Silver, K.; Rowland, T.J.; Bonham, A.J. Rapid and Quantitative Detection of Lung Cancer Biomarker ENOX2 Using a Novel Aptamer in an Electrochemical DNA-Based (E-DNA) Biosensor. Biosensors 2023, 13, 675. https://doi.org/10.3390/bios13070675
Quansah M, Fetter L, Fineran A, Colling HV, Silver K, Rowland TJ, Bonham AJ. Rapid and Quantitative Detection of Lung Cancer Biomarker ENOX2 Using a Novel Aptamer in an Electrochemical DNA-Based (E-DNA) Biosensor. Biosensors. 2023; 13(7):675. https://doi.org/10.3390/bios13070675
Chicago/Turabian StyleQuansah, Mary, Lisa Fetter, Autumn Fineran, Haley V. Colling, Keaton Silver, Teisha J. Rowland, and Andrew J. Bonham. 2023. "Rapid and Quantitative Detection of Lung Cancer Biomarker ENOX2 Using a Novel Aptamer in an Electrochemical DNA-Based (E-DNA) Biosensor" Biosensors 13, no. 7: 675. https://doi.org/10.3390/bios13070675
APA StyleQuansah, M., Fetter, L., Fineran, A., Colling, H. V., Silver, K., Rowland, T. J., & Bonham, A. J. (2023). Rapid and Quantitative Detection of Lung Cancer Biomarker ENOX2 Using a Novel Aptamer in an Electrochemical DNA-Based (E-DNA) Biosensor. Biosensors, 13(7), 675. https://doi.org/10.3390/bios13070675








