Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases
Abstract
1. Introduction
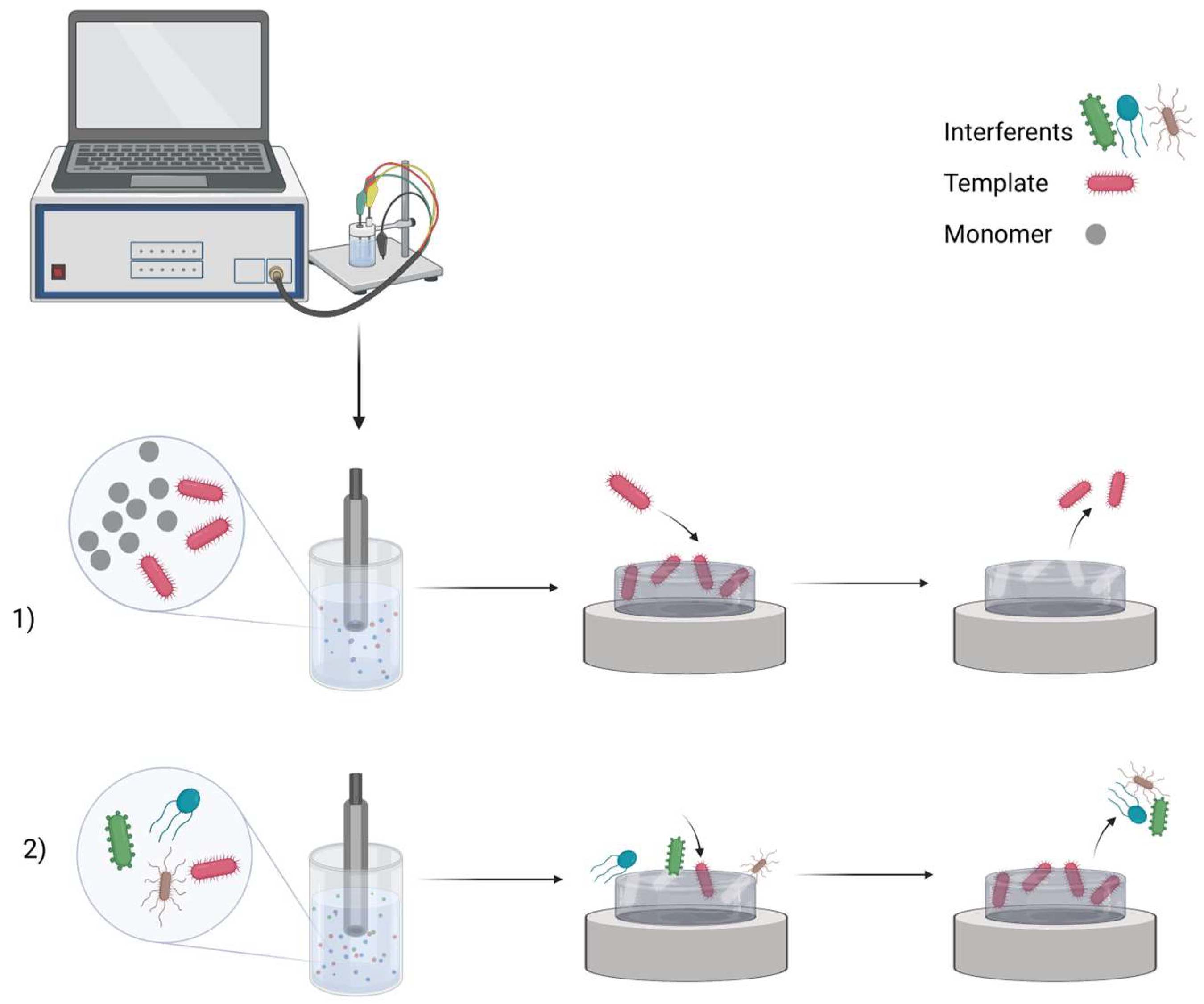
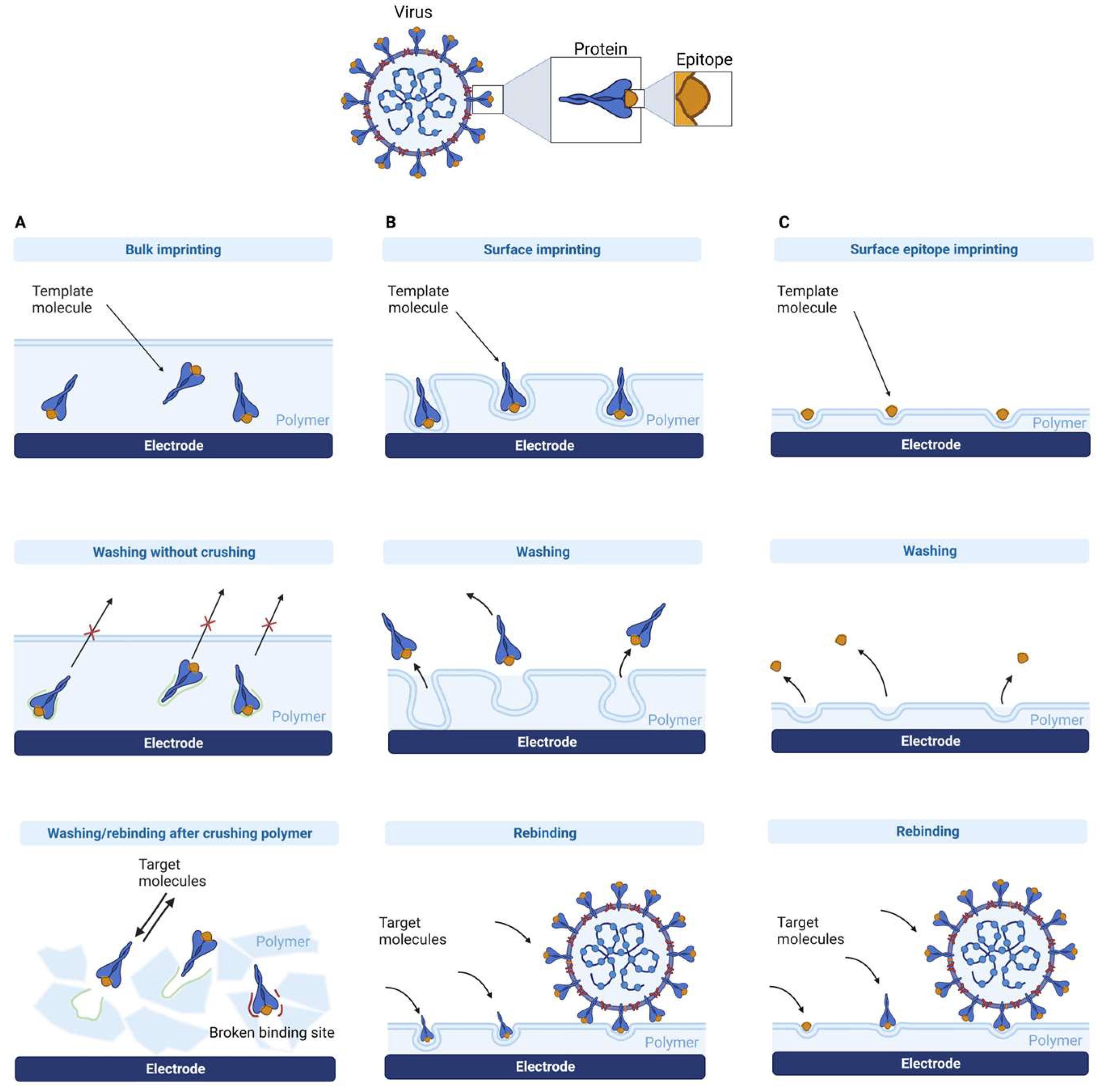
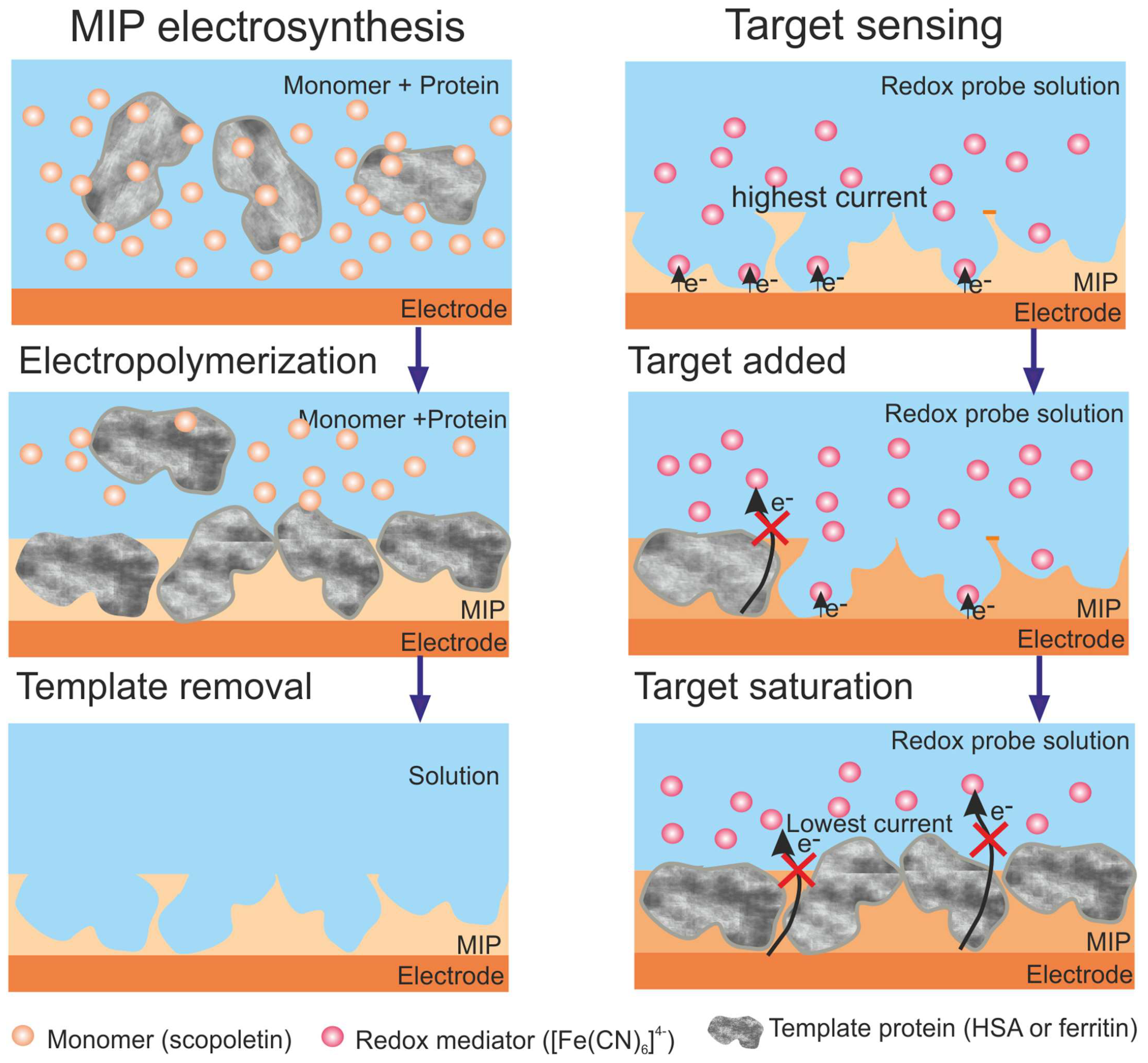
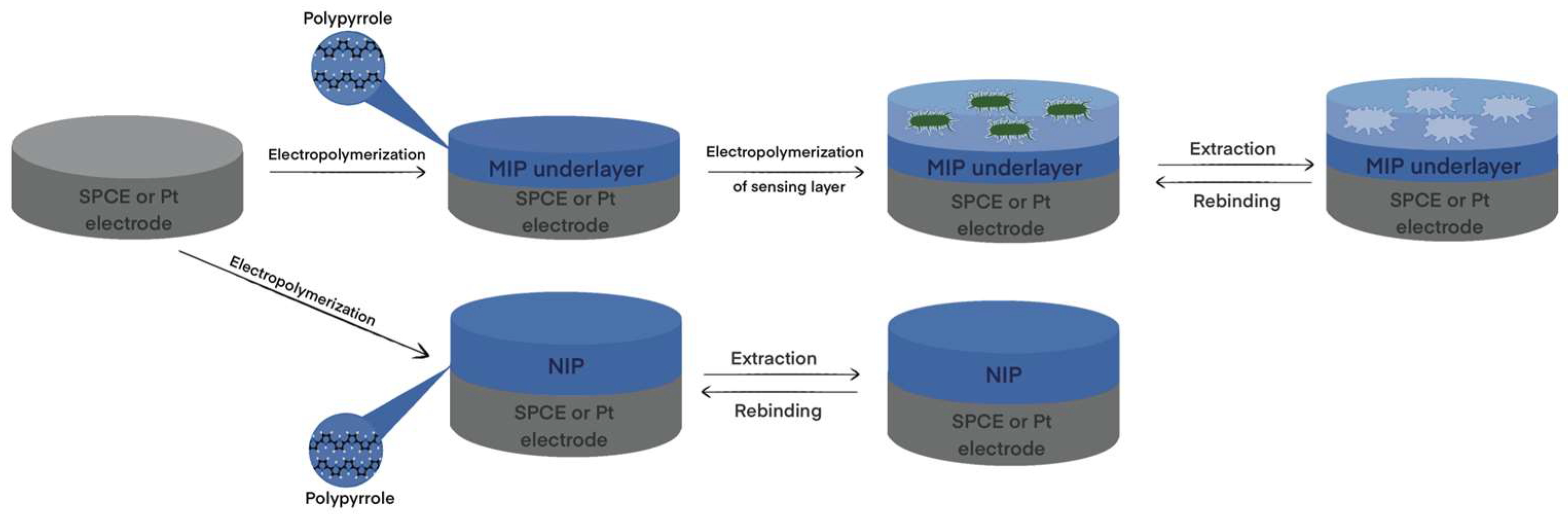
2. MIP Formation Principles
3. MIP Application for Detection of Biomarkers of Inflammation and Sepsis
3.1. Human Serum Albumin (HSA)
3.2. Acute-Phase Proteins
3.2.1. C-Reactive Protein (CRP)
3.2.2. Serum Amyloid-A (SAA)
3.3. Cytokines
3.3.1. Tumor Necrosis Factor (TNF-α)
3.3.2. Interleukin-6 (IL-6)
3.3.3. Interleukin-1β (IL-1β)
3.3.4. Interleukin-2 (IL-2)
4. MIP Application for the Detection of Biomarkers of Infectious Diseases
4.1. HIV-1
4.2. COVID-19
4.3. Dengue Virus
4.4. Hepatitis C Virus
4.5. Nosocomial Infections
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Naseri, M.; Fotouhi, L.; Ehsani, A. Recent progress in the development of conducting polymer-based nanocomposites for electrochemical biosensors applications: A mini-review. Chem. Rec. 2018, 18, 599–618. [Google Scholar] [CrossRef] [PubMed]
- Ramanavicius, S.; Jagminas, A.; Ramanavicius, A. Advances in molecularly imprinted polymers based affinity sensors (review). Polymers 2021, 13, 974. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xu, S.; Li, J. Recent advances in molecular imprinting technology: Current status, challenges and highlighted applications. Chem. Soc. Rev. 2011, 40, 2922–2942. [Google Scholar] [CrossRef]
- Lakard, B. Electrochemical biosensors based on conducting polymers: A review. Appl. Sci. 2020, 10, 6614. [Google Scholar] [CrossRef]
- Li, J.; Sun, D. Molecularly imprinted ratiometric fluorescence nanosensors. Langmuir 2022, 38, 13305–13312. [Google Scholar] [CrossRef]
- Emir, G.; Dilgin, Y.; Ramanaviciene, A.; Ramanavicius, A. Amperometric nonenzymatic glucose biosensor based on graphite rod electrode modified by Ni-nanoparticle/polypyrrole composite. Microchem. J. 2021, 161, 105751. [Google Scholar] [CrossRef]
- Ratautaite, V.; Plausinaitis, D.; Baleviciute, I.; Mikoliunaite, L.; Ramanaviciene, A.; Ramanavicius, A. Characterization of caffeine-imprinted polypyrrole by a quartz crystal microbalance and electrochemical impedance spectroscopy. Sens. Actuator B-Chem. 2015, 212, 63–71. [Google Scholar] [CrossRef]
- Holguín, M.; Rojas Álvarez, O.E.; Arizabaleta, C.A.; Torres, W. Molecular dynamics of the interaction of l-tryptophan with polypyrrole oligomers. Comput. Theor. Chem. 2019, 1147, 29–34. [Google Scholar] [CrossRef]
- Kumar, V.; Mirzaei, A.; Bonyani, M.; Kim, K.-H.; Kim, H.W.; Kim, S.S. Advances in electrospun nanofiber fabrication for polyaniline (PANI)-based chemoresistive sensors for gaseous ammonia. TRAC-Trends Anal. Chem. 2020, 129, 115938. [Google Scholar] [CrossRef]
- Tekbaşoğlu, T.Y.; Soganci, T.; Ak, M.; Koca, A.; Şener, M.K. Enhancing biosensor properties of conducting polymers via copolymerization: Synthesis of EDOT-substituted bis(2-pyridylimino)isoindolato-palladium complex and electrochemical sensing of glucose by its copolymerized film. Biosens. Bioelectron. 2017, 87, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Pontes, K.; Indrusiak, T.; Soares, B.G. Poly(vinylidene fluoride-co-hexafluorpropylene)/polyaniline conductive blends: Effect of the mixing procedure on the electrical properties and electromagnetic interference shielding effectiveness. J. Appl. Polym. Sci. 2021, 138, 49705. [Google Scholar] [CrossRef]
- Samukaite-Bubniene, U.; Valiūnienė, A.; Bucinskas, V.; Genys, P.; Ratautaite, V.; Ramanaviciene, A.; Aksun, E.; Tereshchenko, A.; Zeybek, B.; Ramanavicius, A. Towards supercapacitors: Cyclic voltammetry and fast Fourier transform electrochemical impedance spectroscopy based evaluation of polypyrrole electrochemically deposited on the pencil graphite electrode. Colloid Surf. A 2021, 610, 125750. [Google Scholar] [CrossRef]
- Zhao, Z.; Yu, T.; Miao, Y.; Zhao, X. Chloride ion-doped polyaniline/carbon nanotube nanocomposite materials as new cathodes for chloride ion battery. Electrochim. Acta 2018, 270, 30–36. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, Y.; Liu, Y.; Liu, W.; Zhao, P.; Li, Y.; Dong, Y.; Wang, H.; Yang, J. Urchin-like Ni1/3Co2/3(CO3)0.5OH·0.11H2O anchoring on polypyrrole nanotubes for supercapacitor electrodes. Electrochim. Acta 2019, 295, 989–996. [Google Scholar] [CrossRef]
- Ratautaite, V.; Ramanaviciene, A.; Oztekin, Y.; Voronovic, J.; Balevicius, Z.; Mikoliunaite, L.; Ramanavicius, A. Electrochemical stability and repulsion of polypyrrole film. Colloid Surf. A 2013, 418, 16–21. [Google Scholar] [CrossRef]
- Iroh, J.O.; Su, W. Corrosion performance of polypyrrole coating applied to low carbon steel by an electrochemical process. Electrochim. Acta 2000, 46, 15–24. [Google Scholar] [CrossRef]
- Leonavicius, K.; Ramanaviciene, A.; Ramanavicius, A. Polymerization model for hydrogen peroxide initiated synthesis of polypyrrole nanoparticles. Langmuir 2011, 27, 10970–10976. [Google Scholar] [CrossRef]
- Felix, F.S.; Angnes, L. Electrochemical immunosensors—A powerful tool for analytical applications. Biosens. Bioelectron. 2018, 102, 470–478. [Google Scholar] [CrossRef]
- Ramanavicius, A.; Andriukonis, E.; Stirke, A.; Mikoliunaite, L.; Balevicius, Z.; Ramanaviciene, A. Synthesis of polypyrrole within the cell wall of yeast by redox-cycling of [Fe(CN)6]3−/[Fe(CN)6]4−. Enzym. Microb. Technol. 2016, 83, 40–47. [Google Scholar] [CrossRef]
- Apetrei, R.-M.; Carac, G.; Bahrim, G.; Ramanaviciene, A.; Ramanavicius, A. Modification of Aspergillus niger by conducting polymer, Polypyrrole, and the evaluation of electrochemical properties of modified cells. Bioelectrochemistry 2018, 121, 46–55. [Google Scholar] [CrossRef]
- Apetrei, R.-M.; Carac, G.; Ramanaviciene, A.; Bahrim, G.; Tanase, C.; Ramanavicius, A. Cell-assisted synthesis of conducting polymer-polypyrrole-for the improvement of electric charge transfer through fungal cell wall. Colloids Surf. B Biointerfaces 2019, 175, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Ramanavicius, A.; Kausaite, A.; Ramanaviciene, A. Self-encapsulation of oxidases as a basic approach to tune the upper detection limit of amperometric biosensors. Analyst 2008, 133, 1083–1089. [Google Scholar] [CrossRef]
- Lakard, B.; Magnin, D.; Deschaume, O.; Vanlancker, G.; Glinel, K.; Demoustier-Champagne, S.; Nysten, B.; Jonas, A.M.; Bertrand, P.; Yunus, S. Urea potentiometric enzymatic biosensor based on charged biopolymers and electrodeposited polyaniline. Biosens. Bioelectron. 2011, 26, 4139–4145. [Google Scholar] [CrossRef]
- German, N.; Ramanavicius, A.; Voronovic, J.; Ramanaviciene, A. Glucose biosensor based on glucose oxidase and gold nanoparticles of different sizes covered by polypyrrole layer. Colloid Surf. A 2012, 413, 224–230. [Google Scholar] [CrossRef]
- Ramanavicius, A.; Oztekin, Y.; Ramanaviciene, A. Electrochemical formation of polypyrrole-based layer for immunosensor design. Sens. Actuator B-Chem. 2014, 197, 237–243. [Google Scholar] [CrossRef]
- Ramanaviciene, A.; Ramanavicius, A. Pulsed amperometric detection of DNA with an ssDNA/polypyrrole-modified electrode. Anal. Bioanal. Chem. 2004, 379, 287–293. [Google Scholar] [CrossRef]
- Plikusiene, I.; Balevicius, Z.; Ramanaviciene, A.; Talbot, J.; Mickiene, G.; Balevicius, S.; Stirke, A.; Tereshchenko, A.; Tamosaitis, L.; Zvirblis, G.; et al. Evaluation of affinity sensor response kinetics towards dimeric ligands linked with spacers of different rigidity: Immobilized recombinant granulocyte colony-stimulating factor based synthetic receptor binding with genetically engineered dimeric analyte derivatives. Biosens. Bioelectron. 2020, 156, 112112. [Google Scholar] [CrossRef]
- Baleviciute, I.; Ratautaite, V.; Ramanaviciene, A.; Balevicius, Z.; Broeders, J.; Croux, D.; McDonald, M.; Vahidpour, F.; Thoelen, R.; Ceuninck, W.D.; et al. Evaluation of theophylline imprinted polypyrrole film. Synth. Met. 2015, 209, 206–211. [Google Scholar] [CrossRef]
- Kryscio, D.R.; Peppas, N.A. Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 2012, 8, 461–473. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, G.; Han, K.; Sun, D.; Zhou, N.; Song, Z.; Liu, H.; Li, J.; Li, G. Applications of molecular imprinting technology in the study of traditional Chinese medicine. Molecules 2023, 28, 301. [Google Scholar] [CrossRef]
- Lowdon, J.W.; Diliën, H.; Singla, P.; Peeters, M.; Cleij, T.J.; van Grinsven, B.; Eersels, K. MIPs for commercial application in low-cost sensors and assays—An overview of the current status quo. Sens. Actuator B-Chem. 2020, 325, 128973. [Google Scholar] [CrossRef]
- Mustafa, Y.L.; Keirouz, A.; Leese, H.S. Molecularly imprinted polymers in diagnostics: Accessing analytes in biofluids. J. Mater. Chem. B 2022, 10, 7418–7449. [Google Scholar] [CrossRef]
- Sangiorgi, N.; Sangiorgi, A.; Tarterini, F.; Sanson, A. Molecularly imprinted polypyrrole counter electrode for gel-state dye-sensitized solar cells. Electrochim. Acta 2019, 305, 322–328. [Google Scholar] [CrossRef]
- Syritski, V.; Reut, J.; Öpik, A.; Idla, K. Environmental QCM sensors coated with polypyrrole. Synth. Met. 1999, 102, 1326–1327. [Google Scholar] [CrossRef]
- Rebelo, P.; Costa-Rama, E.; Seguro, I.; Pacheco, J.G.; Nouws, H.P.A.; Cordeiro, M.N.D.S.; Delerue-Matos, C. Molecularly imprinted polymer-based electrochemical sensors for environmental analysis. Biosens. Bioelectron. 2021, 172, 112719. [Google Scholar] [CrossRef]
- Keçili, R.; Yılmaz, E.; Ersöz, A.; Say, R. Chapter 12-imprinted materials: From green chemistry to sustainable engineering. In Sustainable Nanoscale Engineering; Szekely, G., Livingston, A., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 317–350. [Google Scholar]
- Cui, F.; Zhou, Z.; Zhou, H.S. Molecularly imprinted polymers and surface imprinted polymers based electrochemical biosensor for infectious diseases. Sensors 2020, 20, 996. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Dempsey-Hibbert, N.C.; Peeters, M.; Tridente, A.; Banks, C.E. Molecularly imprinted polymer based electrochemical biosensors: Overcoming the challenges of detecting vital biomarkers and speeding up diagnosis. Talanta Open 2020, 2, 100018. [Google Scholar] [CrossRef]
- Crapnell, R.D.; Hudson, A.; Foster, C.W.; Eersels, K.; Grinsven, B.v.; Cleij, T.J.; Banks, C.E.; Peeters, M. Recent advances in electrosynthesized molecularly imprinted polymer sensing platforms for bioanalyte detection. Sensors 2019, 19, 1204. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Ahmad, A.; Mehboob, R. Nosocomial infections and their control strategies. Asian Pac. J. Trop. Biomed. 2015, 5, 509–514. [Google Scholar] [CrossRef]
- Chuang, Y.-C.; Sheng, W.-H.; Li, S.-Y.; Lin, Y.-C.; Wang, J.-T.; Chen, Y.-C.; Chang, S.-C. Influence of genospecies of Acinetobacter baumannii complex on clinical outcomes of patients with Acinetobacter bacteremia. Clin. Infect. Dis. 2011, 52, 352–360. [Google Scholar] [CrossRef] [PubMed]
- McElrath, M.J.; Haynes, B.F. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity 2010, 33, 542–554. [Google Scholar] [CrossRef]
- Manns, M.P.; Buti, M.; Gane, E.; Pawlotsky, J.-M.; Razavi, H.; Terrault, N.; Younossi, Z. Hepatitis C virus infection. Nat. Rev. Dis. Prim. 2017, 3, 17006. [Google Scholar] [CrossRef]
- Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M.; Falzone, L.; Gattuso, G.; Tsatsakis, A.; Spandidos, D.A.; Libra, M.; et al. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int. J. Mol. Med. 2021, 47, 100. [Google Scholar] [CrossRef]
- Ratautaite, V.; Topkaya, S.N.; Mikoliunaite, L.; Ozsoz, M.; Oztekin, Y.; Ramanaviciene, A.; Ramanavicius, A. Molecularly imprinted polypyrrole for DNA determination. Electroanalysis 2013, 25, 1169–1177. [Google Scholar] [CrossRef]
- Ratautaite, V.; Brazys, E.; Ramanaviciene, A.; Ramanavicius, A. Electrochemical sensors based on L-tryptophan molecularly imprinted polypyrrole and polyaniline. J. Electroanal. Chem. 2022, 917, 116389. [Google Scholar] [CrossRef]
- Ratautaite, V.; Janssens, S.D.; Haenen, K.; Nesládek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A. Molecularly imprinted polypyrrole based impedimetric sensor for theophylline determination. Electrochim. Acta 2014, 130, 361–367. [Google Scholar] [CrossRef]
- Ratautaite, V.; Nesladek, M.; Ramanaviciene, A.; Baleviciute, I.; Ramanavicius, A. Evaluation of histamine imprinted polypyrrole deposited on boron doped nanocrystalline diamond. Electroanalysis 2014, 26, 2458–2464. [Google Scholar] [CrossRef]
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Ramanaviciene, A.; Ciplys, E.; Juozapaitis, M.; Slibinskas, R.; Bechelany, M.; Ramanavicius, A. Molecularly imprinted polypyrrole based sensor for the detection of SARS-CoV-2 spike glycoprotein. Electrochim. Acta 2022, 403, 139581. [Google Scholar] [CrossRef]
- Ratautaite, V.; Boguzaite, R.; Brazys, E.; Plausinaitis, D.; Ramanavicius, S.; Samukaite-Bubniene, U.; Bechelany, M.; Ramanavicius, A. Evaluation of the interaction between SARS-CoV-2 spike glycoproteins and the molecularly imprinted polypyrrole. Talanta 2023, 253, 123981. [Google Scholar] [CrossRef]
- Plausinaitis, D.; Ratautaite, V.; Mikoliunaite, L.; Sinkevicius, L.; Ramanaviciene, A.; Ramanavicius, A. Quartz crystal microbalance-based evaluation of the electrochemical formation of an aggregated polypyrrole particle-based layer. Langmuir 2015, 31, 3186–3193. [Google Scholar] [CrossRef]
- Plausinaitis, D.; Sinkevicius, L.; Samukaite-Bubniene, U.; Ratautaite, V.; Ramanavicius, A. Evaluation of electrochemical quartz crystal microbalance based sensor modified by uric acid-imprinted polypyrrole. Talanta 2020, 220, 121414. [Google Scholar] [CrossRef]
- Balciunas, D.; Plausinaitis, D.; Ratautaite, V.; Ramanaviciene, A.; Ramanavicius, A. Towards electrochemical surface plasmon resonance sensor based on the molecularly imprinted polypyrrole for glyphosate sensing. Talanta 2022, 241, 123252. [Google Scholar] [CrossRef] [PubMed]
- Ratautaite, V.; Samukaite-Bubniene, U.; Plausinaitis, D.; Boguzaite, R.; Balciunas, D.; Ramanaviciene, A.; Neunert, G.; Ramanavicius, A. Molecular imprinting technology for determination of uric acid. Int. J. Mol. Sci. 2021, 22, 5032. [Google Scholar] [CrossRef]
- Ramanaviciene, A.; Ramanavicius, A. Molecularly imprinted polypyrrole-based synthetic receptor for direct detection of bovine leukemia virus glycoproteins. Biosens. Bioelectron. 2004, 20, 1076–1082. [Google Scholar] [CrossRef] [PubMed]
- El Sharif, H.F.; Dennison, S.R.; Tully, M.; Crossley, S.; Mwangi, W.; Bailey, D.; Graham, S.P.; Reddy, S.M. Evaluation of electropolymerized molecularly imprinted polymers (E-MIPs) on disposable electrodes for detection of SARS-CoV-2 in saliva. Anal. Chim. Acta 2022, 1206, 339777. [Google Scholar] [CrossRef] [PubMed]
- Pintavirooj, C.; Vongmanee, N.; Sukjee, W.; Sangma, C.; Visitsattapongse, S. Biosensors for Klebsiella pneumoniae with molecularly imprinted polymer (MIP) technique. Sensors 2022, 22, 4638. [Google Scholar] [CrossRef]
- Dabrowski, M.; Lach, P.; Cieplak, M.; Kutner, W. Nanostructured molecularly imprinted polymers for protein chemosensing. Biosens. Bioelectron. 2018, 102, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Ansari, S.; Masoum, S. Molecularly imprinted polymers for capturing and sensing proteins: Current progress and future implications. TRAC-Trends Anal. Chem. 2019, 114, 29–47. [Google Scholar] [CrossRef]
- Stojanovic, Z.; Erdőssy, J.; Keltai, K.; Scheller, F.W.; Gyurcsányi, R.E. Electrosynthesized molecularly imprinted polyscopoletin nanofilms for human serum albumin detection. Anal. Chim. Acta 2017, 977, 1–9. [Google Scholar] [CrossRef]
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sens. Actuator B-Chem. 2022, 353, 131160. [Google Scholar] [CrossRef]
- Land, K.J.; Boeras, D.I.; Chen, X.-S.; Ramsay, A.R.; Peeling, R.W. REASSURED diagnostics to inform disease control strategies, strengthen health systems and improve patient outcomes. Nat. Microbiol. 2019, 4, 46–54. [Google Scholar] [CrossRef]
- Medina-Plaza, C.; García-Hernández, C.; de Saja, J.A.; Fernández-Escudero, J.A.; Barajas, E.; Medrano, G.; García-Cabezón, C.; Martin-Pedrosa, F.; Rodriguez-Mendez, M.L. The advantages of disposable screen-printed biosensors in a bioelectronic tongue for the analysis of grapes. LWT-Food Sci. Technol. 2015, 62, 940–947. [Google Scholar] [CrossRef]
- Wynn, J.L.; Wong, H.R.; Shanley, T.P.; Bizzarro, M.J.; Saiman, L.; Polin, R.A. Time for a neonatal-specific consensus definition for sepsis. Pediatr. Crit. Care Med. 2014, 15, 523–528. [Google Scholar] [CrossRef]
- Kumar, D.; Prasad, B.B. Multiwalled carbon nanotubes embedded molecularly imprinted polymer-modified screen printed carbon electrode for the quantitative analysis of C-reactive protein. Sens. Actuator B-Chem. 2012, 171–172, 1141–1150. [Google Scholar] [CrossRef]
- Balayan, S.; Chauhan, N.; Chandra, R.; Jain, U. Molecular imprinting based electrochemical biosensor for identification of serum amyloid A (SAA), a neonatal sepsis biomarker. Int. J. Biol. Macromol. 2022, 195, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Balayan, S.; Chauhan, N.; Kumar, P.; Chandra, R.; Jain, U. Fabrication of a sensing platform for identification of tumor necrosis factor-alpha: A biomarker for neonatal sepsis. 3 Biotech 2022, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Sener, G.; Ozgur, E.; Rad, A.Y.; Uzun, L.; Say, R.; Denizli, A. Rapid real-time detection of procalcitonin using a microcontact imprinted surface plasmon resonance biosensor. Analyst 2013, 138, 6422–6428. [Google Scholar] [CrossRef]
- Gonçalves, M.d.L.; Truta, L.A.N.; Sales, M.G.F.; Moreira, F.T.C. Electrochemical point-of care (PoC) determination of interleukin-6 (IL-6) using a pyrrole (Py) molecularly imprinted polymer (MIP) on a carbon-screen printed electrode (C-SPE). Anal. Lett. 2021, 54, 2611–2623. [Google Scholar] [CrossRef]
- Cardoso, A.R.; de Sá, M.H.; Sales, M.G.F. An impedimetric molecularly-imprinted biosensor for Interleukin-1β determination, prepared by in-situ electropolymerization on carbon screen-printed electrodes. Bioelectrochemistry 2019, 130, 107287. [Google Scholar] [CrossRef]
- ABIM (American Board of Internal Medicine) Laboratory Test Reference Ranges—January 2022. Available online: https://www.abim.org/Media/bfijryql/laboratory-reference-ranges.pdf (accessed on 21 December 2022).
- Fujii, T.; Tokuda, S.; Nakazawa, Y.; Kurozumi, S.; Obayashi, S.; Yajima, R.; Shirabe, K. Implications of low serum albumin as a prognostic factor of long-term outcomes in patients with breast cancer. Vivo 2020, 34, 2033–2036. [Google Scholar] [CrossRef]
- Yin, M.; Si, L.; Qin, W.; Li, C.; Zhang, J.; Yang, H.; Han, H.; Zhang, F.; Ding, S.; Zhou, M.; et al. Predictive value of serum albumin level for the prognosis of severe sepsis without exogenous human albumin administration: A prospective cohort study. J. Intensive Care Med. 2018, 33, 687–694. [Google Scholar] [CrossRef]
- Zhang, G.; Yu, Y.; Guo, M.; Lin, B.; Zhang, L. A sensitive determination of albumin in urine by molecularly imprinted electrochemical biosensor based on dual-signal strategy. Sens. Actuator B-Chem. 2019, 288, 564–570. [Google Scholar] [CrossRef]
- Cieplak, M.; Szwabinska, K.; Sosnowska, M.; Chandra, B.K.C.; Borowicz, P.; Noworyta, K.; D’Souza, F.; Kutner, W. Selective electrochemical sensing of human serum albumin by semi-covalent molecular imprinting. Biosens. Bioelectron. 2015, 74, 960–966. [Google Scholar] [CrossRef]
- Brenner, D.R.; Scherer, D.; Muir, K.; Schildkraut, J.; Boffetta, P.; Spitz, M.R.; Le Marchand, L.; Chan, A.T.; Goode, E.L.; Ulrich, C.M.; et al. A review of the application of inflammatory biomarkers in epidemiologic cancer research. Cancer Epidemiol. Biomark. Prev. 2014, 23, 1729–1751. [Google Scholar] [CrossRef]
- Anush, M.M.; Ashok, V.K.; Sarma, R.I.; Pillai, S.K. Role of C-reactive protein as an indicator for determining the outcome of sepsis. Indian J. Crit. Care Med. 2019, 23, 11–14. [Google Scholar] [CrossRef] [PubMed]
- Balayan, S.; Chauhan, N.; Rosario, W.; Jain, U. Biosensor development for C-reactive protein detection: A review. Appl. Surf. Sci. Adv. 2022, 12, 100343. [Google Scholar] [CrossRef]
- Balayan, S.; Chauhan, N.; Chandra, R.; Jain, U. Electrochemical based C-reactive protein (CRP) sensing through molecularly imprinted polymer (MIP) pore structure coupled with bi-metallic tuned screen-printed electrode. Biointerface Res. Appl. Chem. 2022, 12, 7697–7714. [Google Scholar] [CrossRef]
- Lee, M.-H.; Liu, K.-H.; Thomas, J.L.; Chen, C.-Y.; Chen, C.-Y.; Yang, C.-H.; Lin, H.-Y. Doping of MXenes enhances the electrochemical response of peptide-imprinted conductive polymers for the recognition of C-Reactive protein. Biosens. Bioelectron. 2022, 200, 113930. [Google Scholar] [CrossRef] [PubMed]
- Cui, M.; Che, Z.; Gong, Y.; Li, T.; Hu, W.; Wang, S. A graphdiyne-based protein molecularly imprinted biosensor for highly sensitive human C-reactive protein detection in human serum. Chem. Eng. J. 2022, 431, 133455. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, S.; Ren, J.; Han, F.; Yu, X.; Tang, F.; Xue, F.; Chen, W.; Yang, J.; Jiang, Y.; et al. Facile construction of a molecularly imprinted polymer–based electrochemical sensor for the detection of milk amyloid A. Microchim. Acta 2020, 187, 642. [Google Scholar] [CrossRef]
- Lv, Y.B.; Wang, F.F.; Li, N.; Wu, R.L.; Li, J.J.; Shen, H.B.; Li, L.S.; Guo, F. Development of dual quantum dots-based fluorescence-linked immunosorbent assay for simultaneous detection on inflammation biomarkers. Sens. Actuator B-Chem. 2019, 301, 127118. [Google Scholar] [CrossRef]
- Liu, X.; Yang, X.; Li, K.; Liu, H.; Xiao, R.; Wang, W.; Wang, C.; Wang, S. Fe3O4@Au SERS tags-based lateral flow assay for simultaneous detection of serum amyloid A and C-reactive protein in unprocessed blood sample. Sens. Actuator B-Chem. 2020, 320, 128350. [Google Scholar] [CrossRef]
- Xia, C.; Li, Y.; Yuan, G.; Guo, Y.; Yu, C. Immunoassay for serum amyloid A using a glassy carbon electrode modified with carboxy-polypyrrole, multiwalled carbon nanotubes, ionic liquid and chitosan. Microchim. Acta 2015, 182, 1395–1402. [Google Scholar] [CrossRef]
- Mercogliano, M.F.; Bruni, S.; Elizalde, P.V.; Schillaci, R. Tumor necrosis factor α blockade: An opportunity to tackle breast cancer. Front. Oncol. 2020, 10, 584. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in Inflammation, Immunity, and Disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Ma, L.; Zhang, H.; Yin, Y.L.; Guo, W.Z.; Ma, Y.Q.; Wang, Y.B.; Shu, C.; Dong, L.Q. Role of interleukin-6 to differentiate sepsis from non-infectious systemic inflammatory response syndrome. Cytokine 2016, 88, 126–135. [Google Scholar] [CrossRef]
- Shahkar, L.; Keshtkar, A.; Mirfazeli, A.; Ahani, A.; Roshandel, G. The role of IL-6 for predicting neonatal sepsis: A systematic review and meta-analysis. Iran. J. Pediatr. 2011, 21, 411. [Google Scholar] [PubMed]
- Guirao, J.J.; Cabrera, C.M.; Jiménez, N.; Rincón, L.; Urra, J.M. High serum IL-6 values increase the risk of mortality and the severity of pneumonia in patients diagnosed with COVID-19. Mol. Immunol. 2020, 128, 64–68. [Google Scholar] [CrossRef]
- Santa Cruz, A.; Mendes-Frias, A.; Oliveira, A.I.; Dias, L.; Matos, A.R.; Carvalho, A.; Capela, C.; Pedrosa, J.; Castro, A.G.; Silvestre, R. Interleukin-6 is a biomarker for the development of fatal severe acute respiratory syndrome coronavirus 2 pneumonia. Front. Immunol. 2021, 12, 613422. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.; Correia, B.P.; Sharma, S.; Moreira, F.T.C. Molecular imprinted polymers on microneedle arrays for Point of Care transdermal sampling and sensing of inflammatory biomarkers. ACS Omega 2022, 7, 39039–39044. [Google Scholar] [CrossRef]
- Lopez-Castejon, G.; Brough, D. Understanding the mechanism of IL-1β secretion. Cytokine Growth Factor Rev. 2011, 22, 189–195. [Google Scholar] [CrossRef]
- Choi, D.Y.; Yang, J.C.; Hong, S.W.; Park, J. Molecularly imprinted polymer-based electrochemical impedimetric sensors on screen-printed carbon electrodes for the detection of trace cytokine IL-1β. Biosens. Bioelectron. 2022, 204, 114073. [Google Scholar] [CrossRef]
- Piloto, A.M.L.; Ribeiro, D.S.M.; Rodrigues, S.S.M.; Santos, J.L.M.; Ferreira Sales, M.G. Label-free quantum dot conjugates for human protein IL-2 based on molecularly imprinted polymers. Sens. Actuator B-Chem. 2020, 304, 127343. [Google Scholar] [CrossRef]
- Jamalipour Soufi, G.; Iravani, S.; Varma, R.S. Molecularly imprinted polymers for the detection of viruses: Challenges and opportunities. Analyst 2021, 146, 3087–3100. [Google Scholar] [CrossRef]
- He, K.; Chen, C.; Liang, C.; Liu, C.; Yang, B.; Chen, X.; Cai, C. Highly selective recognition and fluorescent detection of JEV via virus-imprinted magnetic silicon microspheres. Sens. Actuator B-Chem. 2016, 233, 607–614. [Google Scholar] [CrossRef]
- Yang, B.; Gong, H.; Chen, C.; Chen, X.; Cai, C. A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus. Biosens. Bioelectron. 2017, 87, 679–685. [Google Scholar] [CrossRef]
- Babamiri, B.; Salimi, A.; Hallaj, R. A molecularly imprinted electrochemiluminescence sensor for ultrasensitive HIV-1 gene detection using EuS nanocrystals as luminophore. Biosens. Bioelectron. 2018, 117, 332–339. [Google Scholar] [CrossRef] [PubMed]
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-based impedimetric sensor for detecting Dengue fever biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Shen, X.-L.; Zeng, Q.; Wang, H.-S.; Wang, L.-S. A multi-walled carbon nanotubes based molecularly imprinted polymers electrochemical sensor for the sensitive determination of HIV-p24. Talanta 2017, 164, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.-H.; Zhang, Y.; Tang, S.-F.; Fang, Z.-B.; Yang, H.-H.; Chen, X.; Chen, G.-N. Sensing HIV related protein using epitope imprinted hydrophilic polymer coated quartz crystal microbalance. Biosens. Bioelectron. 2012, 31, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, X.; Si, L.; Lu, L.; Jiang, S. The development of HIV vaccines targeting gp41 membrane-proximal external region (MPER): Challenges and prospects. Protein Cell 2018, 9, 596–615. [Google Scholar] [CrossRef]
- Nandi, S.; Mondal, A.; Roberts, A.; Gandhi, S. Chapter one-Biosensor platforms for rapid HIV detection. In Advances in Clinical Chemistry; Makowski, G.S., Ed.; Elsevier: Amsterdam, The Netherlands, 2020; Volume 98, pp. 1–34. [Google Scholar]
- Yang, K.; Li, S.; Liu, L.; Chen, Y.; Zhou, W.; Pei, J.; Liang, Z.; Zhang, L.; Zhang, Y. Epitope imprinting technology: Progress, applications, and perspectives toward artificial antibodies. Adv. Mater. 2019, 31, 1902048. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, C.; Wang, M.; Wang, L.-S. Sensitive electrochemical detection of gp120 based on the combination of NBD-556 and gp120. Talanta 2019, 196, 486–492. [Google Scholar] [CrossRef]
- Ou, X.; Liu, Y.; Lei, X.; Li, P.; Mi, D.; Ren, L.; Guo, L.; Guo, R.; Chen, T.; Hu, J.; et al. Author correction: Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2021, 12, 2144. [Google Scholar] [CrossRef]
- Hasseb, A.A.; Abdel Ghani, N.d.T.; Shehab, O.R.; El Nashar, R.M. Application of molecularly imprinted polymers for electrochemical detection of some important biomedical markers and pathogens. Curr. Opin. Electrochem. 2022, 31, 100848. [Google Scholar] [CrossRef]
- Zeng, W.; Liu, G.; Ma, H.; Zhao, D.; Yang, Y.; Liu, M.; Mohammed, A.; Zhao, C.; Yang, Y.; Xie, J.; et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem. Biophys. Res. Commun. 2020, 527, 618–623. [Google Scholar] [CrossRef]
- Raziq, A.; Kidakova, A.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Development of a portable MIP-based electrochemical sensor for detection of SARS-CoV-2 antigen. Biosens. Bioelectron. 2021, 178, 113029. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.A.; Bahrani, S.; Mousavi, S.M.; Omidifar, N.; Behbahan, N.G.G.; Arjmand, M.; Ramakrishna, S.; Lankarani, K.B.; Moghadami, M.; Firoozsani, M. Graphene-based femtogram-level sensitive molecularly imprinted polymer of SARS-CoV-2. Adv. Mater. Interfaces 2021, 8, 2101466. [Google Scholar] [CrossRef] [PubMed]
- Nikitina, V.N.; Zaryanov, N.V.; Kochetkov, I.R.; Karyakina, E.E.; Yatsimirsky, A.K.; Karyakin, A.A. Molecular imprinting of boronate functionalized polyaniline for enzyme-free selective detection of saccharides and hydroxy acids. Sens. Actuator B-Chem. 2017, 246, 428–433. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, L.; Zhang, Y. Highly sensitive electrochemical determination of the SARS-COV-2 antigen based on a gold/graphene imprinted poly-arginine sensor. Anal. Methods 2021, 13, 5772–5776. [Google Scholar] [CrossRef]
- Hussein, H.A.; Kandeil, A.; Gomaa, M.; Mohamed El Nashar, R.; El-Sherbiny, I.M.; Hassan, R.Y.A. SARS-CoV-2-impedimetric biosensor: Virus-imprinted chips for early and rapid diagnosis. ACS Sens. 2021, 6, 4098–4107. [Google Scholar] [CrossRef] [PubMed]
- Tabrizi, M.A.; Fernández-Blázquez, J.P.; Medina, D.M.; Acedo, P. An ultrasensitive molecularly imprinted polymer-based electrochemical sensor for the determination of SARS-CoV-2-RBD by using macroporous gold screen-printed electrode. Biosens. Bioelectron. 2021, 196, 113729. [Google Scholar] [CrossRef]
- Drobysh, M.; Liustrovaite, V.; Baradoke, A.; Rucinskiene, A.; Ramanaviciene, A.; Ratautaite, V.; Viter, R.; Chen, C.-F.; Plikusiene, I.; Samukaite-Bubniene, U.; et al. Electrochemical determination of interaction between SARS-CoV-2 spike protein and specific antibodies. Int. J. Mol. Sci. 2022, 23, 6768. [Google Scholar] [CrossRef] [PubMed]
- Liustrovaite, V.; Drobysh, M.; Rucinskiene, A.; Baradoke, A.; Ramanaviciene, A.; Plikusiene, I.; Samukaite-Bubniene, U.; Viter, R.; Chen, C.-F.; Ramanavicius, A. Towards an electrochemical immunosensor for the detection of antibodies against SARS-CoV-2 spike protein. J. Electrochem. Soc. 2022, 169, 037523. [Google Scholar] [CrossRef]
- Rastogi, M.; Sharma, N.; Singh, S.K. Flavivirus NS1: A multifaceted enigmatic viral protein. Virol. J. 2016, 13, 131. [Google Scholar] [CrossRef]
- Buensuceso, C.E.; Tiu, B.D.B.; Lee, L.P.; Sabido, P.M.G.; Nuesca, G.M.; Caldona, E.B.; del Mundo, F.R.; Advincula, R.C. Electropolymerized-molecularly imprinted polymers (E-MIPS) as sensing elements for the detection of dengue infection. Anal. Bioanal. Chem. 2022, 414, 1347–1357. [Google Scholar] [CrossRef] [PubMed]
- Tai, D.-F.; Lin, C.-Y.; Wu, T.-Z.; Huang, J.-H.; Shu, P.-Y. Artificial receptors in serologic tests for the early diagnosis of Dengue virus infection. Clin. Chem. 2006, 52, 1486–1491. [Google Scholar] [CrossRef]
- Tai, D.-F.; Lin, C.-Y.; Wu, T.-Z.; Chen, L.-K. Recognition of Dengue virus protein using epitope-mediated molecularly imprinted film. Anal. Chem. 2005, 77, 5140–5143. [Google Scholar] [CrossRef]
- Siqueira Silva, M.; Moreira Tavares, A.P.; Leomil Coelho, L.F.; Morganti Ferreira Dias, L.E.; Chura-Chambi, R.M.; Guimarães da Fonseca, F.; Ferreira Sales, M.G.; Costa Figueiredo, E. Rational selection of hidden epitopes for a molecularly imprinted electrochemical sensor in the recognition of heat-denatured dengue NS1 protein. Biosens. Bioelectron. 2021, 191, 113419. [Google Scholar] [CrossRef]
- Antipchik, M.; Reut, J.; Ayankojo, A.G.; Öpik, A.; Syritski, V. MIP-based electrochemical sensor for direct detection of hepatitis C virus via E2 envelope protein. Talanta 2022, 250, 123737. [Google Scholar] [CrossRef]
- Ertürk, G.; Mattiasson, B. Molecular imprinting techniques used for the preparation of biosensors. Sensors 2017, 17, 288. [Google Scholar] [CrossRef] [PubMed]
- Ghanbari, K.; Roushani, M. A nanohybrid probe based on double recognition of an aptamer MIP grafted onto a MWCNTs-Chit nanocomposite for sensing hepatitis C virus core antigen. Sens. Actuator B-Chem. 2018, 258, 1066–1071. [Google Scholar] [CrossRef]
- Liu, Y.; Dykstra, G. Recent progress on electrochemical (bio)sensors based on aptamer-molecularly imprinted polymer dual recognition. Sens. Actuators Rep. 2022, 4, 100112. [Google Scholar] [CrossRef]
- Ma, C.; Xie, G.; Zhang, W.; Liang, M.; Liu, B.; Xiang, H. Label-free sandwich type of immunosensor for hepatitis C virus core antigen based on the use of gold nanoparticles on a nanostructured metal oxide surface. Microchim. Acta 2012, 178, 331–340. [Google Scholar] [CrossRef]
- Ma, C.; Liang, M.; Wang, L.; Xiang, H.; Jiang, Y.; Li, Y.; Xie, G. MultisHRP-DNA-coated CMWNTs as signal labels for an ultrasensitive hepatitis C virus core antigen electrochemical immunosensor. Biosens. Bioelectron. 2013, 47, 467–474. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, B.O.; Lingaas, E.; Torfoss, D.; Strøm, E.H.; Nordøy, I. A large outbreak of Listeria monocytogenes infection with short incubation period in a tertiary care hospital. J. Infect. 2010, 61, 465–470. [Google Scholar] [CrossRef]
- Li, X.-P.; Wang, S.-F.; Hou, P.-B.; Liu, J.; Du, P.; Bai, L.; Fanning, S.; Zhang, H.-N.; Chen, Y.-Z.; Zhang, Y.-K.; et al. Nosocomial cross-infection of hypervirulent Listeria monocytogenes sequence type 87 in China. Ann. Transl. Med. 2020, 8, 603. [Google Scholar] [CrossRef]
- Sharma, R.; Lakshmi, G.B.V.S.; Kumar, A.; Solanki, P. Polypyrrole based molecularly imprinted polymer platform for Klebsiella pneumonia detection. ECS Sens. Plus 2022, 1, 010603. [Google Scholar] [CrossRef]
- Sarabaegi, M.; Roushani, M. Rapid and sensitive determination of Pseudomonas aeruginosa by using a glassy carbon electrode modified with gold nanoparticles and aptamer-imprinted polydopamine. Microchem. J. 2021, 168, 106388. [Google Scholar] [CrossRef]
- Tokonami, S.; Nakadoi, Y.; Takahashi, M.; Ikemizu, M.; Kadoma, T.; Saimatsu, K.; Dung, L.Q.; Shiigi, H.; Nagaoka, T. Label-free and selective bacteria detection using a film with transferred bacterial configuration. Anal. Chem. 2013, 85, 4925–4929. [Google Scholar] [CrossRef]
- Liustrovaite, V.; Pogorielov, M.; Boguzaite, R.; Ratautaite, V.; Ramanaviciene, A.; Pilvenyte, G.; Holubnycha, V.; Korniienko, V.; Diedkova, K.; Viter, R.; et al. Towards electrochemical sensor based on molecularly imprinted polypyrrole for the detection of bacteria-Listeria monocytogenes. Polymers 2023, 15, 1597. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Guo, Z.; Qiu, X.; Lu, W.; Yang, W.; Wang, Q.; Wu, Q. Simple electrochemical detection of Listeria monocytogenes based on a surface-imprinted polymer-modified electrode. Anal. Methods 2021, 13, 4864–4870. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Lv, Z.; Ding, W.; Zhang, Y.; Lin, F. Pathogen-imprinted polymer film integrated probe/Ti3C2Tx MXenes electrochemical sensor for highly sensitive determination of Listeria monocytogenes. J. Electrochem. Sci. Technol. 2022, 13, 431–437. [Google Scholar] [CrossRef]
- Roushani, M.; Sarabaegi, M.; Rostamzad, A. Novel electrochemical sensor based on polydopamine molecularly imprinted polymer for sensitive and selective detection of Acinetobacter baumannii. J. Iran. Chem. Soc. 2020, 17, 2407–2413. [Google Scholar] [CrossRef]





| Biomarkers | Polymers and Modifiers | Electrodes | Extraction of the Template | Electrochemical Analysis Methods | LOD, LOQ, LR | Interferents | Reference |
|---|---|---|---|---|---|---|---|
| HSA | |||||||
| HSA | Polyscopoletin | AuE | 10 min in 5 mM NaOH, 5 min in 5% SDS, 10 min in 5 mM NaOH | CV | LOD 3.7 mg/L, LR 20–100 mg/L | Ferritin, avidin, and lysozyme | [60] |
| HSA | poly(thionine-methylene blue), PoPD, hydroquinone, AuNPs | AuE | 1 mol/L NaOH, ethanol/water (v/v, 2/1) under 50 ℃ | DPV, EIS | LOD 0.03 ng/L; LR 0.1–100,000 ng/L | L-glycine, L-glutamate, L-cysteine, L-tryptophan, L-histidine, dopamine, ascorbic acid, hemoglobin, and bovine serum albumin | [74] |
| HSA | Polythiophene | AuE | DPV, EIS | LOD 16.6 ng/mL (DPV); LR 0.8–20 µg/mL LOD 800 ng/mL (EIS); LR 4–80 µg/mL | creatinine, urea, uric acid, and glucose | [75] | |
| Acute-phase proteins (CRP and SAA) | |||||||
| CRP | Poly(AEDP-DMAA), MWCNTs | SPCE | 10% (w/v) SDS and 0.1 N HCl mixture solutions for 4 h, 0.5 M EDTA treatment for 1 h. | DPV | LOD 0.04 μg/mL | BSA, insulin, Hb, and lysozyme | [65] |
| CRP | Poly(MMA, Au-PtNMs | SPCE | Methanol and acetic acid (4:1) for 24 h | EIS | LOD 0.1 nM; LR 0.1 nM–500 nM | glucose, uric acid, ascorbic acid, acetylcholine, cholesterol, TNF-α, and procalcitonin | [79] |
| CRP | Poly(aniline-co-m-amino benzene sulfonic acid), MXene | ITO | 10 mL of 5 vol% ethanol at 130 rpm for 10 min (orbital shaker), deionized water. | CV | LOD 0.1 fg/mL | pR, pK, pI, HSA, and lysozyme | [80] |
| CRP | PDA, GDY, PEG (antifouling additive) | GCE | Acetone for 50 min. | EIS | LOD 4.1 fg/mL; LR 10 fg/mL–1 µg/mL | carcinoembryonic antigen, immunoglobulin G, alpha fetal protein, transferrin | [81] |
| SAA | poly(methyl methacrylate-ethylene glycol dimethacrylate), MWCNTs, MnO2NSs, Co3O4NPs | SPE | Methanol and acetic acid (4:1) for 24 h | CV, DPV, EIS | LOD 0.01 pM; LR 0.01 pM–1 μM | Interferents: ascorbic acid, cholesterol, glucose, uric acid, acetylcholine | [66] |
| Cytokines (TNF-α, IL-6, IL-1β, and IL-2) | |||||||
| TNF-α | Poly(MMA), MoS2NSs, Fe3O4@SiO2NPs | SPE | Methanol and acetic acid | SWV, DPV, EIS | LOD 0.01 pM | glucose, acetylcholine, cholesterol, uric acid, ascorbic acid | [67] |
| IL-6 | Ppy, Ppy-COOH | SPCE | incubation for 3 h in 0.05 M oxalic acid dihydrate, CV | EIS, CV | LOD 0.02 pg/mL; LR 0.02–2 × 106 pg/mL; | [69] | |
| IL-6 | Poly(APBA) | incubation with 20 μL of proteinase K overnight at 40 °C, CV | EIS, CV | LOD 1 pg/mL | [92] | ||
| IL-1β | PEDOT, Poly(EBT) | SPCE | EIS, SWV, CV | LOD 1.5 pM; LR 60 pM–600 nM | Myo, IgG. | [70] | |
| IL-1β | PoPD, poly(chromotrope 2R) | SPCE | EIS | LOD 0.23 pg/mL | IL-6, TNF-α, and IL-1α; | [94] | |
| Biomarkers | Polymers and Modifiers | Electrodes | Extraction of Templates | Electrochemical Analysis Methods | LOD, LOQ, LR | Interfering Molecules | Reference |
|---|---|---|---|---|---|---|---|
| HIV-1 | |||||||
| gp41 | PDA | QCM | 5% acetic acid (in H2O) for five times, DI water | X-ray photoelectron spectrometer (XPS) | LOD 2 ng/mL; LR 5–200 ng/mL | [102] | |
| gp120 | Ppy, CNF-Bi, chitosan | GCE | Hyper pure water; methanol and acetic acid solution for 20 min. | CV, DPV | LOD 0.0003 ng/mL; LR 0.002–200 ng/mL | HIV-1 protein p24, human chorionic gonadotropin, carcinoembryonic antigen | [106] |
| COVID-19 | |||||||
| SARS-CoV-2 nucleocapsid protein | PmPD | AuTFE | Ethanolic solution of 0.1 M 2-mercaptoethanol, 10% acetic acid solution | DPV | LOD 15 fM; LR 2.22–111 fM | S1, BSA, CD48, HCV, E2 | [110] |
| SARS-CoV-2 nucleocapsid protein | P-Arg, gold/graphene nanohybrids | SPCE | Ethanolic solution containing 0.1 M 2-mercaptoethanol; acetic acid (10%) solution | DPV, EIS | LOD 3.0 fM; LR 10–200 fM | cTnI, SARS-CoV-2 spiken, HER2, BSA, CD48, MPT64 | [113] |
| SARS-CoV-2 antigen | Ppy, graphene oxide flakes | GCE | 10 vol% acetic acid, acetone, and ethanol | DPV, amperometry | LOD 0.326 fg/mL (DPV); LOD 11.32 fg/mL (amperometric); LR 0.74–9.03 fg/mL (DPV); LR 13.14–118.9 g/mL (amperometric) | H1N1 influenza virus, H3N2 influenza virus, glucose, lactose, maltose, ascorbic acid, sucrose, fructose, BSA | [111] |
| SARS-CoV-2-S spike glycoprotein | Ppy | Pt | Incubation in 0.05 M H2SO4 for 10 min. | Pulsed Amperometric Detection | BSA | [50] | |
| SARS-CoV-2 spike protein | Poly(aminophenylboronic acid) | SPE | 50 mM dithiothreitol for 30 min; 30 min in 10% acetic acid | SWV, CV | LOD 1.12 pg/mL; LR 0–400 fM | SARS-CoV-2 nucleocapsid protein, E2, HSA, IgG | [61] |
| Dengue virus | |||||||
| NS1 | PDA, polysulfone fibres | SPCE | PBS; 500 μg/mL of proteinase K for 2 h in the dark | EIS, CV | LOD 0.3 ng/mL; LR 1–200 ng/mL | FBS, lysozyme | [100] |
| NS1 | Poly(G03TCOOH), gold | QCM | Potential washing (−0.7 V) 0.1 M tetrabutylammonium hexafluorophosphate in acetonitrile | EIS | LOD 0.056 μg/mL; LR 0.2 to 10 μg/mL | angiotensin II human, glycyl glycine, bovine serum albumin, fibrinogen | [119] |
| Hepatitis C virus | |||||||
| HCV surface protein E2 | PmPD | SPE | PBS with 50 mM dithiothreitol for 30 min, 10% acetic acid solution on vortex for 30 min | DPV | LOD 0.46 pg/mL; LR 0.01–50 ng/mL; LOQ 15.3 × 10−5 ng/mL | HSA, IgG, CD81 | [123] |
| HCV core antigen | PDA, MWCNTs- Chit nanocomposite | GCE | Water, overnight in 5% v/v acetic acid and 1% w/v cetyl trimethyl ammonium bromide in water with stirring | CV, DPV, EIS | LOD 1.67 fg/mL; LR 5.0 fg/mL to 1.0 pg/mL; | [125] | |
| Nosocomial infections | |||||||
| K. pneumoniae | Ppy | ITO | DI water, ethanol | CV, DPV | LOD 1.352 CFU/mL; LR 1–105 CFU/mL | uric acid, K+, Mg++, urea, Lactobacillus, E. coli | [131] |
| K. pneumoniae | Poly(MAM:AAM:NVP), graphene oxide | AuSPE | 10% acetic acid for 30 min, water at 50 °C for 30 min | CV | LOD 0.012 CFU/mL; LOQ 1.61 CFU/mL; LR 101–105 CFU/mL | E. faecalis, P. aeruginosa | [57] |
| P. aeruginosa | PDA, AuNPs | GCE | Solution containing SDS 0.01 M and 5% HNO3 in water | CV, EIS, DPV | LOD 1 CFU/mL; LR 10–107 CFU/mL | Shigella flexneri, Salmonella enteritidis, E. coli, K. pneumonia | [132] |
| L. monocytogenes | Poly(3-thiopheneacetic acid) | GCE | SDS/AA (w/v, 5%) solution | DPV, CV | LOD 6 CFU/mL; LR 10–106 CFU/mL | Staphylococcus aureus, Vibrio parahaemolyticus, Shigella, Salmonella enteritidis, Escherichia | [135] |
| L. monocytogenes | Polythionine, MXenes nanoribbon (Ti3C2TxR) | GCE | 0.5 M HCl | DPV, EIS | LOD 2 CFU/mL; LR 10 to 108 CFU/mL | Escherichia, Vibrio parahaemolyticus, Staphylococcus aureus, Shigella, Salmonella enteriditis | [136] |
| L. monocytogenes | Ppy | SPCE | 10% acetic acid, or sulfuric acid, or L-lysin, or trypsin | PAD | LOD 70 CFU/mL, LOQ 210 CFU/mL, LR 300–6700 CFU/mL. | [134] | |
| A. baumannii | PDA | GCE | 2 h in 0.01 M SDS and 10 mM HNO3 in water with stirring | CV, EIS, DPV | LOD (CFU/mL; LR 102–107 CFU/mL | P. aeruginosa, E. coli, K. pneumonia, S. enteritidis, S. fexneri | [137] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilvenyte, G.; Ratautaite, V.; Boguzaite, R.; Ramanavicius, S.; Chen, C.-F.; Viter, R.; Ramanavicius, A. Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases. Biosensors 2023, 13, 620. https://doi.org/10.3390/bios13060620
Pilvenyte G, Ratautaite V, Boguzaite R, Ramanavicius S, Chen C-F, Viter R, Ramanavicius A. Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases. Biosensors. 2023; 13(6):620. https://doi.org/10.3390/bios13060620
Chicago/Turabian StylePilvenyte, Greta, Vilma Ratautaite, Raimonda Boguzaite, Simonas Ramanavicius, Chien-Fu Chen, Roman Viter, and Arunas Ramanavicius. 2023. "Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases" Biosensors 13, no. 6: 620. https://doi.org/10.3390/bios13060620
APA StylePilvenyte, G., Ratautaite, V., Boguzaite, R., Ramanavicius, S., Chen, C.-F., Viter, R., & Ramanavicius, A. (2023). Molecularly Imprinted Polymer-Based Electrochemical Sensors for the Diagnosis of Infectious Diseases. Biosensors, 13(6), 620. https://doi.org/10.3390/bios13060620









