Recent Advances in Molecular and Immunological Diagnostic Platform for Virus Detection: A Review
Abstract
1. Introduction
2. Virus Detection on Microfluidic Platform
2.1. Immunoassay Methods
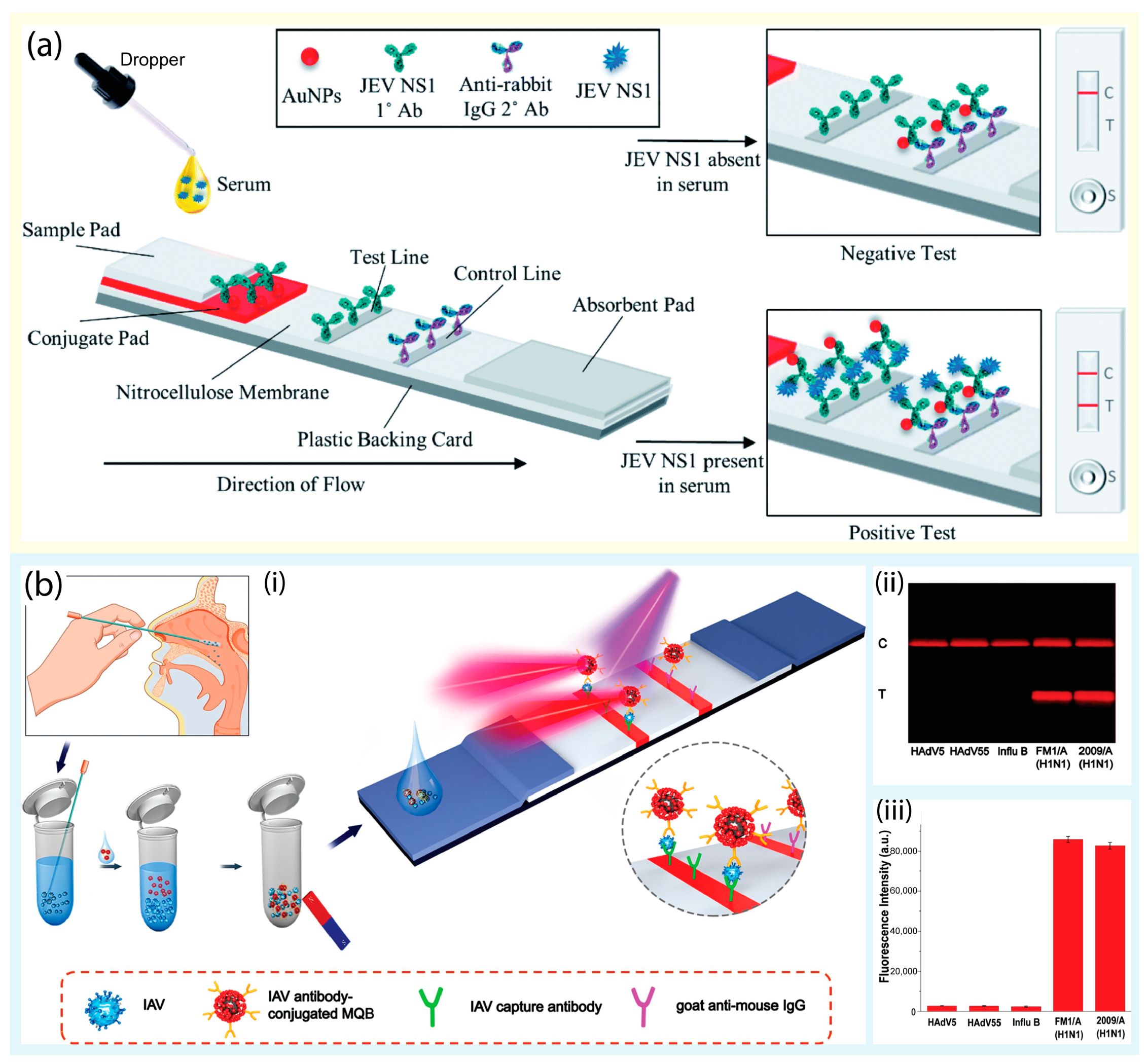
2.1.1. Lateral Flow Assay-Based Tests
2.1.2. Enzyme-Linked Immunosorbent Assay-Based Tests
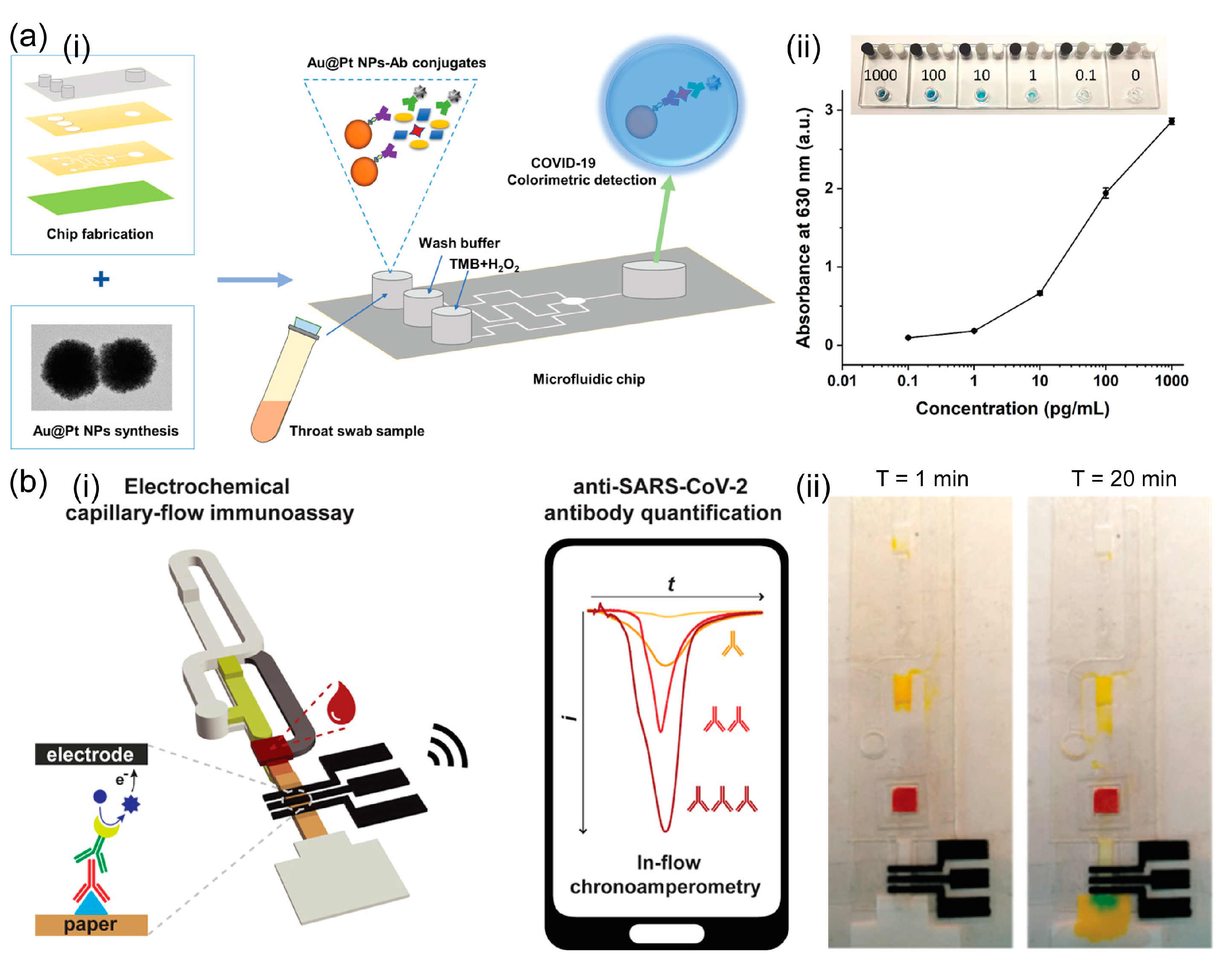
2.1.3. Others
2.2. Molecular Methods
2.2.1. PCR
2.2.2. Isothermal Amplification

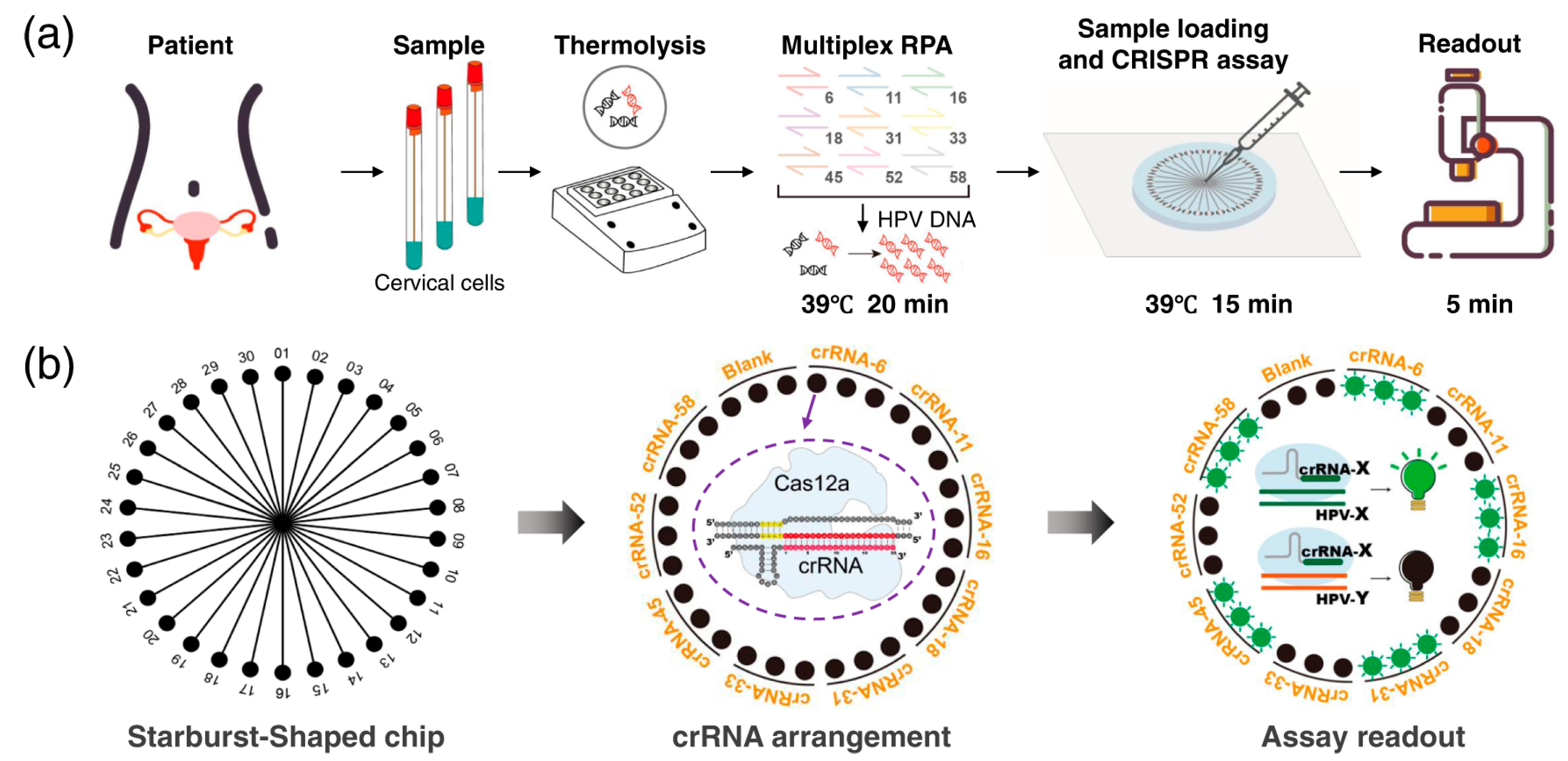
2.2.3. CRISPR-Assisted Method
2.3. Others
3. Microfluidic Assists in the Pandemic Era
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, N.-C.; Wang, C.-H.; Yoshimura, M.; Yeh, Y.-Q.; Guan, H.-H.; Chuankhayan, P.; Lin, C.-C.; Lin, P.-J.; Huang, Y.-C.; Wakatsuki, S.; et al. Structures of Honeybee-Infecting Lake Sinai Virus Reveal Domain Functions and Capsid Assembly with Dynamic Motions. Nat. Commun. 2023, 14, 545. [Google Scholar] [CrossRef] [PubMed]
- Zaeck, L.M.; Lamers, M.M.; Verstrepen, B.E.; Bestebroer, T.M.; van Royen, M.E.; Götz, H.; Shamier, M.C.; van Leeuwen, L.P.M.; Schmitz, K.S.; Alblas, K.; et al. Low Levels of Monkeypox Virus-Neutralizing Antibodies after MVA-BN Vaccination in Healthy Individuals. Nat. Med. 2023, 29, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Sumbria, D.; Berber, E.; Mathayan, M.; Rouse, B.T. Virus Infections and Host Metabolism—Can We Manage the Interactions? Front. Immunol. 2021, 11, 594963. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Naiyer, S.; Mansuri, S.; Soni, N.; Singh, V.; Bhat, K.H.; Singh, N.; Arora, G.; Mansuri, M.S. COVID-19 Diagnosis: A Comprehensive Review of the RT-QPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12, 1503. [Google Scholar] [CrossRef]
- Chowdhury, A.D.; Takemura, K.; Li, T.-C.; Suzuki, T.; Park, E.Y. Electrical Pulse-Induced Electrochemical Biosensor for Hepatitis E Virus Detection. Nat. Commun. 2019, 10, 3737. [Google Scholar] [CrossRef]
- Musso, D.; Gubler, D.J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. [Google Scholar] [CrossRef]
- Alhabbab, R.Y. Lateral Flow Immunoassays for Detecting Viral Infectious Antigens and Antibodies. Micromachines 2022, 13, 1901. [Google Scholar] [CrossRef]
- Fox, J.D. Nucleic Acid Amplification Tests for Detection of Respiratory Viruses. J. Clin. Virol. 2007, 40, S15–S23. [Google Scholar] [CrossRef]
- Pedersen, J.C. Hemagglutination-Inhibition Assay for Influenza Virus Subtype Identification and the Detection and Quantitation of Serum Antibodies to Influenza Virus. In Animal Influenza Virus; Spackman, E., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2014; Volume 1161, pp. 11–25. ISBN 978-1-4939-0757-1. [Google Scholar]
- Xiao, M.; Tian, F.; Liu, X.; Zhou, Q.; Pan, J.; Luo, Z.; Yang, M.; Yi, C. Virus Detection: From State-of-the-Art Laboratories to Smartphone-Based Point-of-Care Testing. Adv. Sci. 2022, 9, 2105904. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Wang, J.; Ren, L.; Li, L.; Liu, W.; Zhou, J.; Yu, W.; Tong, D.; Chen, S. Microfluidics: A New Cosset for Neurobiology. Lab Chip 2009, 9, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Nishat, S.; Jafry, A.T.; Martinez, A.W.; Awan, F.R. Paper-Based Microfluidics: Simplified Fabrication and Assay Methods. Sens. Actuators B Chem. 2021, 336, 129681. [Google Scholar] [CrossRef]
- Yang, Y.; Chen, Y.; Tang, H.; Zong, N.; Jiang, X. Microfluidics for Biomedical Analysis. Small Methods 2020, 4, 1900451. [Google Scholar] [CrossRef]
- Vigneshvar, S.; Sudhakumari, C.C.; Senthilkumaran, B.; Prakash, H. Recent Advances in Biosensor Technology for Potential Applications—An Overview. Front. Bioeng. Biotechnol. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Lin, B.; Tian, T.; Xu, X.; Wang, W.; Ruan, Q.; Guo, J.; Zhu, Z.; Yang, C. Recent Progress in Microfluidics-Based Biosensing. Anal. Chem. 2019, 91, 388–404. [Google Scholar] [CrossRef]
- Saez, J.; Catalan-Carrio, R.; Owens, R.M.; Basabe-Desmonts, L.; Benito-Lopez, F. Microfluidics and Materials for Smart Water Monitoring: A Review. Anal. Chim. Acta 2021, 1186, 338392. [Google Scholar] [CrossRef]
- Chen, G.; Zheng, J.; Liu, L.; Xu, L. Application of Microfluidics in Wearable Devices. Small Methods 2019, 3, 1900688. [Google Scholar] [CrossRef]
- Nilghaz, A.; Mousavi, S.M.; Li, M.; Tian, J.; Cao, R.; Wang, X. Paper-Based Microfluidics for Food Safety and Quality Analysis. Trends Food Sci. Technol. 2021, 118, 273–284. [Google Scholar] [CrossRef]
- Wang, J.; Jiang, C.; Jin, J.; Huang, L.; Yu, W.; Su, B.; Hu, J. Ratiometric Fluorescent Lateral Flow Immunoassay for Point-of-Care Testing of Acute Myocardial Infarction. Angew. Chem. Int. Ed. 2021, 60, 13042–13049. [Google Scholar] [CrossRef]
- Sena-Torralba, A.; Álvarez-Diduk, R.; Parolo, C.; Piper, A.; Merkoçi, A. Toward Next Generation Lateral Flow Assays: Integration of Nanomaterials. Chem. Rev. 2022, 122, 14881–14910. [Google Scholar] [CrossRef]
- Hwang, S.G.; Ha, K.; Guk, K.; Lee, D.K.; Eom, G.; Song, S.; Kang, T.; Park, H.; Jung, J.; Lim, E.-K. Rapid and Simple Detection of Tamiflu-Resistant Influenza Virus: Development of Oseltamivir Derivative-Based Lateral Flow Biosensor for Point-of-Care (POC) Diagnostics. Sci. Rep. 2018, 8, 12999. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chai, Y.; Hu, Z.; Xu, Z.; Li, M.; Chen, X.; Yang, C.; Liu, J. Recent Progress on Rapid Lateral Flow Assay-Based Early Diagnosis of COVID-19. Front. Bioeng. Biotechnol. 2022, 10, 866368. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-T.; Song, S.; Park, S.; Joo, C. Recent Advances in High-Sensitivity Detection Methods for Paper-Based Lateral-Flow Assay. Biosens. Bioelectron. 2020, 152, 112015. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.; Prakashan, D.; Dhanze, H.; Gandham, R.K.; Gandhi, S.; Sharma, G.T. Immuno-Chromatic Probe Based Lateral Flow Assay for Point-of-Care Detection of Japanese Encephalitis Virus NS1 Protein Biomarker in Clinical Samples Using a Smartphone-Based Approach. Nanoscale Adv. 2022, 4, 3966–3977. [Google Scholar] [CrossRef] [PubMed]
- Le, T.T.; Chang, P.; Benton, D.J.; McCauley, J.W.; Iqbal, M.; Cass, A.E.G. Dual Recognition Element Lateral Flow Assay toward Multiplex Strain Specific Influenza Virus Detection. Anal. Chem. 2017, 89, 6781–6786. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Wei, H.; Yang, X.; Zhu, Y.; Peng, Y.; Yang, J.; Wang, C.; Rong, Z.; Wang, S. Rapid Enrichment and Ultrasensitive Detection of Influenza A Virus in Human Specimen Using Magnetic Quantum Dot Nanobeads Based Test Strips. Sens. Actuators B Chem. 2020, 325, 128780. [Google Scholar] [CrossRef]
- Wiriyachaiporn, N.; Sirikett, H.; Maneeprakorn, W.; Dharakul, T. Carbon Nanotag Based Visual Detection of Influenza A Virus by a Lateral Flow Immunoassay. Microchim. Acta 2017, 184, 1827–1835. [Google Scholar] [CrossRef]
- Wang, D.; He, S.; Wang, X.; Yan, Y.; Liu, J.; Wu, S.; Liu, S.; Lei, Y.; Chen, M.; Li, L.; et al. Rapid Lateral Flow Immunoassay for the Fluorescence Detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. 2020, 4, 1150–1158. [Google Scholar] [CrossRef]
- Yue, W.; Xia, Z.; Zeng, Z.; Chen, Z.; Qiao, L.; Li, P.; He, Y.; Luo, X. In Situ Surface-Enhanced Raman Scattering Detection of a SARS-CoV-2 Biomarker Using Flexible and Transparent Polydimethylsiloxane Films with Embedded Au Nanoplates. ACS Appl. Nano Mater. 2022, 5, 12897–12906. [Google Scholar] [CrossRef]
- Tabarov, A.; Vitkin, V.; Andreeva, O.; Shemanaeva, A.; Popov, E.; Dobroslavin, A.; Kurikova, V.; Kuznetsova, O.; Grigorenko, K.; Tzibizov, I.; et al. Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors 2022, 12, 1065. [Google Scholar] [CrossRef]
- Driskell, J.D.; Zhu, Y.; Kirkwood, C.D.; Zhao, Y.; Dluhy, R.A.; Tripp, R.A. Rapid and Sensitive Detection of Rotavirus Molecular Signatures Using Surface Enhanced Raman Spectroscopy. PLoS ONE 2010, 5, e10222. [Google Scholar] [CrossRef] [PubMed]
- Nasir, S.; Majeed, M.I.; Nawaz, H.; Rashid, N.; Ali, S.; Farooq, S.; Kashif, M.; Rafiq, S.; Bano, S.; Ashraf, M.N.; et al. Surface Enhanced Raman Spectroscopy of RNA Samples Extracted from Blood of Hepatitis C Patients for Quantification of Viral Loads. Photodiagnosis Photodyn. Ther. 2021, 33, 102152. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Yang, S.C.; Choo, J. Early Diagnosis of Influenza Virus A Using Surface-Enhanced Raman Scattering-Based Lateral Flow Assay: Early Diagnosis of Influenza Virus A Using SERS LFA. Bull. Korean Chem. Soc. 2016, 37, 2019–2024. [Google Scholar] [CrossRef]
- Xiao, M.; Xie, K.; Dong, X.; Wang, L.; Huang, C.; Xu, F.; Xiao, W.; Jin, M.; Huang, B.; Tang, Y. Ultrasensitive Detection of Avian Influenza A (H7N9) Virus Using Surface-Enhanced Raman Scattering-Based Lateral Flow Immunoassay Strips. Anal. Chim. Acta 2019, 1053, 139–147. [Google Scholar] [CrossRef]
- Lu, M.; Joung, Y.; Jeon, C.S.; Kim, S.; Yong, D.; Jang, H.; Pyun, S.H.; Kang, T.; Choo, J. Dual-Mode SERS-Based Lateral Flow Assay Strips for Simultaneous Diagnosis of SARS-CoV-2 and Influenza a Virus. Nano Converg. 2022, 9, 39. [Google Scholar] [CrossRef] [PubMed]
- Joung, Y.; Kim, K.; Lee, S.; Chun, B.-S.; Lee, S.; Hwang, J.; Choi, S.; Kang, T.; Lee, M.-K.; Chen, L.; et al. Rapid and Accurate On-Site Immunodiagnostics of Highly Contagious Severe Acute Respiratory Syndrome Coronavirus 2 Using Portable Surface-Enhanced Raman Scattering-Lateral Flow Assay Reader. ACS Sens. 2022, 7, 3470–3480. [Google Scholar] [CrossRef]
- Yue, H.; Huang, M.; Tian, T.; Xiong, E.; Zhou, X. Advances in Clustered, Regularly Interspaced Short Palindromic Repeats (CRISPR)-Based Diagnostic Assays Assisted by Micro/Nanotechnologies. ACS Nano 2021, 15, 7848–7859. [Google Scholar] [CrossRef]
- Shao, N.; Han, X.; Song, Y.; Zhang, P.; Qin, L. CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip. Anal. Chem. 2019, 91, 12384–12391. [Google Scholar] [CrossRef]
- Wang, L.; Shen, X.; Wang, T.; Chen, P.; Qi, N.; Yin, B.-C.; Ye, B.-C. A Lateral Flow Strip Combined with Cas9 Nickase-Triggered Amplification Reaction for Dual Food-Borne Pathogen Detection. Biosens. Bioelectron. 2020, 165, 112364. [Google Scholar] [CrossRef]
- Zai, J.; Yi, K.; Xie, L.; Zhu, J.; Feng, X.; Li, Y. Dual Monoclonal Antibody-Based Sandwich ELISA for Detection of in vitro Packaged Ebola Virus. Diagn. Pathol. 2018, 13, 96. [Google Scholar] [CrossRef]
- Farre, C.; Viezzi, S.; Wright, A.; Robin, P.; Lejal, N.; Manzano, M.; Vidic, J.; Chaix, C. Specific and Sensitive Detection of Influenza A Virus Using a Biotin-Coated Nanoparticle Enhanced Immunomagnetic Assay. Anal. Bioanal. Chem. 2022, 414, 265–276. [Google Scholar] [CrossRef]
- MacMullan, M.A.; Ibrayeva, A.; Trettner, K.; Deming, L.; Das, S.; Tran, F.; Moreno, J.R.; Casian, J.G.; Chellamuthu, P.; Kraft, J.; et al. ELISA Detection of SARS-CoV-2 Antibodies in Saliva. Sci. Rep. 2020, 10, 20818. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-K.; Huang, H.-Y.; Chen, W.-R.; Nishie, W.; Ujiie, H.; Natsuga, K.; Fan, S.-T.; Wang, H.-K.; Lee, J.Y.-Y.; Tsai, W.-L.; et al. Paper-Based ELISA for the Detection of Autoimmune Antibodies in Body Fluid—The Case of Bullous Pemphigoid. Anal. Chem. 2014, 86, 4605–4610. [Google Scholar] [CrossRef] [PubMed]
- Hoy, C.F.O.; Kushiro, K.; Yamaoka, Y.; Ryo, A.; Takai, M. Rapid Multiplex Microfiber-Based Immunoassay for Anti-MERS-CoV Antibody Detection. Sens. Bio-Sens. Res. 2019, 26, 100304. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, H.; Wang, Y.; Zhang, P.; Zhu, Z.; Yang, C. Dissolution-Enhanced Luminescence Enhanced Digital Microfluidics Immunoassay for Sensitive and Automated Detection of H5N1. ACS Appl. Mater. Interfaces 2023, 15, 6526–6535. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Liu, L.; Wu, X.; Kuang, H.; Xu, C.; Xu, L. A Colorimetric Paper-Based Sensor for Toltrazuril and Its Metabolites in Feed, Chicken, and Egg Samples. Food Chem. 2019, 276, 707–713. [Google Scholar] [CrossRef]
- Coarsey, C.; Coleman, B.; Kabir, M.A.; Sher, M.; Asghar, W. Development of a Flow-Free Magnetic Actuation Platform for an Automated Microfluidic ELISA. RSC Adv. 2019, 9, 8159–8168. [Google Scholar] [CrossRef]
- Yamaoka, Y.; Miyakawa, K.; Jeremiah, S.S.; Funabashi, R.; Okudela, K.; Kikuchi, S.; Katada, J.; Wada, A.; Takei, T.; Nishi, M.; et al. Highly Specific Monoclonal Antibodies and Epitope Identification against SARS-CoV-2 Nucleocapsid Protein for Antigen Detection Tests. Cell Rep. Med. 2021, 2, 100311. [Google Scholar] [CrossRef]
- Shan, Y.; Feng, Y.; Li, J.; Yi, W.; Ge, M.; Huang, H.; Yan, K.; Wang, S.; Liu, F. Rapid On-Site PEDV Detection Using Homogeneous Fluorescence Resonance Energy Transfer-Based ELISA. Sens. Actuators B Chem. 2023, 378, 133138. [Google Scholar] [CrossRef]
- Wu, F.; Mao, M.; Cai, L.; Lin, Q.; Guan, X.; Shi, X.; Ma, L. Platinum-Decorated Gold Nanoparticle-Based Microfluidic Chip Immunoassay for Ultrasensitive Colorimetric Detection of SARS-CoV-2 Nucleocapsid Protein. ACS Biomater. Sci. Eng. 2022, 8, 3924–3932. [Google Scholar] [CrossRef]
- Ma, L.; Abugalyon, Y.; Li, X. Multicolorimetric ELISA Biosensors on a Paper/Polymer Hybrid Analytical Device for Visual Point-of-Care Detection of Infection Diseases. Anal. Bioanal. Chem. 2021, 413, 4655–4663. [Google Scholar] [CrossRef]
- Kasetsirikul, S.; Umer, M.; Soda, N.; Sreejith, K.R.; Shiddiky, M.J.A.; Nguyen, N.-T. Detection of the SARS-CoV-2 Humanized Antibody with Paper-Based ELISA. Analyst 2020, 145, 7680–7686. [Google Scholar] [CrossRef]
- Ozefe, F.; Yildiz, A.A. Fabrication and Development of a Microfluidic Paper-Based Immunosorbent Assay Platform (μPISA) for Colorimetric Detection of Hepatitis C. Analyst 2023, 148, 898–905. [Google Scholar] [CrossRef]
- Song, Y.; Ye, Y.; Su, S.-H.; Stephens, A.; Cai, T.; Chung, M.-T.; Han, M.K.; Newstead, M.W.; Yessayan, L.; Frame, D.; et al. A Digital Protein Microarray for COVID-19 Cytokine Storm Monitoring. Lab Chip 2021, 21, 331–343. [Google Scholar] [CrossRef]
- Clark, K.M.; Schenkel, M.S.; Pittman, T.W.; Samper, I.C.; Anderson, L.B.R.; Khamcharoen, W.; Elmegerhi, S.; Perera, R.; Siangproh, W.; Kennan, A.J.; et al. Electrochemical Capillary Driven Immunoassay for Detection of SARS-CoV-2. ACS Meas. Sci. Au 2022, 2, 584–594. [Google Scholar] [CrossRef]
- Samper, I.C.; Sánchez-Cano, A.; Khamcharoen, W.; Jang, I.; Siangproh, W.; Baldrich, E.; Geiss, B.J.; Dandy, D.S.; Henry, C.S. Electrochemical Capillary-Flow Immunoassay for Detecting Anti-SARS-CoV-2 Nucleocapsid Protein Antibodies at the Point of Care. ACS Sens. 2021, 6, 4067–4075. [Google Scholar] [CrossRef] [PubMed]
- Samper, I.C.; McMahon, C.J.; Schenkel, M.S.; Clark, K.M.; Khamcharoen, W.; Anderson, L.B.R.; Terry, J.S.; Gallichotte, E.N.; Ebel, G.D.; Geiss, B.J.; et al. Electrochemical Immunoassay for the Detection of SARS-CoV-2 Nucleocapsid Protein in Nasopharyngeal Samples. Anal. Chem. 2022, 94, 4712–4719. [Google Scholar] [CrossRef]
- Wang, C.; Wang, C.; Qiu, J.; Gao, J.; Liu, H.; Zhang, Y.; Han, L. Ultrasensitive, High-Throughput, and Rapid Simultaneous Detection of SARS-CoV-2 Antigens and IgG/IgM Antibodies within 10 Min through an Immunoassay Biochip. Microchim. Acta 2021, 188, 262. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Wei, H.; Qi, J.; Ma, H.; Liu, L.; Weng, J.; Zheng, X.; Li, Q.; Zhao, D.; Fang, H.; et al. Pulling-Force Spinning Top for Serum Separation Combined with Paper-Based Microfluidic Devices in COVID-19 ELISA Diagnosis. ACS Sens. 2021, 6, 2709–2719. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-A.; Yuan, H.; Chen, C.-W.; Chien, Y.-S.; Sheng, W.-H.; Chen, C.-F. An Electricity- and Instrument-Free Infectious Disease Sensor Based on a 3D Origami Paper-Based Analytical Device. Lab Chip 2021, 21, 1908–1915. [Google Scholar] [CrossRef]
- Ke, G.; Su, D.; Li, Y.; Zhao, Y.; Wang, H.; Liu, W.; Li, M.; Yang, Z.; Xiao, F.; Yuan, Y.; et al. An Accurate, High-Speed, Portable Bifunctional Electrical Detector for COVID-19. Sci. China Mater. 2021, 64, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Ramadan, S.; Rosa, B.G.; Zhang, Y.; Yin, T.; Torres, E.; Shaforost, O.; Panagiotopoulos, A.; Li, B.; Kerherve, G.; et al. On-Chip Integrated Graphene Aptasensor with Portable Readout for Fast and Label-Free COVID-19 Detection in Virus Transport Medium. Sens. Diagn. 2022, 1, 719–730. [Google Scholar] [CrossRef]
- Manimekala, T.; Sivasubramanian, R.; Dharmalingam, G. Nanomaterial-Based Biosensors Using Field-Effect Transistors: A Review. J. Electron. Mater. 2022, 51, 1950–1973. [Google Scholar] [CrossRef]
- Seo, G.; Lee, G.; Kim, M.J.; Baek, S.-H.; Choi, M.; Ku, K.B.; Lee, C.-S.; Jun, S.; Park, D.; Kim, H.G.; et al. Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor. ACS Nano 2020, 14, 5135–5142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ren, R.; Pu, H.; Guo, X.; Chang, J.; Zhou, G.; Mao, S.; Kron, M.; Chen, J. Field-Effect Transistor Biosensor for Rapid Detection of Ebola Antigen. Sci. Rep. 2017, 7, 10974. [Google Scholar] [CrossRef] [PubMed]
- Wasfi, A.; Awwad, F.; Qamhieh, N.; Al Murshidi, B.; Palakkott, A.R.; Gelovani, J.G. Real-Time COVID-19 Detection via Graphite Oxide-Based Field-Effect Transistor Biosensors Decorated with Pt/Pd Nanoparticles. Sci. Rep. 2022, 12, 18155. [Google Scholar] [CrossRef]
- Fathi-Hafshejani, P.; Azam, N.; Wang, L.; Kuroda, M.A.; Hamilton, M.C.; Hasim, S.; Mahjouri-Samani, M. Two-Dimensional-Material-Based Field-Effect Transistor Biosensor for Detecting COVID-19 Virus (SARS-CoV-2). ACS Nano 2021, 15, 11461–11469. [Google Scholar] [CrossRef]
- Park, S.; Su Jeon, C.; Choi, N.; Moon, J.-I.; Min Lee, K.; Hyun Pyun, S.; Kang, T.; Choo, J. Sensitive and Reproducible Detection of SARS-CoV-2 Using SERS-Based Microdroplet Sensor. Chem. Eng. J. 2022, 446, 137085. [Google Scholar] [CrossRef]
- Akter, N.; Hasan, M.M.; Pala, N. A Review of THz Technologies for Rapid Sensing and Detection of Viruses Including SARS-CoV-2. Biosensors 2021, 11, 349. [Google Scholar] [CrossRef]
- Di Fabrizio, M.; Lupi, S.; D’Arco, A. Virus Recognition with Terahertz Radiation: Drawbacks and Potentialities. J. Phys. Photonics 2021, 3, 032001. [Google Scholar] [CrossRef]
- Mancini, T.; Marcelli, A.; Lupi, S.; D’Arco, A. New Frontier in Terahertz Technologies for Virus Sensing. Electronics 2022, 12, 135. [Google Scholar] [CrossRef]
- Lee, D.K.; Kang, J.H.; Kwon, J.; Lee, J.S.; Lee, S.; Woo, D.H.; Kim, J.H.; Song, C.-S.; Park, Q.-H.; Seo, M. Nano Metamaterials for Ultrasensitive Terahertz Biosensing. Sci. Rep. 2017, 7, 8146. [Google Scholar] [CrossRef] [PubMed]
- Ahmadivand, A.; Gerislioglu, B.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Rapid Detection of Infectious Envelope Proteins by Magnetoplasmonic Toroidal Metasensors. ACS Sens. 2017, 2, 1359–1368. [Google Scholar] [CrossRef]
- Ahmadivand, A.; Gerislioglu, B.; Tomitaka, A.; Manickam, P.; Kaushik, A.; Bhansali, S.; Nair, M.; Pala, N. Extreme Sensitive Metasensor for Targeted Biomarkers Identification Using Colloidal Nanoparticles-Integrated Plasmonic Unit Cells. Biomed. Opt. Express 2018, 9, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Fan, F.; Li, S.; Zhang, Z.; Liu, H.; Wang, X.; Chang, S. Terahertz Immunosensing Assisted by Functionalized Au NPs Based on All-Dielectric Metasurface. Sens. Actuators B Chem. 2022, 362, 131777. [Google Scholar] [CrossRef]
- Chen, H.; Kim, S.; Hardie, J.M.; Thirumalaraju, P.; Gharpure, S.; Rostamian, S.; Udayakumar, S.; Lei, Q.; Cho, G.; Kanakasabapathy, M.K.; et al. Deep Learning-Assisted Sensitive Detection of Fentanyl Using a Bubbling-Microchip. Lab Chip 2022, 22, 4531–4540. [Google Scholar] [CrossRef]
- Gao, Z.; Song, Y.; Hsiao, T.Y.; He, J.; Wang, C.; Shen, J.; MacLachlan, A.; Dai, S.; Singer, B.H.; Kurabayashi, K.; et al. Machine-Learning-Assisted Microfluidic Nanoplasmonic Digital Immunoassay for Cytokine Storm Profiling in COVID-19 Patients. ACS Nano 2021, 15, 18023–18036. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tontisirin, S.; Jiraseree-amornkun, A.; Chuaypen, N.; Tangkijvanich, P.; Henry, C.S.; Ngamrojanavanich, N.; Chailapakul, O. NFC-Enabling Smartphone-Based Portable Amperometric Immunosensor for Hepatitis B Virus Detection. Sens. Actuators B Chem. 2021, 326, 128825. [Google Scholar] [CrossRef]
- Saylan, Y.; Erdem, Ö.; Ünal, S.; Denizli, A. An Alternative Medical Diagnosis Method: Biosensors for Virus Detection. Biosensors 2019, 9, 65. [Google Scholar] [CrossRef]
- Zhu, H.; Fohlerová, Z.; Pekárek, J.; Basova, E.; Neužil, P. Recent Advances in Lab-on-a-Chip Technologies for Viral Diagnosis. Biosens. Bioelectron. 2020, 153, 112041. [Google Scholar] [CrossRef]
- Paniz-Mondolfi, A.; Guerra, S.; Muñoz, M.; Luna, N.; Hernandez, M.M.; Patino, L.H.; Reidy, J.; Banu, R.; Shrestha, P.; Liggayu, B.; et al. Evaluation and Validation of an RT-PCR Assay for Specific Detection of Monkeypox Virus (MPXV). J. Med. Virol. 2023, 95, e28247. [Google Scholar] [CrossRef] [PubMed]
- Gupta, N.; Augustine, S.; Narayan, T.; O’Riordan, A.; Das, A.; Kumar, D.; Luong, J.H.T.; Malhotra, B.D. Point-of-Care PCR Assays for COVID-19 Detection. Biosensors 2021, 11, 141. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, K.J.; Read, S.J.; Peto, T.E.; Mayon-White, R.T.; Bangham, C.R. Diagnosis of Viral Infections of the Central Nervous System: Clinical Interpretation of PCR Results. Lancet 1997, 349, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Trinh, K.T.L.; Stabler, R.A.; Lee, N.Y. Fabrication of a Foldable All-in-One Point-of-Care Molecular Diagnostic Microdevice for the Facile Identification of Multiple Pathogens. Sens. Actuators B Chem. 2020, 314, 128057. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Trinh, K.T.L.; Yoon, W.J.; Lee, N.Y.; Ju, H. Integration of a Microfluidic Polymerase Chain Reaction Device and Surface Plasmon Resonance Fiber Sensor into an Inline All-in-One Platform for Pathogenic Bacteria Detection. Sens. Actuators B Chem. 2017, 242, 1–8. [Google Scholar] [CrossRef]
- Trick, A.Y.; Chen, F.; Chen, L.; Lee, P.; Hasnain, A.C.; Mostafa, H.H.; Carroll, K.C.; Wang, T. Point-of-Care Platform for Rapid Multiplexed Detection of SARS-CoV-2 Variants and Respiratory Pathogens. Adv. Mater. Technol. 2022, 7, 2101013. [Google Scholar] [CrossRef]
- Ngo, H.T.; Jin, M.; Trick, A.Y.; Chen, F.-E.; Chen, L.; Hsieh, K.; Wang, T.-H. Sensitive and Quantitative Point-of-Care HIV Viral Load Quantification from Blood Using a Power-Free Plasma Separation and Portable Magnetofluidic Polymerase Chain Reaction Instrument. Anal. Chem. 2023, 95, 1159–1168. [Google Scholar] [CrossRef]
- Poritz, M.A.; Blaschke, A.J.; Byington, C.L.; Meyers, L.; Nilsson, K.; Jones, D.E.; Thatcher, S.A.; Robbins, T.; Lingenfelter, B.; Amiott, E.; et al. FilmArray, an Automated Nested Multiplex PCR System for Multi-Pathogen Detection: Development and Application to Respiratory Tract Infection. PLoS ONE 2011, 6, e26047. [Google Scholar] [CrossRef]
- Jiang, Y.; Manz, A.; Wu, W. Fully Automatic Integrated Continuous-Flow Digital PCR Device for Absolute DNA Quantification. Anal. Chim. Acta 2020, 1125, 50–56. [Google Scholar] [CrossRef]
- Feng, J.; Xue, G.; Cui, X.; Du, B.; Feng, Y.; Cui, J.; Zhao, H.; Gan, L.; Fan, Z.; Fu, T.; et al. Development of a Loop-Mediated Isothermal Amplification Method for Rapid and Visual Detection of Monkeypox Virus. Microbiol. Spectr. 2022, 10, e02714-22. [Google Scholar] [CrossRef]
- Wu, W.; Yin, C.; Yue, A.; Niu, J.; Du, W.; Liu, D.; Zhao, J. Rapid and Visual Detection of Soybean Mosaic Virus SC7 with a Loop-Mediated Isothermal Amplification Strategy. Sens. Actuators B Chem. 2022, 373, 132733. [Google Scholar] [CrossRef]
- Ye, X.; Zhou, H.; Guo, X.; Liu, D.; Li, Z.; Sun, J.; Huang, J.; Liu, T.; Zhao, P.; Xu, H.; et al. Argonaute-Integrated Isothermal Amplification for Rapid, Portable, Multiplex Detection of SARS-CoV-2 and Influenza Viruses. Biosens. Bioelectron. 2022, 207, 114169. [Google Scholar] [CrossRef]
- Trinh, T.N.D.; Lee, N.Y. Advances in Nucleic Acid Amplification-Based Microfluidic Devices for Clinical Microbial Detection. Chemosensors 2022, 10, 123. [Google Scholar] [CrossRef]
- Xia, X.; Yang, H.; Cao, J.; Zhang, J.; He, Q.; Deng, R. Isothermal Nucleic Acid Amplification for Food Safety Analysis. TrAC Trends Anal. Chem. 2022, 153, 116641. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Li, Q.; Wang, L.; Fan, C. Isothermal Amplification of Nucleic Acids. Chem. Rev. 2015, 115, 12491–12545. [Google Scholar] [CrossRef] [PubMed]
- Atceken, N.; Munzer Alseed, M.; Dabbagh, S.R.; Yetisen, A.K.; Tasoglu, S. Point-of-Care Diagnostic Platforms for Loop-Mediated Isothermal Amplification. Adv. Eng. Mater. 2023, 25, 2201174. [Google Scholar] [CrossRef]
- Deng, H.; Jayawardena, A.; Chan, J.; Tan, S.M.; Alan, T.; Kwan, P. An Ultra-Portable, Self-Contained Point-of-Care Nucleic Acid Amplification Test for Diagnosis of Active COVID-19 Infection. Sci. Rep. 2021, 11, 15176. [Google Scholar] [CrossRef]
- Davidson, J.L.; Wang, J.; Maruthamuthu, M.K.; Dextre, A.; Pascual-Garrigos, A.; Mohan, S.; Putikam, S.V.S.; Osman, F.O.I.; McChesney, D.; Seville, J.; et al. A Paper-Based Colorimetric Molecular Test for SARS-CoV-2 in Saliva. Biosens. Bioelectron. X 2021, 9, 100076. [Google Scholar] [CrossRef]
- Wang, H.; Xu, J.; Li, S.; Wang, X.; Liu, G.; Yang, S.; Zhao, F.; Liu, Q.; Chen, X.; He, C.; et al. An integrated dual-layer microfluidic platform for multiple respiratory viruses screening. Anal. Chim. Acta 2023, 1242, 340812. [Google Scholar] [CrossRef] [PubMed]
- Lobato, I.M.; O’Sullivan, C.K. Recombinase Polymerase Amplification: Basics, Applications and Recent Advances. TrAC Trends Anal. Chem. 2018, 98, 19–35. [Google Scholar] [CrossRef]
- Kim, H.E.; Schuck, A.; Lee, S.H.; Lee, Y.; Kang, M.; Kim, Y.-S. Sensitive Electrochemical Biosensor Combined with Isothermal Amplification for Point-of-Care COVID-19 Tests. Biosens. Bioelectron. 2021, 182, 113168. [Google Scholar] [CrossRef] [PubMed]
- Kong, M.; Li, Z.; Wu, J.; Hu, J.; Sheng, Y.; Wu, D.; Lin, Y.; Li, M.; Wang, X.; Wang, S. A Wearable Microfluidic Device for Rapid Detection of HIV-1 DNA Using Recombinase Polymerase Amplification. Talanta 2019, 205, 120155. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.-K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling Circle Amplification: A Versatile Tool for Chemical Biology, Materials Science and Medicine. Chem. Soc. Rev. 2014, 43, 3324. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Abbas, N.; Shin, S. A Rapid Diagnosis of SARS-CoV-2 Using DNA Hydrogel Formation on Microfluidic Pores. Biosens. Bioelectron. 2021, 177, 113005. [Google Scholar] [CrossRef]
- Kaarj, K.; Akarapipad, P.; Yoon, J.-Y. Simpler, Faster, and Sensitive Zika Virus Assay Using Smartphone Detection of Loop-Mediated Isothermal Amplification on Paper Microfluidic Chips. Sci. Rep. 2018, 8, 12438. [Google Scholar] [CrossRef] [PubMed]
- Alamillo, J.M.; López, C.M.; Martínez Rivas, F.J.; Torralbo, F.; Bulut, M.; Alseekh, S. Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-Associated Protein and Hairy Roots: A Perfect Match for Gene Functional Analysis and Crop Improvement. Curr. Opin. Biotechnol. 2023, 79, 102876. [Google Scholar] [CrossRef]
- Azimzadeh, M.; Mousazadeh, M.; Jahangiri-Manesh, A.; Khashayar, P.; Khashayar, P. CRISPR-Powered Microfluidics in Diagnostics: A Review of Main Applications. Chemosensors 2021, 10, 3. [Google Scholar] [CrossRef]
- Nafian, F.; Nafian, S.; Kamali Doust Azad, B.; Hashemi, M. CRISPR-Based Diagnostics and Microfluidics for COVID-19 Point-of-Care Testing: A Review of Main Applications. Mol. Biotechnol. 2023, 65, 497–508. [Google Scholar] [CrossRef]
- Tian, T.; Qiu, Z.; Jiang, Y.; Zhu, D.; Zhou, X. Exploiting the Orthogonal CRISPR-Cas12a/Cas13a Trans-Cleavage for Dual-Gene Virus Detection Using a Handheld Device. Biosens. Bioelectron. 2022, 196, 113701. [Google Scholar] [CrossRef]
- Li, Y.; Man, S.; Ye, S.; Liu, G.; Ma, L. CRISPR-Cas-based Detection for Food Safety Problems: Current Status, Challenges, and Opportunities. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3770–3798. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An Updated Evolutionary Classification of CRISPR–Cas Systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Kaminski, M.M.; Abudayyeh, O.O.; Gootenberg, J.S.; Zhang, F.; Collins, J.J. CRISPR-Based Diagnostics. Nat. Biomed. Eng. 2021, 5, 643–656. [Google Scholar] [CrossRef] [PubMed]
- Feng, W.; Newbigging, A.M.; Tao, J.; Cao, Y.; Peng, H.; Le, C.; Wu, J.; Pang, B.; Li, J.; Tyrrell, D.L.; et al. CRISPR Technology Incorporating Amplification Strategies: Molecular Assays for Nucleic Acids, Proteins, and Small Molecules. Chem. Sci. 2021, 12, 4683–4698. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Shen, Z.; Wang, G.; Gu, W.; Zhao, S.; Lin, Z.; Liu, W.; Cai, Y.; Mushtaq, G.; Jia, J.; et al. Research Progress of CRISPR-Based Biosensors and Bioassays for Molecular Diagnosis. Front. Bioeng. Biotechnol. 2022, 10, 986233. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, D.; Li, T.; Yan, J.; Zhu, J.; He, T.; Hu, R.; Li, Y.; Yang, Y.; Liu, M. Microfluidic Space Coding for Multiplexed Nucleic Acid Detection via CRISPR-Cas12a and Recombinase Polymerase Amplification. Nat. Commun. 2022, 13, 6480. [Google Scholar] [CrossRef]
- Nguyen, P.Q.; Soenksen, L.R.; Donghia, N.M.; Angenent-Mari, N.M.; de Puig, H.; Huang, A.; Lee, R.; Slomovic, S.; Galbersanini, T.; Lansberry, G.; et al. Wearable Materials with Embedded Synthetic Biology Sensors for Biomolecule Detection. Nat. Biotechnol. 2021, 39, 1366–1374. [Google Scholar] [CrossRef]
- Qin, P.; Park, M.; Alfson, K.J.; Tamhankar, M.; Carrion, R.; Patterson, J.L.; Griffiths, A.; He, Q.; Yildiz, A.; Mathies, R.; et al. Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a. ACS Sens. 2019, 4, 1048–1054. [Google Scholar] [CrossRef]
- Talwar, C.S.; Park, K.-H.; Ahn, W.-C.; Kim, Y.-S.; Kwon, O.S.; Yong, D.; Kang, T.; Woo, E. Detection of Infectious Viruses Using CRISPR-Cas12-Based Assay. Biosensors 2021, 11, 301. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, Z.; He, L.; Wang, Z.; Zhang, T.; Hu, T.; Huang, F.; Chen, D.; Li, Y.; Yang, Y.; et al. Coupling CRISPR/Cas12a and Recombinase Polymerase Amplification on a Stand-Alone Microfluidics Platform for Fast and Parallel Nucleic Acid Detection. Anal. Chem. 2023, 95, 3379–3389. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, D.; Xu, Z.; Li, T.; Zhu, J.; Hu, R.; Xu, G.; Li, Y.; Yang, Y.; Liu, M. Integrating CRISPR-Cas12a into a Microfluidic Dual-Droplet Device Enables Simultaneous Detection of HPV16 and HPV18. Anal. Chem. 2023, 95, 3476–3485. [Google Scholar] [CrossRef]
- Park, B.J.; Park, M.S.; Lee, J.M.; Song, Y.J. Specific Detection of Influenza A and B Viruses by CRISPR-Cas12a-Based Assay. Biosensors 2021, 11, 88. [Google Scholar] [CrossRef]
- Yin, K.; Ding, X.; Li, Z.; Sfeir, M.M.; Ballesteros, E.; Liu, C. Autonomous Lab-on-Paper for Multiplexed, CRISPR-Based Diagnostics of SARS-CoV-2. Lab Chip 2021, 21, 2730–2737. [Google Scholar] [CrossRef]
- Welch, N.L.; Zhu, M.; Hua, C.; Weller, J.; Mirhashemi, M.E.; Nguyen, T.G.; Mantena, S.; Bauer, M.R.; Shaw, B.M.; Ackerman, C.M.; et al. Multiplexed CRISPR-Based Microfluidic Platform for Clinical Testing of Respiratory Viruses and Identification of SARS-CoV-2 Variants. Nat. Med. 2022, 28, 1083–1094. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Zhang, L.; Lu, Y.; Wang, X.; Shen, M.; Li, N.; Feng, L.; Jing, J.; Cao, B.; et al. Sensitive and Rapid Diagnosis of Respiratory Virus Coinfection Using a Microfluidic Chip-Powered CRISPR/Cas12a System. Small 2022, 18, 2200854. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Deng, R.; Wang, Y.; Wu, C.; Zhang, K.; Wang, C.; Gong, N.; Ledesma-Amaro, R.; Teng, X.; Yang, C.; et al. A Paper-Based Assay for the Colorimetric Detection of SARS-CoV-2 Variants at Single-Nucleotide Resolution. Nat. Biomed. Eng. 2022, 6, 957–967. [Google Scholar] [CrossRef] [PubMed]
- Du, K.; Cai, H.; Park, M.; Wall, T.A.; Stott, M.A.; Alfson, K.J.; Griffiths, A.; Carrion, R.; Patterson, J.L.; Hawkins, A.R.; et al. Multiplexed Efficient On-Chip Sample Preparation and Sensitive Amplification-Free Detection of Ebola Virus. Biosens. Bioelectron. 2017, 91, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.; Li, Y. An Impedance Aptasensor with Microfluidic Chips for Specific Detection of H5N1 Avian Influenza Virus. Sensors 2015, 15, 18565–18578. [Google Scholar] [CrossRef]
- Wang, L.; Wang, X.; Wu, Y.; Guo, M.; Gu, C.; Dai, C.; Kong, D.; Wang, Y.; Zhang, C.; Qu, D.; et al. Rapid and Ultrasensitive Electromechanical Detection of Ions, Biomolecules and SARS-CoV-2 RNA in Unamplified Samples. Nat. Biomed. Eng. 2022, 6, 276–285. [Google Scholar] [CrossRef]
- Naseri, M.; Ziora, Z.M.; Simon, G.P.; Batchelor, W. ASSURED-compliant Point-of-care Diagnostics for the Detection of Human Viral Infections. Rev. Med. Virol. 2022, 32, e2263. [Google Scholar] [CrossRef]
- Otoo, J.A.; Schlappi, T.S. REASSURED Multiplex Diagnostics: A Critical Review and Forecast. Biosensors 2022, 12, 124. [Google Scholar] [CrossRef] [PubMed]
- Microfluidic Market by Product (Devices, Components (Chip, Sensor, Pump, Valve)), Application (IVD (POC, Clinical, Veterinary), Research, Manufacturing, Therapeutics), End User (Hospital, Diagnostic Center, Academic Institutes) & Region–Global Forecast to 2026. Available online: https://www.marketsandmarkets.com/MarketReports/microfluidiccomponentsmarket223516809.html?gclid=Cj0KCQjwocShBhCOARIsAFVYq0hasSvef7ZI99HMun9GGQei_1LrTOZ20E3yLcwmU9rq66VKA2j7saAsIdEALw_wcB (accessed on 7 April 2023).
- Sachdeva, S.; Davis, R.W.; Saha, A.K. Microfluidic Point-of-Care Testing: Commercial Landscape and Future Directions. Front. Bioeng. Biotechnol. 2021, 8, 602659. [Google Scholar] [CrossRef] [PubMed]
- Benda, A.; Zerajic, L.; Ankita, A.; Cleary, E.; Park, Y.; Pandey, S. COVID-19 Testing and Diagnostics: A Review of Commercialized Technologies for Cost, Convenience and Quality of Tests. Sensors 2021, 21, 6581. [Google Scholar] [CrossRef] [PubMed]
- Elecsys® Anti-SARS-CoV-2—Instruction for Use. Available online: https://www.fda.gov/media/137605/download (accessed on 21 February 2023).
- Cobas® SARS-CoV-2—Instruction for Use. Available online: https://www.fda.gov/media/136049/download (accessed on 21 February 2023).
- ID NOW™ COVID-19—Instruction for Use. Available online: https://www.fda.gov/media/136525/download (accessed on 21 February 2023).
- Xpert® Xpress CoV-2 plus—Instruction for Use. Available online: https://www.fda.gov/media/158407/download (accessed on 21 February 2023).
- 1copy COVID-19 qPCR Multi Kit—Instructions for Use. Available online: https://www.fda.gov/media/137935/download (accessed on 7 April 2023).
- Lucira COVID-19 All-In-One Test Kit + PDF Report (Good For Travel)—Plus PDF Results (Good for Travel). Available online: https://www.meenta.io/product/lucira-covid-19-all-in-one-test/ (accessed on 7 April 2023).
- TEST COVID-19—Biosynex. Available online: https://www.biosynex.com/en/pharmacie-para-test-covid-19/ (accessed on 7 April 2023).





| Company | Main Products |
|---|---|
| Roche Diagnostics | Rapid antigens test cobas® SARS-CoV-2 & Influenza A/B test |
| Cepheid | GeneXpert System Xpert® Xpress SARS-CoV-2 |
| Caliper Life Sciences | LabChip system |
| Abbott Laboratories | ID Now™ Determine™ HIV-1/2 AG/AB Combo |
| Standard BioTools Inc. | BioMark HD System X9™ Real-Time PCR System |
| Agilent Technologies | The AriaDx instrument |
| Platform | Manufacturer | Assay | Target | Analysis Time | Ref. |
|---|---|---|---|---|---|
| Elecsys® Anti-SARS-CoV-2 | Roche | Immunoassay | SARS-CoV-2 antibodies | 18 min | [135] |
| Cobas® SARS-CoV-2 | Roche | RT-PCR | ORF1a/b non-structural region, E gene | 20 min | [136] |
| ID NOW™ COVID-19 | Abbott | NEAR | RdRp gene | 15 min | [137] |
| Xpert® Xpress CoV-2 plus | Cepheid | RT-PCR | N, E gene | 45 min | [138] |
| 1copy COVID-19 qPCR Kit | 1drop Inc | RT-PCR | E, RdRp gene | 22 min | [139] |
| Lucira COVID-19 All-In-One Test Kit | Lucira Health | RT-LAMP | N Gene | 30 min | [140] |
| Biosynex COVID-19 Ag+ BSS Rapid Test | BIOSYNEX S.A. | RT-PCR | N-protein | 10 min | [141] |
| Detection Method | Sensing Platform | Virus | Target Analytes | LOD | Assay Time | Year of Publication | Ref. |
|---|---|---|---|---|---|---|---|
| Immunoassay methods | Immuno-chromatic probe based LFA | Japanese Encephalitis | Non-structural 1 (NS1) secretory protein | 10 pg/mL | 10 min | 2022 | [25] |
| Magnetic quantum dot nanobeads based LFA | Infuenza A | Virus particle | 22 PFU/mL | 35 min | 2020 | [27] | |
| SER-based LFA | Influenza A | Virus particle | 1.9 × 104 PFU/mL | - | 2016 | [34] | |
| Portable SERS-LFA system | SARS-CoV-2 | Nucleocapsid protein | 3.53 PFU/mL | 15 min | 2022 | [37] | |
| ELISA-based microfluidic chip | SARS-CoV-2 | Nucleocapsid (N) protein | 0.1 pg/mL | 40 min | 2022 | [51] | |
| Multicolorimetric ELISA integrated with microfluidic device | Hepatitis C | Core antigen | 9.1 ng/µL | 50 min | 2021 | [52] | |
| Electrochemical immunoassay | SARS-CoV-2 | Antigen (Nasopharyngeal samples) | 50 PFU/mL | 70 min | 2022 | [57] | |
| ELISA-based microfluidic immunoassay | SARS-CoV-2 | Antigens and antibodies (Serum sample) | 0.3 pg/mL | 10 min | 2021 | [59] | |
| Graphene-based FET biosensing | SARS-CoV-2 | Spike protein (Nasopharyngeal samples) | 1 fg/mL | - | 2020 | [65] | |
| Molecular methods | Portable Magnetofluidic real-time RT-PCR instrument | HIV | RNA extraction (Whole blood sample) | 500 copies/mL | 15 min | 2023 | [88] |
| “FilmArray”: Nested multiplex PCR | 21 common viral and bacterial respiratory pathogens | DNA/RNA extraction | Coronavirus HKU1 (1.9 × 106 RNA copies/mL) | 60 min | 2011 | [89] | |
| Continuous-flow digital PCR device | Hepatitis B | DNA extraction (Serum sample) | 103 to 105 IU/mL | - | 2020 | [90] | |
| Pocket-size Reverse transcription (RT)-LAMP device | SARS-CoV-2 | RNA extraction (Oropharyngeal swab samples) | 300 RNA copies/reaction | 35 min | 2021 | [98] | |
| Paper-based colorimetric LAMP device | SARS-CoV-2 | Virus particle | 200 genomic copies/μL (Saliva sample) | 60 min | 2021 | [99] | |
| Wearable RPA microfluidic device | HIV-1 | DNA | 100 copies/mL | 24 min | 2019 | [103] | |
| RT-RPA and CRISPR-based paper microdevice | SARS-CoV-2 | Nucleoprotein (N) gene and spike (S) gene | 102 copies viral RNA/test | 60 min | 2021 | [123] | |
| Microfluidic Chip-Powered CRISPR/Cas12a System | Multiple respiratory viruses | DNA/RNA extraction (Swab samples) | 50–200 copies/mL | <40 min | 2022 | [125] | |
| Graphene-based FET biosensing | SARS-CoV-2 | Unamplified nucleic acids (Nasopharyngeal samples) | 1–2 copies/100 μL | <4 min | 2022 | [129] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trinh, K.T.L.; Do, H.D.K.; Lee, N.Y. Recent Advances in Molecular and Immunological Diagnostic Platform for Virus Detection: A Review. Biosensors 2023, 13, 490. https://doi.org/10.3390/bios13040490
Trinh KTL, Do HDK, Lee NY. Recent Advances in Molecular and Immunological Diagnostic Platform for Virus Detection: A Review. Biosensors. 2023; 13(4):490. https://doi.org/10.3390/bios13040490
Chicago/Turabian StyleTrinh, Kieu The Loan, Hoang Dang Khoa Do, and Nae Yoon Lee. 2023. "Recent Advances in Molecular and Immunological Diagnostic Platform for Virus Detection: A Review" Biosensors 13, no. 4: 490. https://doi.org/10.3390/bios13040490
APA StyleTrinh, K. T. L., Do, H. D. K., & Lee, N. Y. (2023). Recent Advances in Molecular and Immunological Diagnostic Platform for Virus Detection: A Review. Biosensors, 13(4), 490. https://doi.org/10.3390/bios13040490







