Advancing Luciferase-Based Antibody Immunoassays to Next-Generation Mix and Read Testing
Abstract
1. Introduction
2. Luciferase Antigen Fusion Proteins for LIPS Antibody Detection
3. LIPS Autoantibody Profiling of Autoimmune and Infectious Diseases
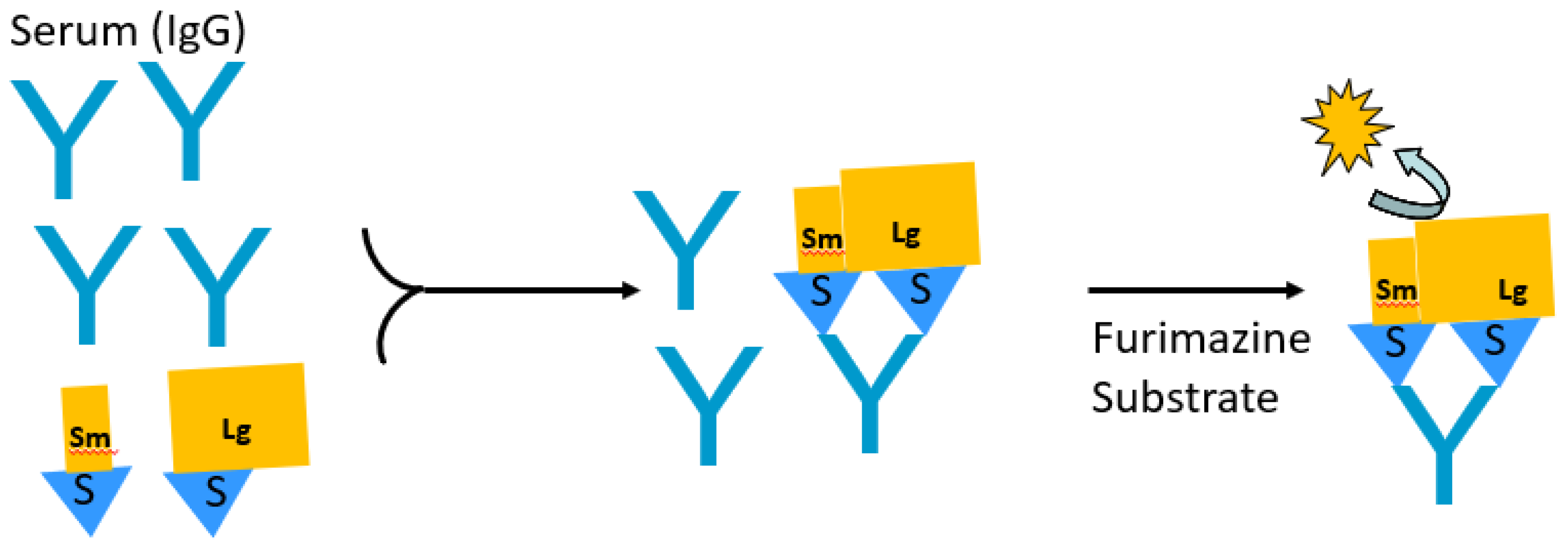
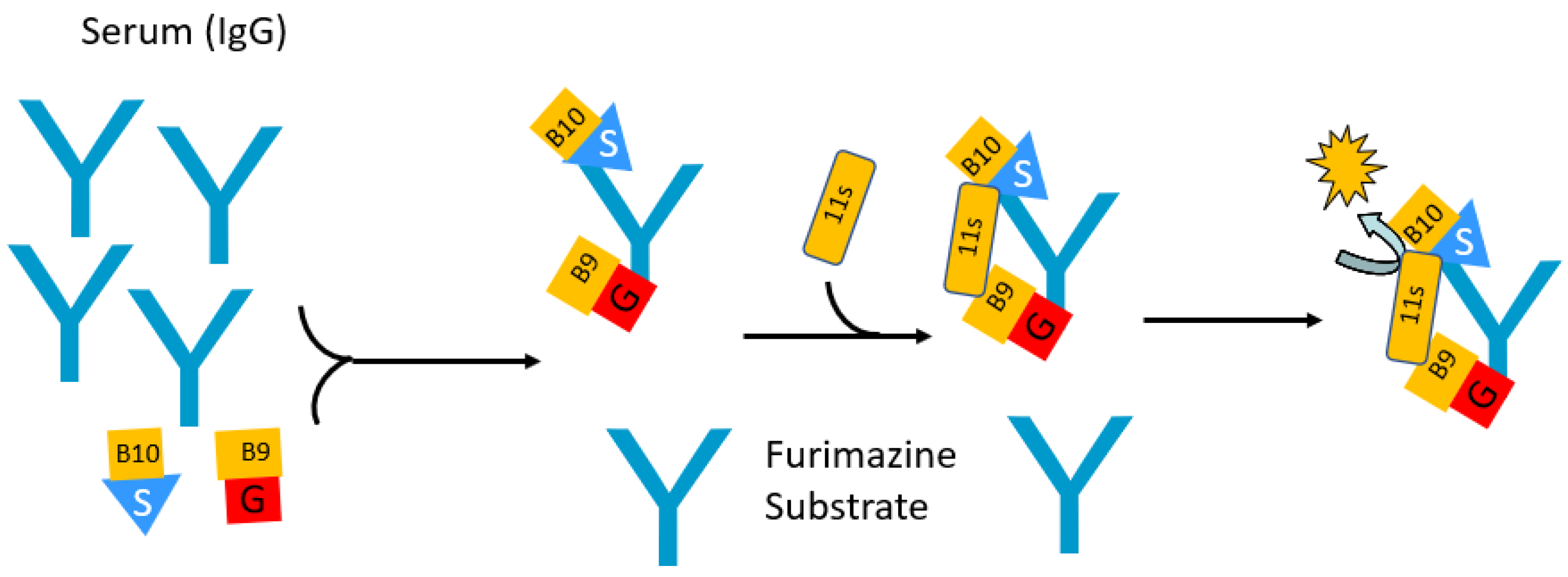
4. Split Nanoluciferase-Based Immunoassays for Antibody Detection
5. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dimech, W. The Standardization and Control of Serology and Nucleic Acid Testing for Infectious Diseases. Clin. Microbiol. Rev. 2021, 34, e0003521. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.X.; Miller, J.S.; Zheng, S.G. An updated advance of autoantibodies in autoimmune diseases. Autoimmun. Rev. 2021, 20, 102743. [Google Scholar] [CrossRef] [PubMed]
- Fritzler, M.J. Advances and applications of multiplexed diagnostic technologies in autoimmune diseases. Lupus 2006, 15, 422–427. [Google Scholar] [CrossRef] [PubMed]
- Ayoglu, B.; Schwenk, J.M.; Nilsson, P. Antigen arrays for profiling autoantibody repertoires. Bioanalysis 2016, 8, 1105–1126. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef]
- Xu, G.J.; Kula, T.; Xu, Q.; Li, M.Z.; Vernon, S.D.; Ndung’u, T.; Ruxrungtham, K.; Sanchez, J.; Brander, C.; Chung, R.T.; et al. Viral immunology. Comprehensive serological profiling of human populations using a synthetic human virome. Science 2015, 348, aaa0698. [Google Scholar] [CrossRef]
- Vogl, T.; Klompus, S.; Leviatan, S.; Kalka, I.N.; Weinberger, A.; Wijmenga, C.; Fu, J.; Zhernakova, A.; Weersma, R.K.; Segal, E. Population-wide diversity and stability of serum antibody epitope repertoires against human microbiota. Nat. Med. 2021, 27, 1442–1450. [Google Scholar] [CrossRef]
- Larman, H.B.; Zhao, Z.; Laserson, U.; Li, M.Z.; Ciccia, A.; Gakidis, M.A.; Church, G.M.; Kesari, S.; Leproust, E.M.; Solimini, N.L.; et al. Autoantigen discovery with a synthetic human peptidome. Nat. Biotechnol. 2011, 29, 535–541. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Lebovitz, E.E.; Notkins, A.L. Luciferase immunoprecipitation systems for measuring antibodies in autoimmune and infectious diseases. Transl. Res. 2015, 165, 325–335. [Google Scholar] [CrossRef]
- Sharifian, S.; Homaei, A.; Hemmati, R.; Khajeh, K. Light emission miracle in the sea and preeminent applications of bioluminescence in recent new biotechnology. J. Photochem. Photobiol. B 2017, 172, 115–128. [Google Scholar] [CrossRef]
- Markova, S.V.; Larionova, M.D.; Vysotski, E.S. Shining Light on the Secreted Luciferases of Marine Copepods: Current Knowledge and Applications. Photochem. Photobiol. 2019, 95, 705–721. [Google Scholar] [CrossRef]
- Inouye, S.; Watanabe, K.; Nakamura, H.; Shimomura, O. Secretional luciferase of the luminous shrimp Oplophorus gracilirostris: cDNA cloning of a novel imidazopyrazinone luciferase(1). FEBS Lett. 2000, 481, 19–25. [Google Scholar] [CrossRef]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef]
- Elledge, S.K.; Zhou, X.X.; Byrnes, J.R.; Martinko, A.J.; Lui, I.; Pance, K.; Lim, S.A.; Glasgow, J.E.; Glasgow, A.A.; Turcios, K.; et al. Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection. Nat. Biotechnol. 2021, 39, 928–935. [Google Scholar] [CrossRef]
- Liberati, D.; Wyatt, R.C.; Brigatti, C.; Marzinotto, I.; Ferrari, M.; Bazzigaluppi, E.; Bosi, E.; Gillard, B.T.; Gillespie, K.M.; Gorus, F.; et al. A novel LIPS assay for insulin autoantibodies. Acta Diabetol. 2018, 55, 263–270. [Google Scholar] [CrossRef]
- Secchi, M.; Bazzigaluppi, E.; Brigatti, C.; Marzinotto, I.; Tresoldi, C.; Rovere-Querini, P.; Poli, A.; Castagna, A.; Scarlatti, G.; Zangrillo, A.; et al. COVID-19 survival associates with the immunoglobulin response to the SARS-CoV-2 spike receptor binding domain. J. Clin. Investig. 2020, 130, 6366–6378. [Google Scholar] [CrossRef]
- Wang, H.; Cai, Q.; Liang, Y.; Shui, J.; Tang, S. A simple and high-throughput luciferase immunosorbent assay for both qualitative and semi-quantitative detection of anti-HIV-1 antibodies. Virus Res. 2019, 263, 9–15. [Google Scholar] [CrossRef]
- Wang, T.; Zhan, Y.; Wu, D.; Chen, Z.; Wu, W.; Deng, Y.; Wang, W.; Tan, W.; Tang, S. Development and Evaluation of a Universal and Supersensitive NS1-Based Luciferase Immunosorbent Assay to Detect Zika Virus-Specific IgG. Virol. Sin. 2020, 35, 93–102. [Google Scholar] [CrossRef]
- Fishman, D.; Kisand, K.; Hertel, C.; Rothe, M.; Remm, A.; Pihlap, M.; Adler, P.; Vilo, J.; Peet, A.; Meloni, A.; et al. Autoantibody Repertoire in APECED Patients Targets Two Distinct Subgroups of Proteins. Front. Immunol. 2017, 8, 976. [Google Scholar] [CrossRef] [PubMed]
- Yamakawa, M.; Mukaino, A.; Kimura, A.; Nagasako, Y.; Kitazaki, Y.; Maeda, Y.; Higuchi, O.; Takamatsu, K.; Watari, M.; Yoshikura, N.; et al. Antibodies to the alpha3 subunit of the ganglionic-type nicotinic acetylcholine receptors in patients with autoimmune encephalitis. J. Neuroimmunol. 2020, 349, 577399. [Google Scholar] [CrossRef] [PubMed]
- Hoang, T.T.T.; Ichinose, K.; Morimoto, S.; Furukawa, K.; Le, L.H.T.; Kawakami, A. Measurement of anti-suprabasin antibodies, multiple cytokines and chemokines as potential predictive biomarkers for neuropsychiatric systemic lupus erythematosus. Clin. Immunol. 2022, 237, 108980. [Google Scholar] [CrossRef] [PubMed]
- Aso, K.; Kono, M.; Abe, N.; Fujieda, Y.; Kato, M.; Atsumi, T. Anti-ganglionic nicotinic acetylcholine receptor alpha3 subunit antibody as a potential biomarker associated with lupus enteritis. Mod. Rheumatol. 2023, 33, 154–159. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Leahy, H.P.; Issa, A.T.; Groot, S.; Baraniuk, J.N.; Nikolov, N.P.; Illei, G.G.; Iadarola, M.J. Sensitive and robust luminescent profiling of anti-La and other autoantibodies in Sjogren’s syndrome. Autoimmunity 2009, 42, 515–524. [Google Scholar] [CrossRef]
- Mukaino, A.; Nakane, S.; Higuchi, O.; Nakamura, H.; Miyagi, T.; Shiroma, K.; Tokashiki, T.; Fuseya, Y.; Ochi, K.; Umeda, M.; et al. Insights from the ganglionic acetylcholine receptor autoantibodies in patients with Sjogren’s syndrome. Mod. Rheumatol. 2016, 26, 708–715. [Google Scholar] [CrossRef]
- Volchenkov, R.; Jonsson, R.; Appel, S. Anti-Ro and anti-La autoantibody profiling in Norwegian patients with primary Sjogren’s syndrome using luciferase immunoprecipitation systems (LIPS). Scand. J. Rheumatol. 2012, 41, 314–315. [Google Scholar] [CrossRef]
- Saare, M.; Hamarik, U.; Venta, R.; Panarina, M.; Zucchelli, C.; Pihlap, M.; Remm, A.; Kisand, K.; Toots, U.; Moll, K.; et al. SP140L, an Evolutionarily Recent Member of the SP100 Family, Is an Autoantigen in Primary Biliary Cirrhosis. J. Immunol. Res. 2015, 2015, 526518. [Google Scholar] [CrossRef]
- Maeda, Y.; Nakane, S.; Higuchi, O.; Nakamura, H.; Komori, A.; Migita, K.; Mukaino, A.; Umeda, M.; Ichinose, K.; Tamai, M.; et al. Ganglionic acetylcholine receptor autoantibodies in patients with autoimmune diseases including primary biliary cirrhosis. Mod. Rheumatol. 2017, 27, 664–668. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Gordon, S.M.; Waldman, M.; Edison, J.D.; Little, D.J.; Stitt, R.S.; Bailey, W.T.; Hughes, J.B.; Olson, S.W. Autoantibodies are present before the clinical diagnosis of systemic sclerosis. PLoS ONE 2019, 14, e0214202. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Beck, L.H., Jr.; Waldman, M. Detection and monitoring PLA2R autoantibodies by LIPS in membranous nephropathy. J. Immunol. Methods 2017, 444, 17–23. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Joshi, M.; Chaturvedi, A.; Little, D.J.; Thurlow, J.S.; Waldman, M.; Olson, S.W. Detection of PLA2R Autoantibodies before the Diagnosis of Membranous Nephropathy. J. Am. Soc. Nephrol. 2020, 31, 208–217. [Google Scholar] [CrossRef]
- Lahner, E.; Brigatti, C.; Marzinotto, I.; Carabotti, M.; Scalese, G.; Davidson, H.W.; Wenzlau, J.M.; Bosi, E.; Piemonti, L.; Annibale, B.; et al. Luminescent Immunoprecipitation System (LIPS) for Detection of Autoantibodies against ATP4A and ATP4B Subunits of Gastric Proton Pump H+,K+-ATPase in Atrophic Body Gastritis Patients. Clin. Transl. Gastroenterol. 2017, 8, e215. [Google Scholar] [CrossRef] [PubMed]
- Lahner, E.; Dilaghi, E.; Cingolani, S.; Pivetta, G.; Dottori, L.; Esposito, G.; Marzinotto, I.; Lampasona, V.; Buzzetti, R.; Annibale, B. Gender-sex differences in autoimmune atrophic gastritis. Transl. Res. 2022, 248, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Marzinotto, I.; Dottori, L.; Baldaro, F.; Dilaghi, E.; Brigatti, C.; Bazzigaluppi, E.; Esposito, G.; Davidson, H.W.; Piemonti, L.; Lampasona, V.; et al. Intrinsic factor autoantibodies by luminescent immuno-precipitation system in patients with corpus atrophic gastritis. J. Transl. Autoimmun. 2021, 4, 100131. [Google Scholar] [CrossRef]
- McLaughlin, K.A.; Richardson, C.C.; Ravishankar, A.; Brigatti, C.; Liberati, D.; Lampasona, V.; Piemonti, L.; Morgan, D.; Feltbower, R.G.; Christie, M.R. Identification of Tetraspanin-7 as a Target of Autoantibodies in Type 1 Diabetes. Diabetes 2016, 65, 1690–1698. [Google Scholar] [CrossRef]
- Shi, X.J.; Zheng, P.L.; Wang, Z.; Li, X.; Huang, G.; Zhou, Z.G. The establishment and application of testing methods of tetraspanin 7 autoantibody in type 1 diabetes. Zhonghua Yi Xue Za Zhi 2021, 101, 243–248. [Google Scholar] [CrossRef]
- Pan, S.; Wu, T.; Shi, X.; Xie, Z.; Huang, G.; Zhou, Z. Organ-specific autoantibodies in Chinese patients newly diagnosed with type 1 diabetes mellitus. Endocr. J. 2020, 67, 793–802. [Google Scholar] [CrossRef]
- Walther, D.; Eugster, A.; Jergens, S.; Gavrisan, A.; Weinzierl, C.; Telieps, T.; Winkler, C.; Ziegler, A.G.; Bonifacio, E. Tetraspanin 7 autoantibodies in type 1 diabetes. Diabetologia 2016, 59, 1973–1976. [Google Scholar] [CrossRef]
- Muller, D.; Telieps, T.; Eugster, A.; Weinzierl, C.; Jolink, M.; Ziegler, A.G.; Bonifacio, E. Novel minor HLA DR associated antigens in type 1 diabetes. Clin. Immunol. 2018, 194, 87–91. [Google Scholar] [CrossRef]
- Ling, Y.; Jiang, P.; Li, N.; Yan, Q.; Wang, X. A luciferase immunoprecipitation assay for the detection of proinsulin/insulin autoantibodies. Clin. Biochem. 2018, 54, 51–55. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Browne, S.K.; Sampaio, E.P.; Giaccone, G.; Zaman, R.; Kristosturyan, E.; Rajan, A.; Ding, L.; Ching, K.H.; Berman, A.; et al. Anti-cytokine autoantibodies are associated with opportunistic infection in patients with thymic neoplasia. Blood 2010, 116, 4848–4858. [Google Scholar] [CrossRef]
- Browne, S.K.; Burbelo, P.D.; Chetchotisakd, P.; Suputtamongkol, Y.; Kiertiburanakul, S.; Shaw, P.A.; Kirk, J.L.; Jutivorakool, K.; Zaman, R.; Ding, L.; et al. Adult-onset immunodeficiency in Thailand and Taiwan. N. Engl. J. Med. 2012, 367, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Karner, J.; Pihlap, M.; Ranki, A.; Krohn, K.; Trebusak Podkrajsek, K.; Bratanic, N.; Battelino, T.; Willcox, N.; Peterson, P.; Kisand, K. IL-6-specific autoantibodies among APECED and thymoma patients. Immun. Inflamm. Dis. 2016, 4, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Erdos, M.; Karner, J.; Ranki, A.; Kisand, K.; Marodi, L. Persistently Increased Anti-cytokine Antibodies without Clinical Disease in a Boy with APS1 Genotype. J. Clin. Immunol. 2022, 42, 433–436. [Google Scholar] [CrossRef]
- Meyer, S.; Woodward, M.; Hertel, C.; Vlaicu, P.; Haque, Y.; Karner, J.; Macagno, A.; Onuoha, S.C.; Fishman, D.; Peterson, H.; et al. AIRE-Deficient Patients Harbor Unique High-Affinity Disease-Ameliorating Autoantibodies. Cell 2016, 166, 582–595. [Google Scholar] [CrossRef]
- Roberts, M.S.; Burbelo, P.D.; Egli-Spichtig, D.; Perwad, F.; Romero, C.J.; Ichikawa, S.; Farrow, E.; Econs, M.J.; Guthrie, L.C.; Collins, M.T.; et al. Autoimmune hyperphosphatemic tumoral calcinosis in a patient with FGF23 autoantibodies. J. Clin. Investig. 2018, 128, 5368–5373. [Google Scholar] [CrossRef]
- Mandl, A.; Burbelo, P.D.; Di Pasquale, G.; Tay, Y.S.; Welch, J.; Lionakis, M.S.; Rosenzweig, S.D.; Waldman, M.A.; Warner, B.M.; Walitt, B.; et al. Parathyroid Hormone Resistance and Autoantibodies to the PTH1 Receptor. N. Engl. J. Med. 2021, 385, 1974–1980. [Google Scholar] [CrossRef]
- Steffen, I.; Lu, K.; Yamamoto, L.K.; Hoff, N.A.; Mulembakani, P.; Wemakoy, E.O.; Muyembe-Tamfum, J.J.; Ndembi, N.; Brennan, C.A.; Hackett, J., Jr.; et al. Serologic Prevalence of Ebola Virus in Equatorial Africa. Emerg. Infect. Dis. 2019, 25, 911–918. [Google Scholar] [CrossRef]
- Aye, K.M.; Nagayasu, E.; Baba, M.; Yoshida, A.; Takashima, Y.; Maruyama, H. Evaluation of LIPS (luciferase immunoprecipitation system) for serodiagnosis of Toxoplasmosis. J. Immunol. Methods 2018, 462, 91–100. [Google Scholar] [CrossRef]
- Aye, K.M.; Nagayasu, E.; Nyunt, M.H.; Zaw, N.N.; Thant, K.Z.; Kyaw, M.P.; Maruyama, H. Seroprevalence of toxoplasmosis among reproductive-aged women in Myanmar and evaluation of luciferase immunoprecipitation system assay. BMC Infect. Dis. 2020, 20, 906. [Google Scholar] [CrossRef]
- Tin, C.M.; Sosnovtsev, S.V. Detection of Human Norovirus-Specific Antibodies Using the Luciferase Immunoprecipitation System (LIPS) Assay. Methods Mol. Biol. 2019, 2024, 137–152. [Google Scholar] [CrossRef]
- Ding, J.; Yang, J.; Jiang, D.; Zhou, Y.; Li, C.; Li, Y. Development of a highly sensitive Gaussia luciferase immunoprecipitation assay for the detection of antibodies against African swine fever virus. Front. Cell. Infect. Microbiol. 2022, 12, 988355. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; He, P.; Meng, F.; Jiang, M.; Xiong, J.; Li, J.; Yu, J.; Wei, H. A Semiautomated Luciferase Immunoprecipitation Assay for Rapid and Easy Detection of African Swine Fever Virus Antibody. J. Clin. Microbiol. 2021, 59, e0099021. [Google Scholar] [CrossRef] [PubMed]
- Fuery, A.; Pursell, T.; Tan, J.; Peng, R.; Burbelo, P.D.; Hayward, G.S.; Ling, P.D. Lethal Hemorrhagic Disease and Clinical Illness Associated with Elephant Endotheliotropic Herpesvirus 1 Are Caused by Primary Infection: Implications for the Detection of Diagnostic Proteins. J. Virol. 2020, 94, e01528-19. [Google Scholar] [CrossRef]
- Konenkamp, L.; Ziegler, U.; Naucke, T.; Groschup, M.H.; Steffen, I. Antibody ratios against NS1 antigens of tick-borne encephalitis and West Nile viruses support differential flavivirus serology in dogs. Transbound. Emerg. Dis. 2022, 69, e2789–e2799. [Google Scholar] [CrossRef]
- Luong, H.Q.; Lai, H.T.L.; Vu, H.L.X. Evaluation of Antibody Response Directed against Porcine Reproductive and Respiratory Syndrome Virus Structural Proteins. Vaccines 2020, 8, 533. [Google Scholar] [CrossRef]
- Chen, S.; Xu, M.; Wu, X.; Bai, Y.; Shi, J.; Zhou, M.; Wu, Q.; Tang, S.; Deng, F.; Qin, B.; et al. A new luciferase immunoprecipitation system assay provided serological evidence for missed diagnosis of severe fever with thrombocytopenia syndrome. Virol. Sin. 2022, 37, 107–114. [Google Scholar] [CrossRef]
- Matsuu, A.; Hamakubo, E.; Yabuki, M. Seroprevalence of severe fever with thrombocytopenia syndrome virus in animals in Kagoshima Prefecture, Japan, and development of Gaussia luciferase immunoprecipitation system to detect specific IgG antibodies. Ticks Tick Borne Dis. 2021, 12, 101771. [Google Scholar] [CrossRef]
- Zhou, P.; Fan, H.; Lan, T.; Yang, X.L.; Shi, W.F.; Zhang, W.; Zhu, Y.; Zhang, Y.W.; Xie, Q.M.; Mani, S.; et al. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 2018, 556, 255–258. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Riedo, F.X.; Morishima, C.; Rawlings, S.; Smith, D.; Das, S.; Strich, J.R.; Chertow, D.S.; Davey, R.T.; Cohen, J.I. Sensitivity in Detection of Antibodies to Nucleocapsid and Spike Proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in Patients with Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 206–213. [Google Scholar] [CrossRef]
- Haljasmagi, L.; Remm, A.; Rumm, A.P.; Krassohhina, E.; Sein, H.; Tamm, A.; Kisand, K.; Peterson, P. LIPS method for the detection of SARS-CoV-2 antibodies to spike and nucleocapsid proteins. Eur. J. Immunol. 2020, 50, 1234–1236. [Google Scholar] [CrossRef]
- Grzelak, L.; Temmam, S.; Planchais, C.; Demeret, C.; Tondeur, L.; Huon, C.; Guivel-Benhassine, F.; Staropoli, I.; Chazal, M.; Dufloo, J.; et al. A comparison of four serological assays for detecting anti-SARS-CoV-2 antibodies in human serum samples from different populations. Sci. Transl. Med. 2020, 12, eabc3103. [Google Scholar] [CrossRef] [PubMed]
- Hachim, A.; Kavian, N.; Cohen, C.A.; Chin, A.W.H.; Chu, D.K.W.; Mok, C.K.P.; Tsang, O.T.Y.; Yeung, Y.C.; Perera, R.; Poon, L.L.M.; et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020, 21, 1293–1301. [Google Scholar] [CrossRef] [PubMed]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Beziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef] [PubMed]
- Dispinseri, S.; Secchi, M.; Pirillo, M.F.; Tolazzi, M.; Borghi, M.; Brigatti, C.; De Angelis, M.L.; Baratella, M.; Bazzigaluppi, E.; Venturi, G.; et al. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat. Commun. 2021, 12, 2670. [Google Scholar] [CrossRef]
- Hippich, M.; Holthaus, L.; Assfalg, R.; Zapardiel-Gonzalo, J.; Kapfelsperger, H.; Heigermoser, M.; Haupt, F.; Ewald, D.A.; Welzhofer, T.C.; Marcus, B.A.; et al. A Public Health Antibody Screening Indicates a 6-Fold Higher SARS-CoV-2 Exposure Rate than Reported Cases in Children. Med 2021, 2, 149–163. [Google Scholar] [CrossRef]
- Hachim, A.; Gu, H.; Kavian, O.; Mori, M.; Kwan, M.Y.W.; Chan, W.H.; Yau, Y.S.; Chiu, S.S.; Tsang, O.T.Y.; Hui, D.S.C.; et al. SARS-CoV-2 accessory proteins reveal distinct serological signatures in children. Nat. Commun. 2022, 13, 2951. [Google Scholar] [CrossRef]
- Delmonte, O.M.; Bergerson, J.R.E.; Burbelo, P.D.; Durkee-Shock, J.R.; Dobbs, K.; Bosticardo, M.; Keller, M.D.; McDermott, D.H.; Rao, V.K.; Dimitrova, D.; et al. Antibody responses to the SARS-CoV-2 vaccine in individuals with various inborn errors of immunity. J. Allergy Clin. Immunol. 2021, 148, 1192–1197. [Google Scholar] [CrossRef]
- Cattaneo, C.; Cancelli, V.; Imberti, L.; Dobbs, K.; Sottini, A.; Pagani, C.; Belotti, A.; Re, A.; Anastasia, A.; Quaresima, V.; et al. Production and persistence of specific antibodies in COVID-19 patients with hematologic malignancies: Role of rituximab. Blood Cancer J. 2021, 11, 151. [Google Scholar] [CrossRef]
- Atanackovic, D.; Kreitman, R.J.; Cohen, J.; Hardy, N.M.; Omili, D.; Iraguha, T.; Burbelo, P.D.; Gebru, E.; Fan, X.; Baddley, J.; et al. T cell responses against SARS-CoV-2 and its Omicron variant in a patient with B cell lymphoma after multiple doses of a COVID-19 mRNA vaccine. J. Immunother. Cancer 2022, 10, e004953. [Google Scholar] [CrossRef]
- Kaskova, Z.M.; Tsarkova, A.S.; Yampolsky, I.V. 1001 lights: Luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine. Chem. Soc. Rev. 2016, 45, 6048–6077. [Google Scholar] [CrossRef]
- Dixon, A.S.; Schwinn, M.K.; Hall, M.P.; Zimmerman, K.; Otto, P.; Lubben, T.H.; Butler, B.L.; Binkowski, B.F.; Machleidt, T.; Kirkland, T.A.; et al. NanoLuc Complementation Reporter Optimized for Accurate Measurement of Protein Interactions in Cells. ACS Chem. Biol. 2016, 11, 400–408. [Google Scholar] [CrossRef]
- Dixon, A.S.; Kim, S.J.; Baumgartner, B.K.; Krippner, S.; Owen, S.C. A Tri-part Protein Complementation System Using Antibody-Small Peptide Fusions Enables Homogeneous Immunoassays. Sci. Rep. 2017, 7, 8186. [Google Scholar] [CrossRef]
- Ohmuro-Matsuyama, Y.; Ueda, H. Homogeneous Noncompetitive Luminescent Immunodetection of Small Molecules by Ternary Protein Fragment Complementation. Anal. Chem. 2018, 90, 3001–3004. [Google Scholar] [CrossRef]
- Azad, T.; Rezaei, R.; Singaravelu, R.; Jamieson, T.R.; Crupi, M.J.F.; Surendran, A.; Poutou, J.; Taklifi, P.; Cowan, J.; Cameron, D.W.; et al. A High-Throughput NanoBiT-Based Serological Assay Detects SARS-CoV-2 Seroconversion. Nanomaterials 2021, 11, 807. [Google Scholar] [CrossRef]
- Elledge, S.K.; Eigl, I.; Phelps, M.; McClinton, K.; Zhou, X.X.; Leung, K.K.; Tato, C.M.; Wells, J.A. Using Split Luminescent Biosensors for SARS-CoV-2 Antibody Detection in Serum, Plasma, and Blood Samples. Curr. Protoc. 2022, 2, e521. [Google Scholar] [CrossRef]
- Peluso, M.J.; Takahashi, S.; Hakim, J.; Kelly, J.D.; Torres, L.; Iyer, N.S.; Turcios, K.; Janson, O.; Munter, S.E.; Thanh, C.; et al. SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay. Sci. Adv. 2021, 7, eabh3409. [Google Scholar] [CrossRef]
- Ni, Y.; Rosier, B.; van Aalen, E.A.; Hanckmann, E.T.L.; Biewenga, L.; Pistikou, A.M.; Timmermans, B.; Vu, C.; Roos, S.; Arts, R.; et al. A plug-and-play platform of ratiometric bioluminescent sensors for homogeneous immunoassays. Nat. Commun. 2021, 12, 4586. [Google Scholar] [CrossRef]
- Yao, Z.; Drecun, L.; Aboualizadeh, F.; Kim, S.J.; Li, Z.; Wood, H.; Valcourt, E.J.; Manguiat, K.; Plenderleith, S.; Yip, L.; et al. A homogeneous split-luciferase assay for rapid and sensitive detection of anti-SARS CoV-2 antibodies. Nat. Commun. 2021, 12, 1806. [Google Scholar] [CrossRef]
- Kim, S.J.; Dixon, A.S.; Adamovich, P.C.; Robinson, P.D.; Owen, S.C. Homogeneous Immunoassay Using a Tri-Part Split-Luciferase for Rapid Quantification of Anti-TNF Therapeutic Antibodies. ACS Sens. 2021, 6, 1807–1814. [Google Scholar] [CrossRef]
- Higuchi, O.; Nakane, S.; Sakai, W.; Maeda, Y.; Niino, M.; Takahashi, T.; Fukazawa, T.; Kikuchi, S.; Fujihara, K.; Matsuo, H. Lack of KIR4.1 autoantibodies in Japanese patients with MS and NMO. Neurol. Neuroimmunol. Neuroinflamm. 2016, 3, e263. [Google Scholar] [CrossRef]
- Marzinotto, I.; Liberati, D.; Brigatti, C.; Bonfanti, R.; Stabilini, A.; Monti, P.; Bosi, E.; Piemonti, L.; Lampasona, V. Autoantibody binding in liquid phase to IL-2 in human sera is not type 1 diabetes specific. Diabetologia 2017, 60, 1834–1835. [Google Scholar] [CrossRef] [PubMed]
- Liberati, D.; Marzinotto, I.; Brigatti, C.; Dugnani, E.; Pasquale, V.; Reni, M.; Balzano, G.; Falconi, M.; Piemonti, L.; Lampasona, V. No evidence of pancreatic ductal adenocarcinoma specific autoantibodies to Ezrin in a liquid phase LIPS immunoassay. Cancer Biomark. 2018, 22, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Mangioni, D.; Peri, A.M.; Rossolini, G.M.; Viaggi, B.; Perno, C.F.; Gori, A.; Bandera, A. Toward Rapid Sepsis Diagnosis and Patient Stratification: What’s New from Microbiology and Omics Science. J. Infect. Dis. 2020, 221, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
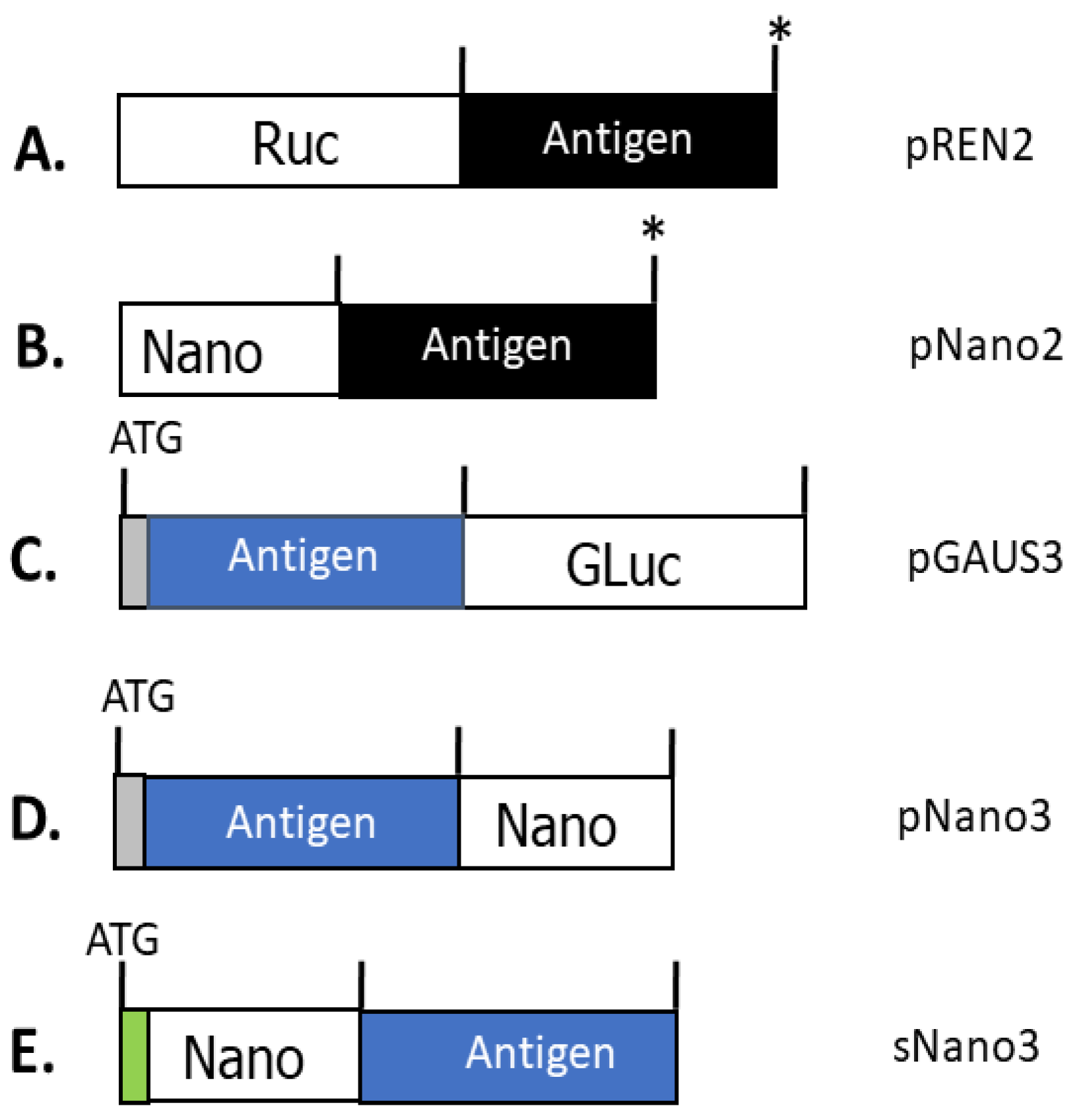


| Luciferase | Renilla Luciferase (Ruc) | Gaussia Luciferase (GLuc) | Nanoluciferase (Nano) |
|---|---|---|---|
| Size | 36 kDa | 20 kDa | 19 kDa |
| Substrate | Coelenterazine | Coelenterazine | Furimazine |
| Signal Type | Flash | Flash | Glow |
| Location of Antigen Target Fusion | C-terminal | N- and C-terminal | N- and C-terminal |
| Adapted to Hand-held Luminometer | No | No | Yes |
| New Autoimmune Disease | Significance |
|---|---|
| Opportunistic Infection (OI) in patients with Thymoma Cancer | Detection of autoantibodies against IFN-α1, IL-12 and other cytokines associated with OI pts [40] |
| Autoimmune associated disseminated non-tuberculosis mycobacterial infection (dNTM) | Autoantibodies against IFN-γ associated with mycobacterial infection [41] |
| Autoimmune hyperphosphatemia | First case of hyperphosphatemia due to autoantibodies against FGF23 [45] |
| Autoimmune hypoparathyroidism | Blocking autoantibodies against PTH1R [46] |
| Features | LIPS | Split Fab | SATiN |
|---|---|---|---|
| Luciferase used | Ruc, Gluc, and Nano | Split Nano | Split Nano |
| Number of protein components | One | Two | Three |
| Assay development | Simple | Complex | Complex |
| Recombinant Expression | Crude cell extract | Purified | Purified |
| Protein size constraints | No | Yes | Yes |
| Hands on time per 96 samples | ~30 min | ~15 min | ~30 min |
| Washing steps | Yes | No | No |
| Antigen discovery | Yes | No | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Burbelo, P.D.; Ji, Y.; Iadarola, M.J. Advancing Luciferase-Based Antibody Immunoassays to Next-Generation Mix and Read Testing. Biosensors 2023, 13, 303. https://doi.org/10.3390/bios13030303
Burbelo PD, Ji Y, Iadarola MJ. Advancing Luciferase-Based Antibody Immunoassays to Next-Generation Mix and Read Testing. Biosensors. 2023; 13(3):303. https://doi.org/10.3390/bios13030303
Chicago/Turabian StyleBurbelo, Peter D., Youngmi Ji, and Michael J. Iadarola. 2023. "Advancing Luciferase-Based Antibody Immunoassays to Next-Generation Mix and Read Testing" Biosensors 13, no. 3: 303. https://doi.org/10.3390/bios13030303
APA StyleBurbelo, P. D., Ji, Y., & Iadarola, M. J. (2023). Advancing Luciferase-Based Antibody Immunoassays to Next-Generation Mix and Read Testing. Biosensors, 13(3), 303. https://doi.org/10.3390/bios13030303





