A Label-Free Carbohydrate-Based Electrochemical Sensor to Detect Escherichia coli Pathogenic Bacteria Using D-mannose on a Glassy Carbon Electrode
Abstract
1. Introduction
2. Experimental Methods
2.1. Materials
2.2. Apparatus
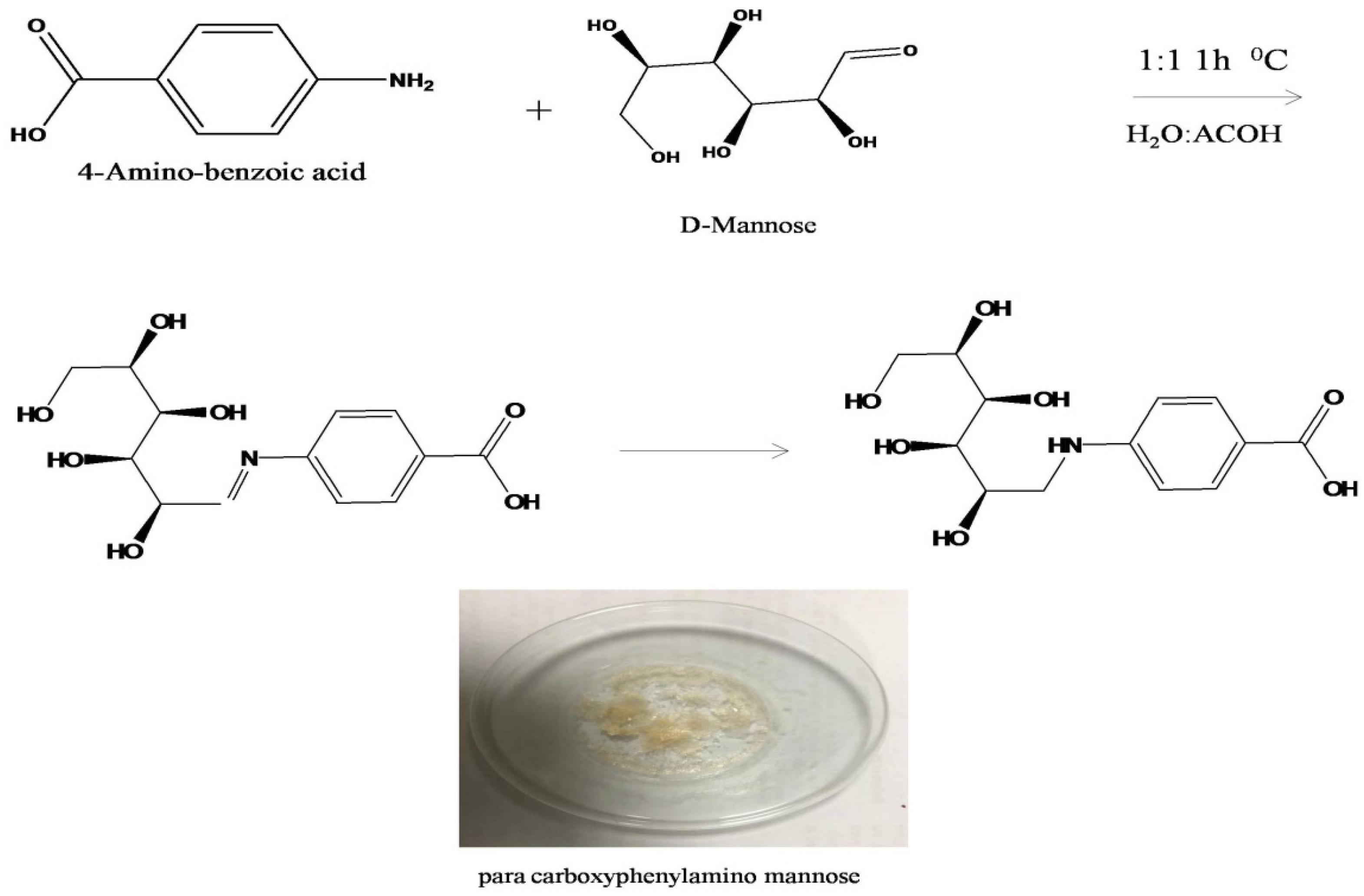
2.3. Synthesis of Biorecognition Element
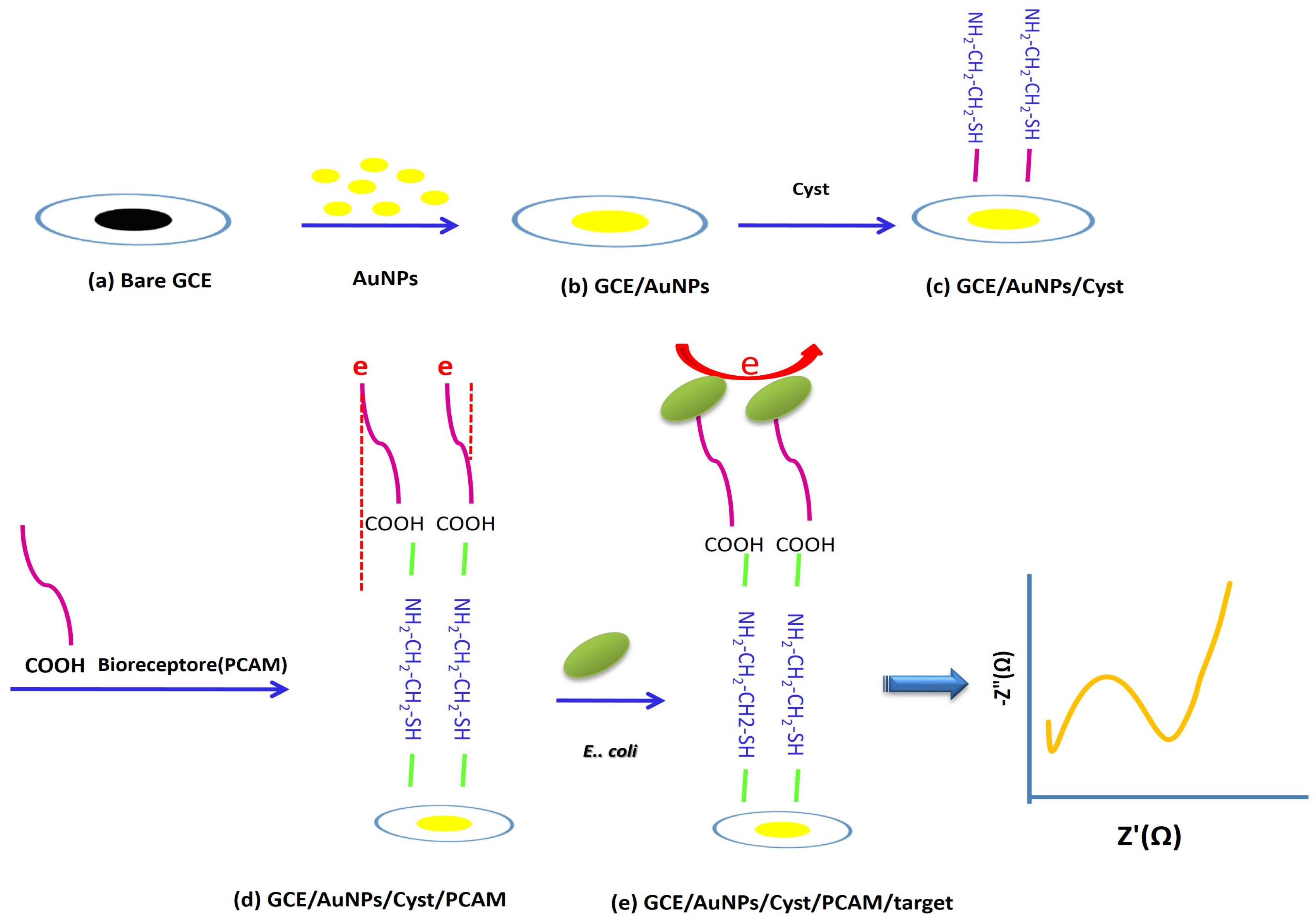
2.4. Sensor Fabrication
2.5. Electroanalytical Measurements
2.6. Real Sample Preparation
2.7. Bacterial Culture and Counting Methods
2.8. Sensor Concept and Design of Modified Mannose
2.9. Material Selection
3. Results and Discussion
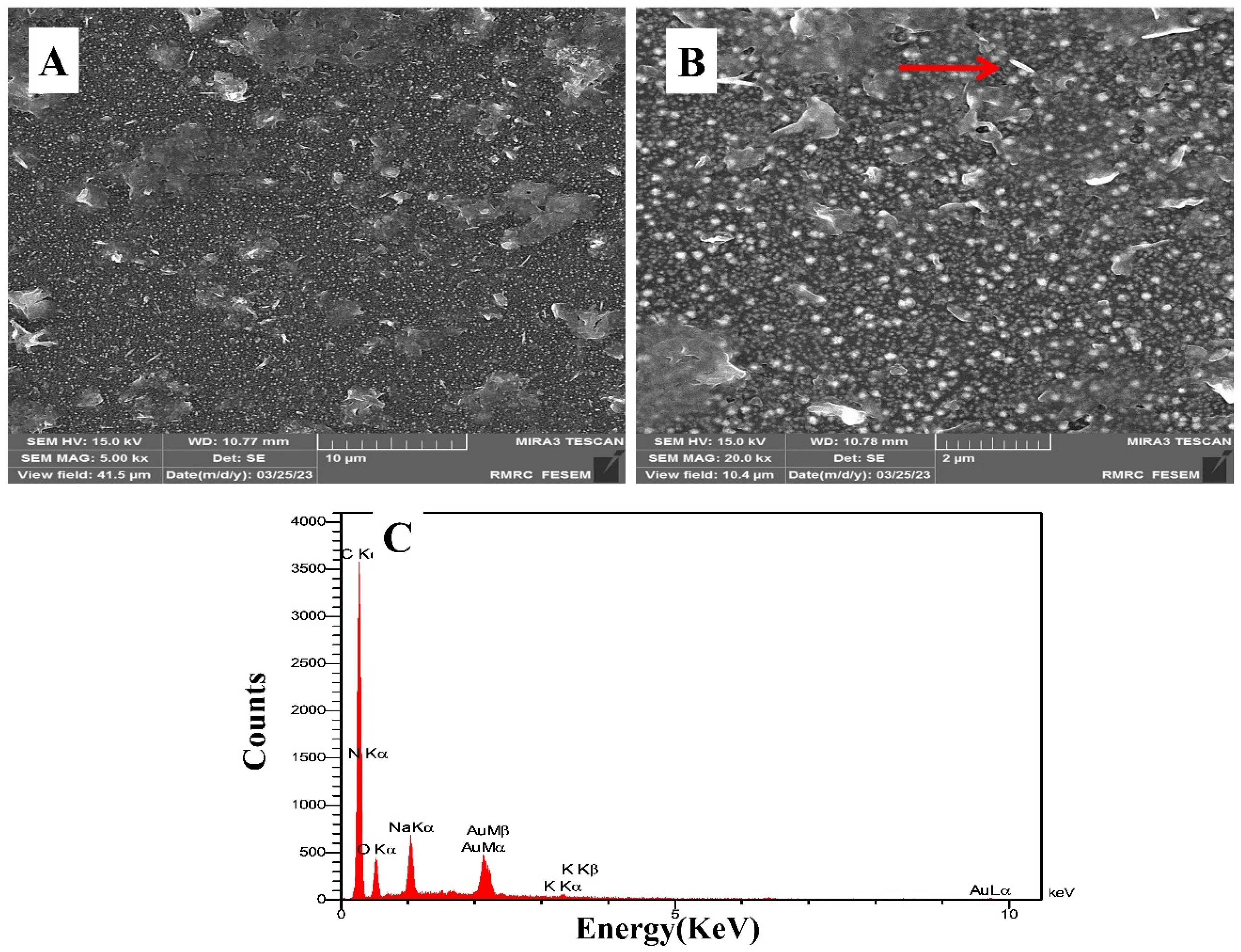
3.1. Surface Characterization
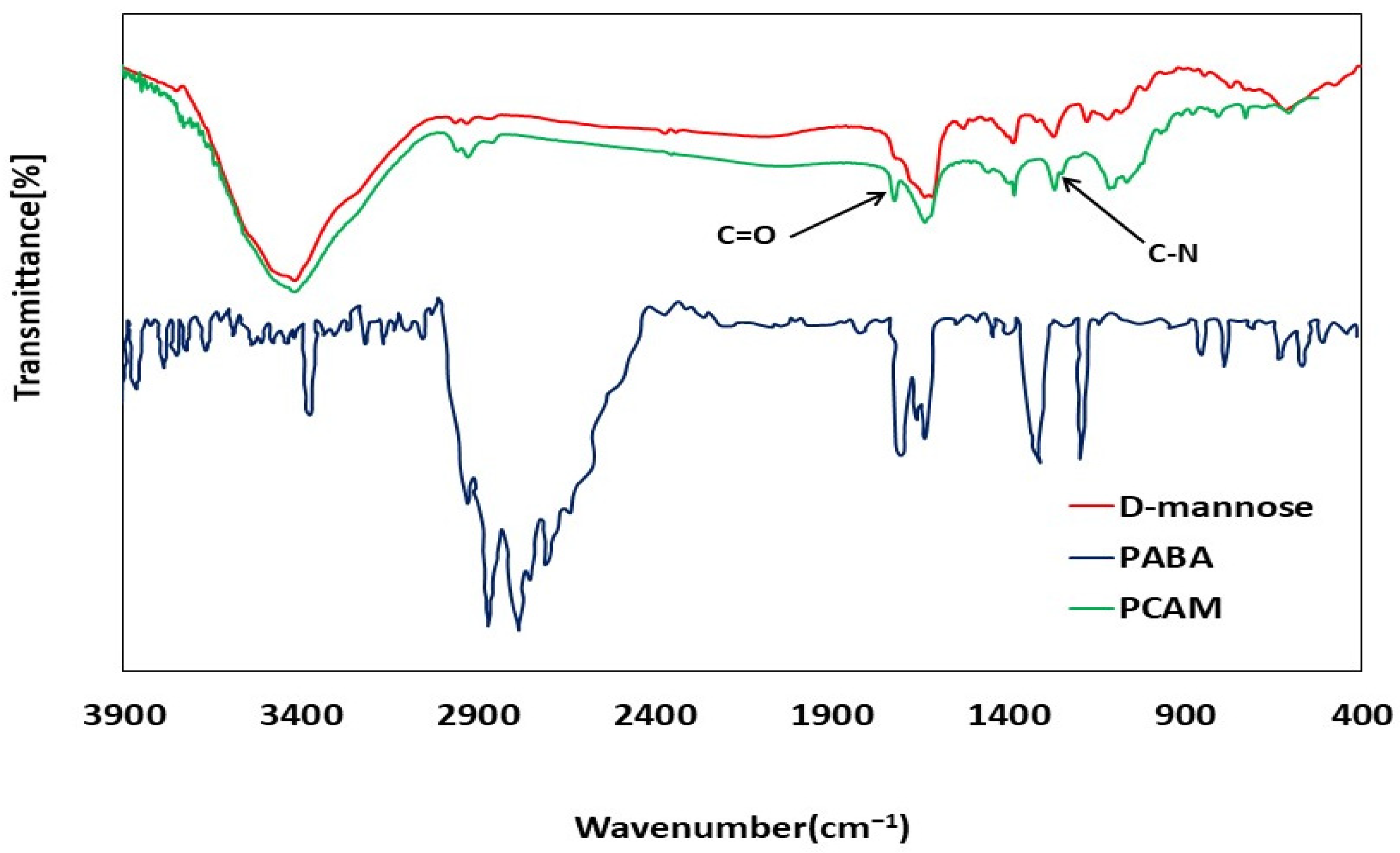
3.2. FTIR Characterizations
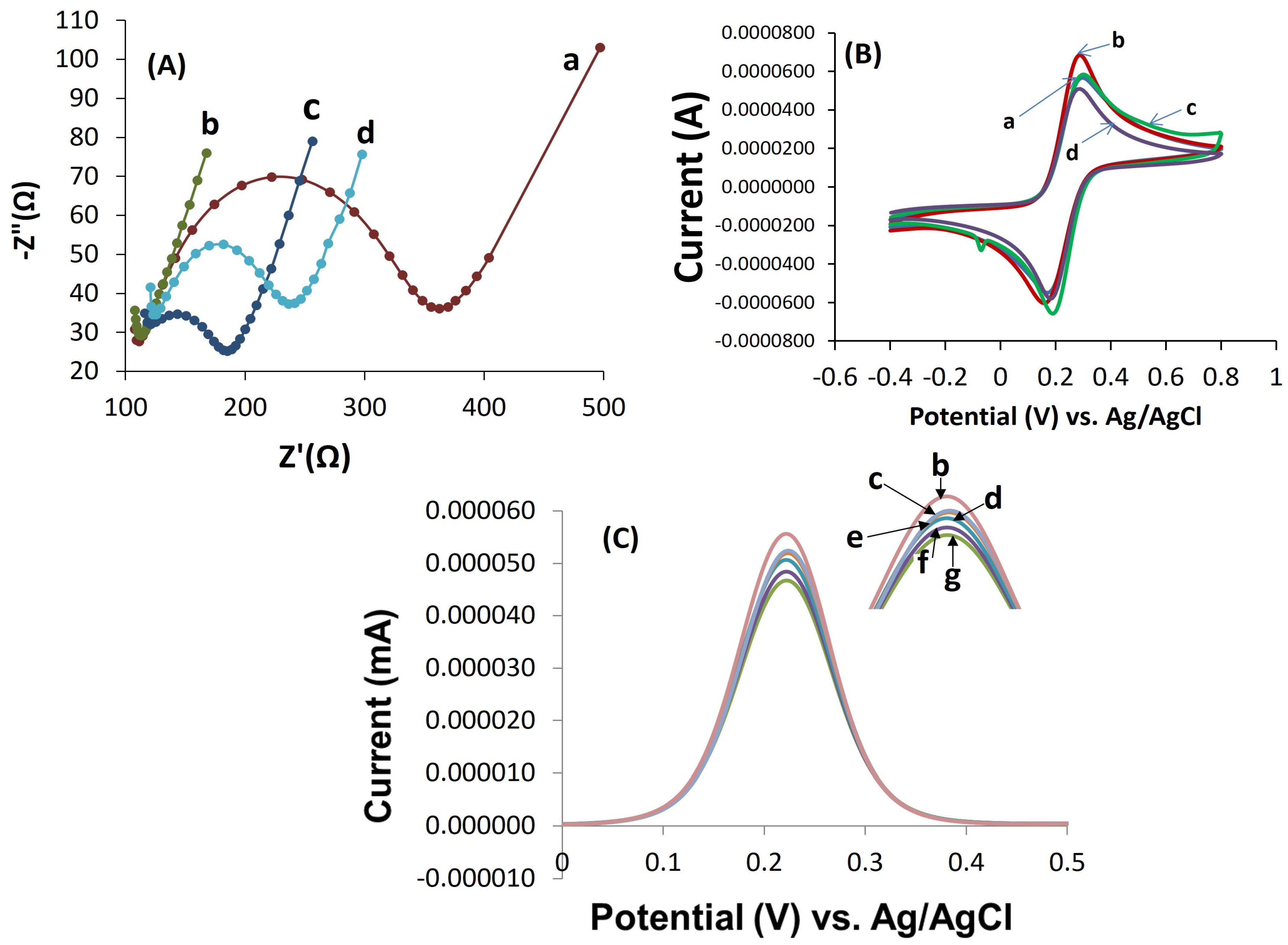
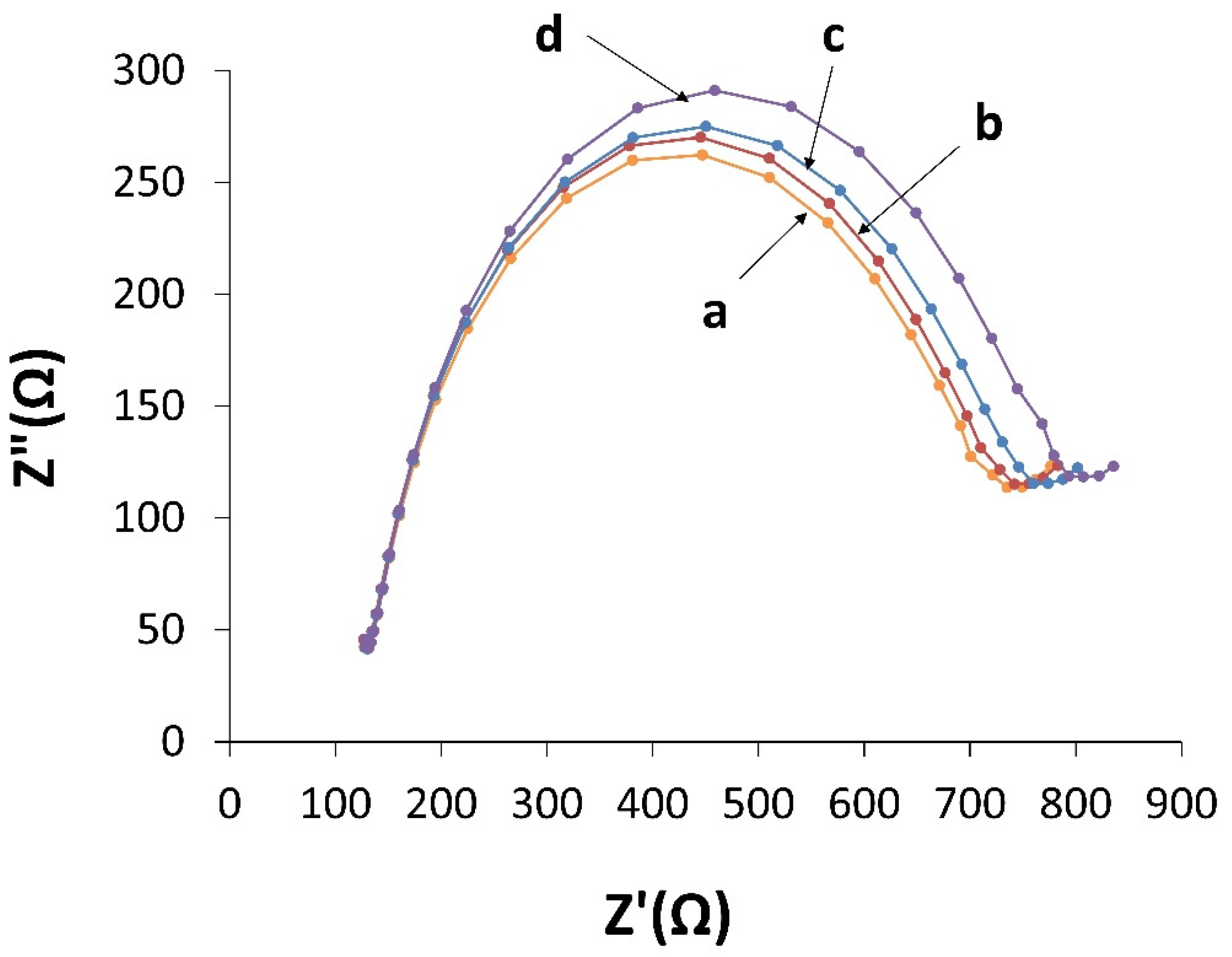
3.3. Electrochemical Characterization
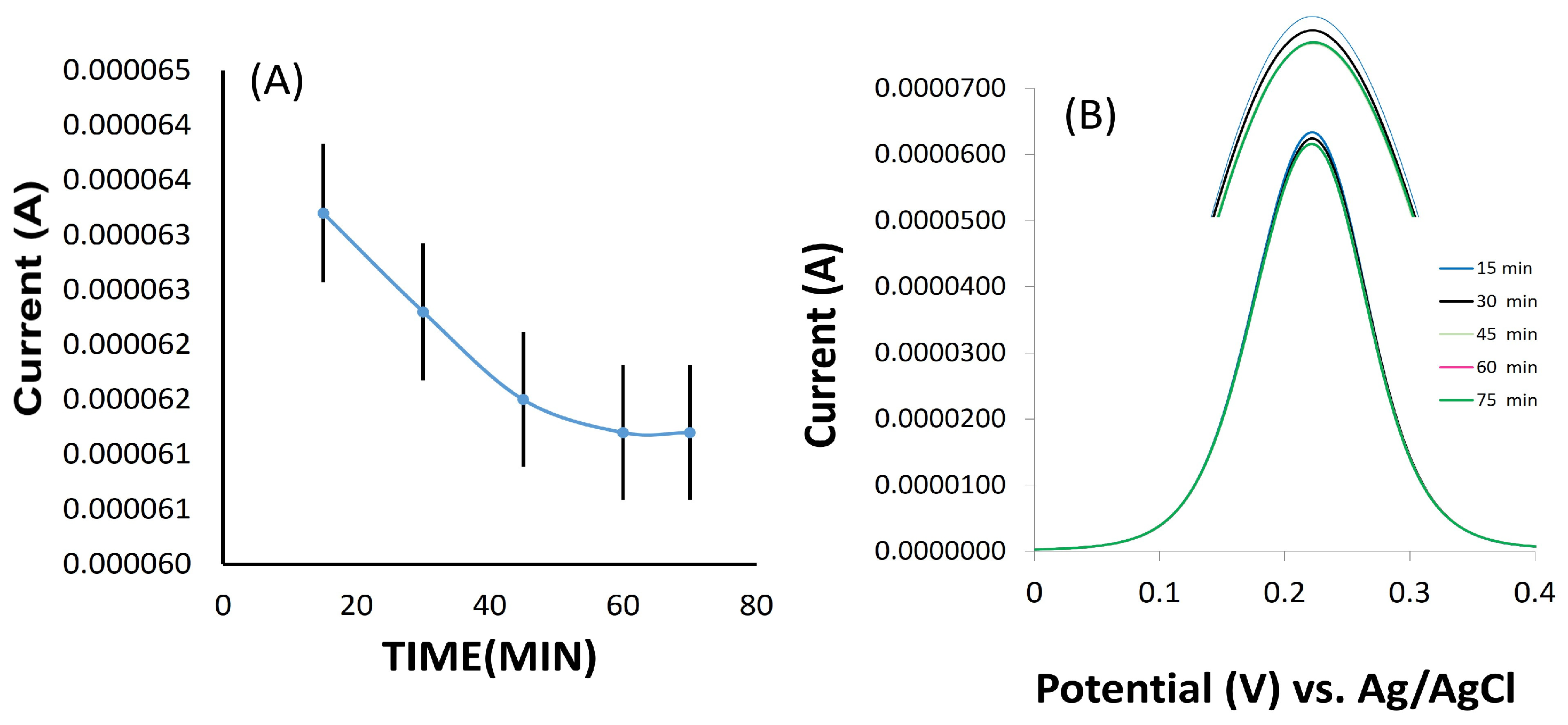
3.4. Optimization of Incubation Time
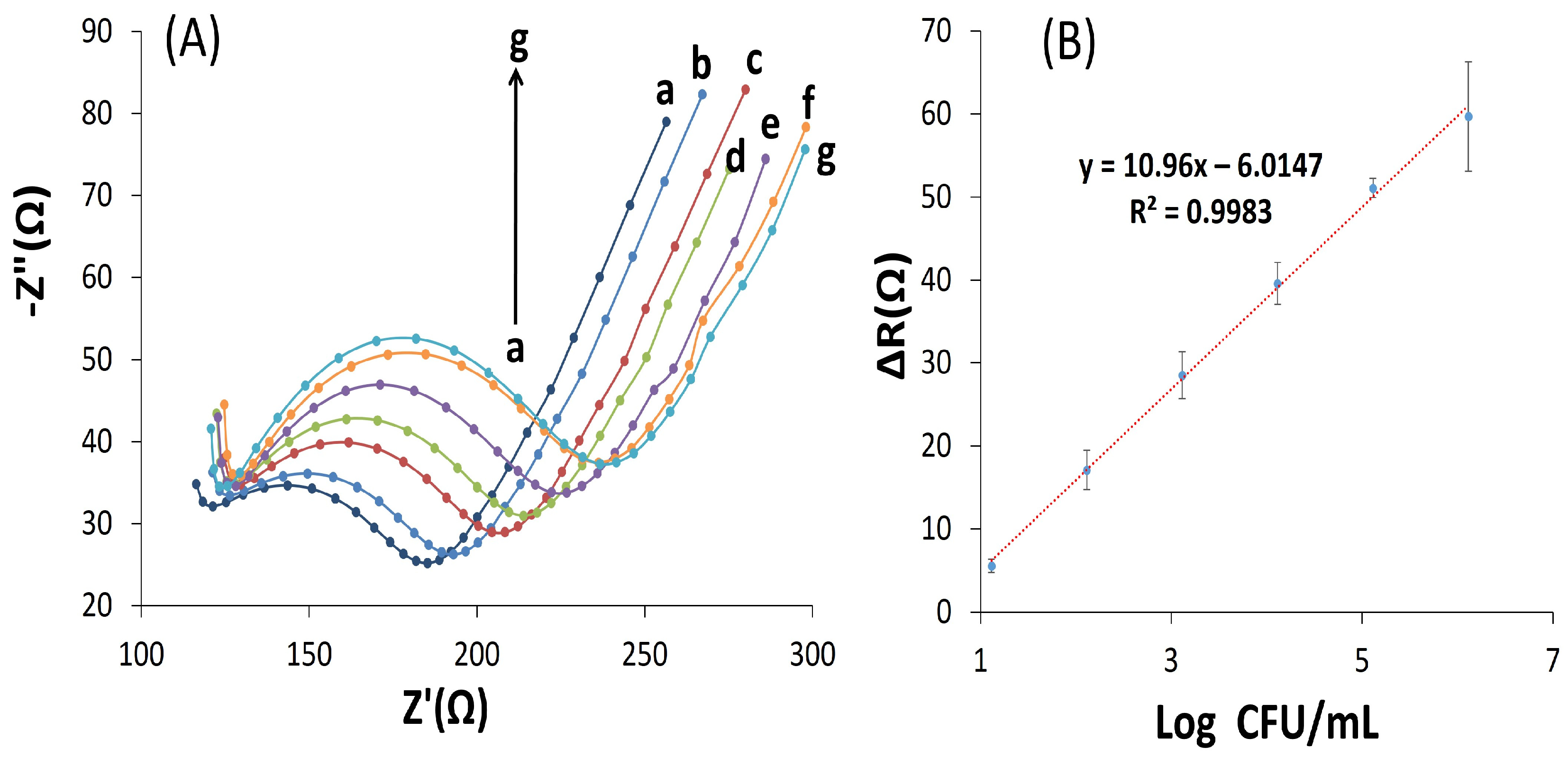
3.5. Biosensor Calibration Curve and Limit of Detection
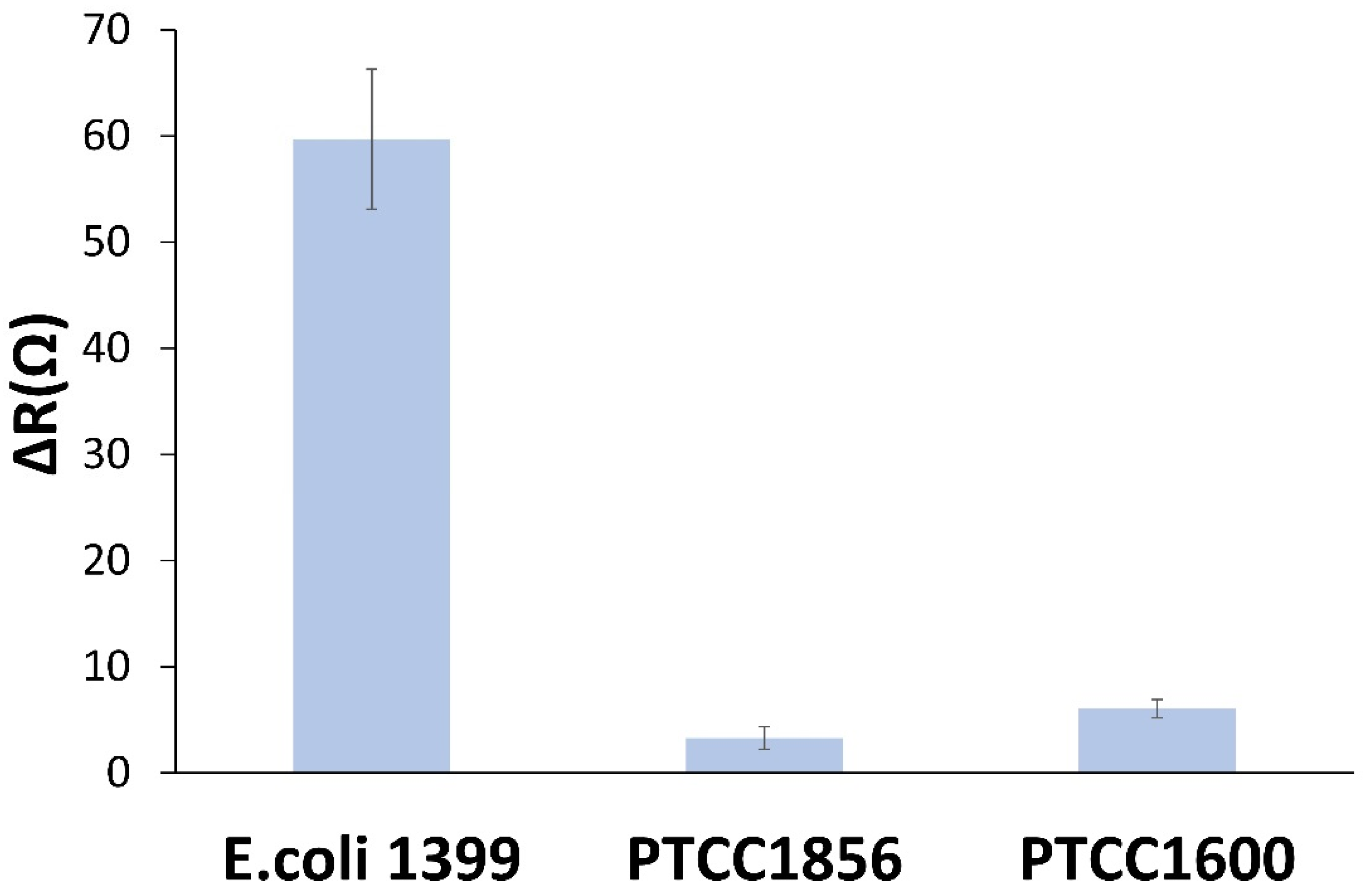
3.6. Selectivity of the Biosensor
3.7. Real Sample Measurement
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, M.P. Food Safety: Bacterial Contamination. In Encyclopedia of Human Nutrition, 3rd ed.; Caballero, B., Ed.; Academic Press: Waltham, MA, USA, 2013; pp. 322–330. [Google Scholar]
- Mead, P.S.; Slutsker, L.; Dietz, V.; McCaig, L.F.; Bresee, J.S.; Shapiro, C.; Griffin, P.M.; Tauxe, R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999, 5, 607. [Google Scholar] [CrossRef]
- Thomas, L.V.; Rowe, B.; McConnell, M.M. In strains of Escherichia coli O167 a single plasmid encodes for the coli surface antigens CS5 and CS6 of putative colonization factor PCF8775, heat-stable enterotoxin, and colicin Ia. Infect. Immun. 1987, 55, 1929–1931. [Google Scholar] [CrossRef]
- Krohn, M.A.; Thwin, S.S.; Rabe, L.K.; Brown, Z.; Hillier, S.L. Vaginal colonization by Escherichia coli as a risk factor for very low birth weight delivery and other perinatal complications. J. Infect. Dis. 1997, 175, 606–610. [Google Scholar] [CrossRef]
- Banatvala, N.; Griffin, P.M.; Greene, K.D.; Barrett, T.J.; Bibb, W.F.; Green, J.H.; Wells, J.G. The United States National Prospective Hemolytic Uremic Syndrome Study: Microbiologic, Serologic, Clinical, and Epidemiologic Findings. J. Infect. Dis. 2001, 183, 1063–1070. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Konoval, H.M.; Marecek, S.; Lathrop, A.A.; Feng, S.; Pokharel, S. Control of Escherichia coli O157: H7 using lytic bacteriophage and lactic acid on marinated and tenderized raw pork loins. Meat Sci. 2023, 196, 109030. [Google Scholar] [CrossRef] [PubMed]
- Dester, E.; Kao, K.; Alocilja, E.C. Detection of unamplified E. coli O157 DNA extracted from large food samples using a gold nanoparticle colorimetric biosensor. Biosensors 2022, 12, 274–288. [Google Scholar] [CrossRef] [PubMed]
- Chan, M.Y.; Smith, M.A. Infections in pregnancy. Compr. Toxicol. 2018, 856, 232–249. [Google Scholar]
- Mayer, K.; Eris, D.; Schwardt, O.; Sager, C.P.; Rabbani, S.; Kleeb, S.; Ernst, B. Urinary tract infection: Which conformation of the bacterial lectin FimH is therapeutically relevant? J. Med. Chem. 2017, 60, 5646–5662. [Google Scholar] [CrossRef]
- Firon, N.; Ofek, I.; Sharon, N. Carbohydrate-binding sites of the mannose-specific fimbrial lectins of enterobacteria. Infect. Immun. 1984, 43, 1088–1090. [Google Scholar] [CrossRef]
- McClure, E.M.; Goldenberg, R.L. Infection and Stillbirth; Elsevier: Amsterdam, The Netherlands, 2009; pp. 182–189. [Google Scholar]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef]
- Spagnolo, S.; De La Franier, B.; Davoudian, K.; Hianik, T.; Thompson, M. Detection of E. coli bacteria in milk by an acoustic wave aptasensor with an anti-fouling coating. Sensors 2022, 22, 1853–1865. [Google Scholar] [CrossRef] [PubMed]
- Sobhan, A.; Jia, F.; Kelso, L.C.; Biswas, S.K.; Muthukumarappan, K.; Cao, C.; Wei, L.; Li, Y. A Novel Activated Biochar-Based Immunosensor for Rapid Detection of E. coli O157: H7. Biosensors 2022, 12, 908–922. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Rehman, A.; Liu, H.; Zhang, J.; Zhu, S.; Zeng, X. Glycosylation of quinone-fused polythiophene for reagentless and label-free detection of E. coli. Anal. Chem. 2015, 87, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Kaur, J.; Choudhary, S.; Chaudhari, R.; Jayant, R.D.; Joshi, A. 9—Enzyme-based biosensors. In Bioelectronics and Medical Devices; Pal, K., Kraatz, H.-B., Khasnobish, A., Bag, S., Banerjee, I., Kuruganti, U., Eds.; Woodhead Publishing: Sawston, UK, 2019; pp. 211–240. [Google Scholar]
- Yao, W.; Shi, J.; Ling, J.; Guo, Y.; Ding, C.; Ding, Y. SiC-functionalized fluorescent aptasensor for determination of Proteus mirabilis. Microchim. Acta 2020, 187, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Yang, X.; Wu, J.; Liu, Q.; Li, D.; Huang, S.; Xie, H.; Yu, Z.; Gan, N. Reusable electrochemical biosensing platform based on egg yolk antibody-labeled magnetic covalent organic framework for on-site detection of Escherichia coli in foods. Sens. Actuators B Chem. 2022, 369, 132320–132338. [Google Scholar] [CrossRef]
- Zhang, G.; Hu, H.; Deng, S.; Xiao, X.; Xiong, Y.; Peng, J.; Lai, W. An integrated colorimetric and photothermal lateral flow immunoassay based on bimetallic Ag–Au urchin-like hollow structures for the sensitive detection of E. coli O157: H7. Biosens. Bioelectron. 2023, 225, 115090–115109. [Google Scholar] [CrossRef]
- Gupta, A.; Garg, M.; Singh, S.; Deep, A.; Sharma, A.L. Highly sensitive optical detection of Escherichia coli using terbium-based metal–organic framework. ACS Appl. Mater. Interfaces 2020, 12, 48198–48205. [Google Scholar] [CrossRef]
- Ngo, V.K.T.; Nguyen, D.G.; Nguyen, H.P.U.; Nguyen, T.K.M.; Huynh, T.P.; Lam, Q.V.; Huynh, T.D.; Truong, T.N.L. Quartz crystal microbalance (QCM) as biosensor for the detecting of Escherichia coli O157: H7. Adv. Nat. Sci. Nanosci. Nanotechnol. 2014, 5, 045004–045013. [Google Scholar]
- Kaur, K.; Chelangat, W.; Druzhinin, S.I.; Karuri, N.W.; Müller, M.; Schönherr, H. Quantitative E. coli enzyme detection in reporter hydrogel-coated paper using a smartphone camera. Biosensors 2021, 11, 25–42. [Google Scholar] [CrossRef]
- Wang, H.; Zhao, Y.; Bie, S.; Suo, T.; Jia, G.; Liu, B.; Ye, R.; Li, Z. Development of an electrochemical biosensor for rapid and effective detection of pathogenic Escherichia coli in licorice extract. Appl. Sci. 2019, 9, 295–310. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, J.; Zhang, X.; He, F. Rapid detection of Escherichia coli based on 16S rDNA nanogap network electrochemical biosensor. Biosens. Bioelectron. 2018, 118, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zhou, H.; Hao, H.; Gong, Q.; Nie, K. Detection of Escherichia coli with a label-free impedimetric biosensor based on lectin functionalized mixed self-assembled monolayer. Sens. Actuators B Chem. 2016, 229, 297–304. [Google Scholar] [CrossRef]
- Yazgan, I.; Noah, N.M.; Toure, O.; Zhang, S.; Sadik, O.A. Biosensor for selective detection of E. coli in spinach using the strong affinity of derivatized mannose with fimbrial lectin. Biosens. Bioelectron. 2014, 61, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Roushani, M.; Shahdost-Fard, F. Fabrication of an ultrasensitive ibuprofen nanoaptasensor based on covalent attachment of aptamer to electrochemically deposited gold-nanoparticles on glassy carbon electrode. Talanta 2015, 144, 510–516. [Google Scholar] [CrossRef]
- Guo, X.; Kulkarni, A.; Doepke, A.; Halsall, H.B.; Iyer, S.; Heineman, W.R. Carbohydrate-based label-free detection of Escherichia coli ORN 178 using electrochemical impedance spectroscopy. Anal. Chem. 2012, 84, 241–246. [Google Scholar] [CrossRef]
- Shaibani, P.M.; Etayash, H.; Jiang, K.; Sohrabi, A.; Hassanpourfard, M.; Naicker, S.; Sadrzadeh, M.; Thundat, T. Portable nanofiber-light addressable potentiometric sensor for rapid Escherichia coli detection in orange juice. ACS Sens. 2018, 3, 815–822. [Google Scholar] [CrossRef]
- Shen, Z.; Huang, M.; Xiao, C.; Zhang, Y.; Zeng, X.; Wang, P.G. Nonlabeled quartz crystal microbalance biosensor for bacterial detection using carbohydrate and lectin recognitions. Anal. Chem. 2007, 79, 2312–2319. [Google Scholar] [CrossRef]

| Method | Linear Dynamic Range | LOD | Reference |
|---|---|---|---|
| Electrochemical and quartz crystal microbalance polythiophene interface biosensor using Concanavalin A | 1.0 × 102–5.0 × 103 cells. mL−1 | 25 cell.mL−1 for electrochemical sensor and 50 cells.mL−1 for QCM sensor | [15] |
| Lectin-based impedimetric biosensor | 102–105 cells. mL−1 | 75 cells. mL−1 | [25] |
| Carbohydrate-based label-free biosensor for detection of E. coli ORN 178 | 102–103 CFU·mL−1 | 102 CFU·mL−1 | [28] |
| Portable nanofiber-light addressable potentiometric sensor for rapid detection of E. coli | - | 102 CFU·mL−1 | [29] |
| QCM Biosensor using carbohydrate and lectin | 7.5 × 102 to 7.5 × 107 cells. mL−1 | 7.5 × 102 cells.mL−1 | [30] |
| Electrochemical carbohydrate-based biosensor | 1.3–1.3 × 106 CFU·mL−1 | 2 CFU·mL−1 | This work |
| Sample | Concentration (CFU·mL−1) | ΔR in Real Sample | ΔR in Calibration Curve | Recovery (%) | RSD (%) |
|---|---|---|---|---|---|
| Tap Water | 104 | 43.791 | 45.099 | 97.10 | 3.84 |
| 105 | 71.491 | 67.231 | 106.34 | 2.37 | |
| 106 | 109.891 | 92.221 | 119.16 | 4.13 |
| Slope in Milk | Slope in Calibration Curve | Deviation from the Calibration Curve |
|---|---|---|
| 11.893 | 10.96 | 7.844 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hargol Zadeh, S.; Kashanian, S.; Nazari, M. A Label-Free Carbohydrate-Based Electrochemical Sensor to Detect Escherichia coli Pathogenic Bacteria Using D-mannose on a Glassy Carbon Electrode. Biosensors 2023, 13, 619. https://doi.org/10.3390/bios13060619
Hargol Zadeh S, Kashanian S, Nazari M. A Label-Free Carbohydrate-Based Electrochemical Sensor to Detect Escherichia coli Pathogenic Bacteria Using D-mannose on a Glassy Carbon Electrode. Biosensors. 2023; 13(6):619. https://doi.org/10.3390/bios13060619
Chicago/Turabian StyleHargol Zadeh, Sakineh, Soheila Kashanian, and Maryam Nazari. 2023. "A Label-Free Carbohydrate-Based Electrochemical Sensor to Detect Escherichia coli Pathogenic Bacteria Using D-mannose on a Glassy Carbon Electrode" Biosensors 13, no. 6: 619. https://doi.org/10.3390/bios13060619
APA StyleHargol Zadeh, S., Kashanian, S., & Nazari, M. (2023). A Label-Free Carbohydrate-Based Electrochemical Sensor to Detect Escherichia coli Pathogenic Bacteria Using D-mannose on a Glassy Carbon Electrode. Biosensors, 13(6), 619. https://doi.org/10.3390/bios13060619





