Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models
Abstract
1. Introduction
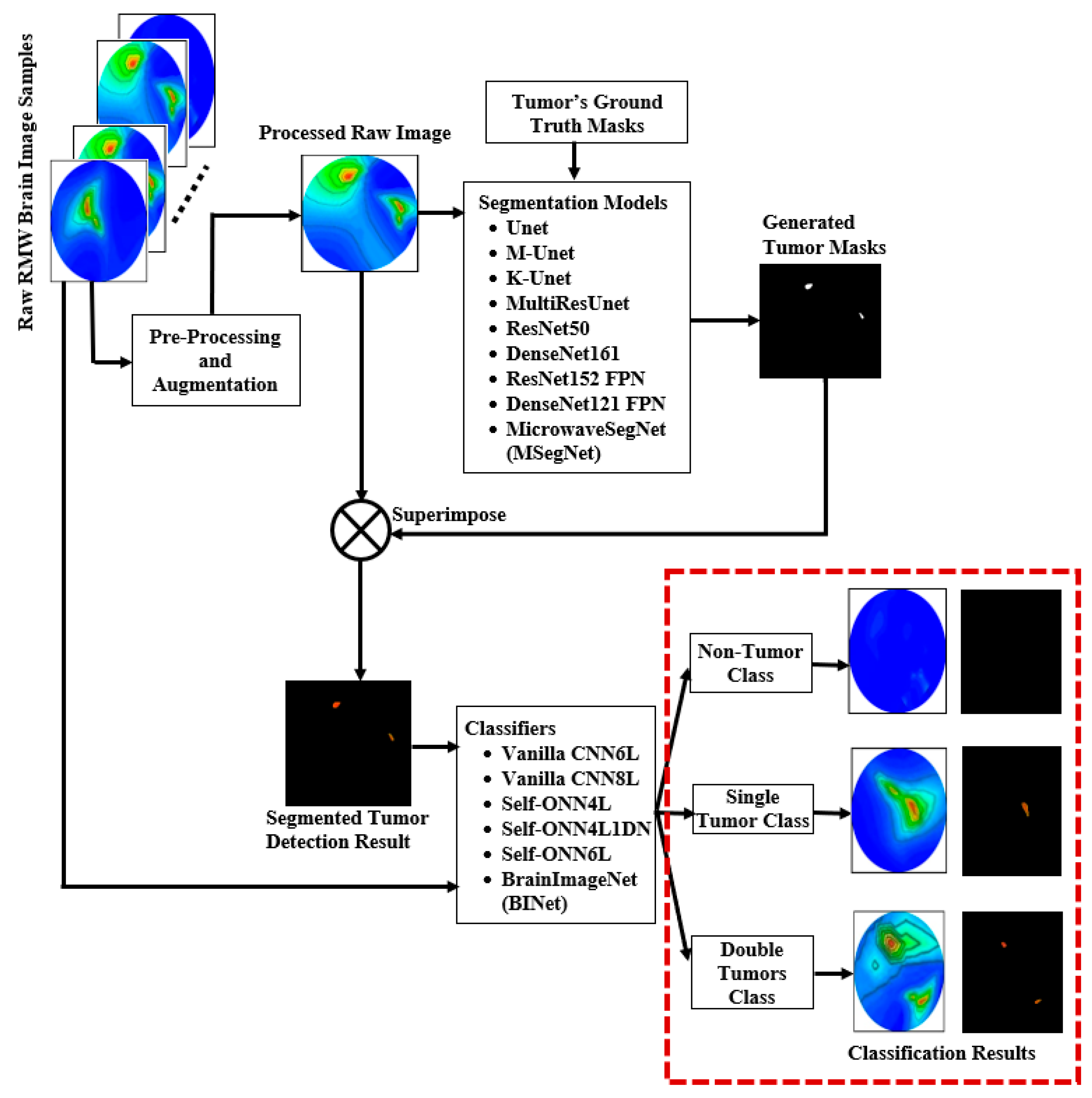
- To the best of our knowledge, this is the first paper to propose a lightweight segmentation model called MicrowaveSegNet (MSegNet) that can automatically segment the desired brain tumors in RMW brain images from the sensors-based MBI system.
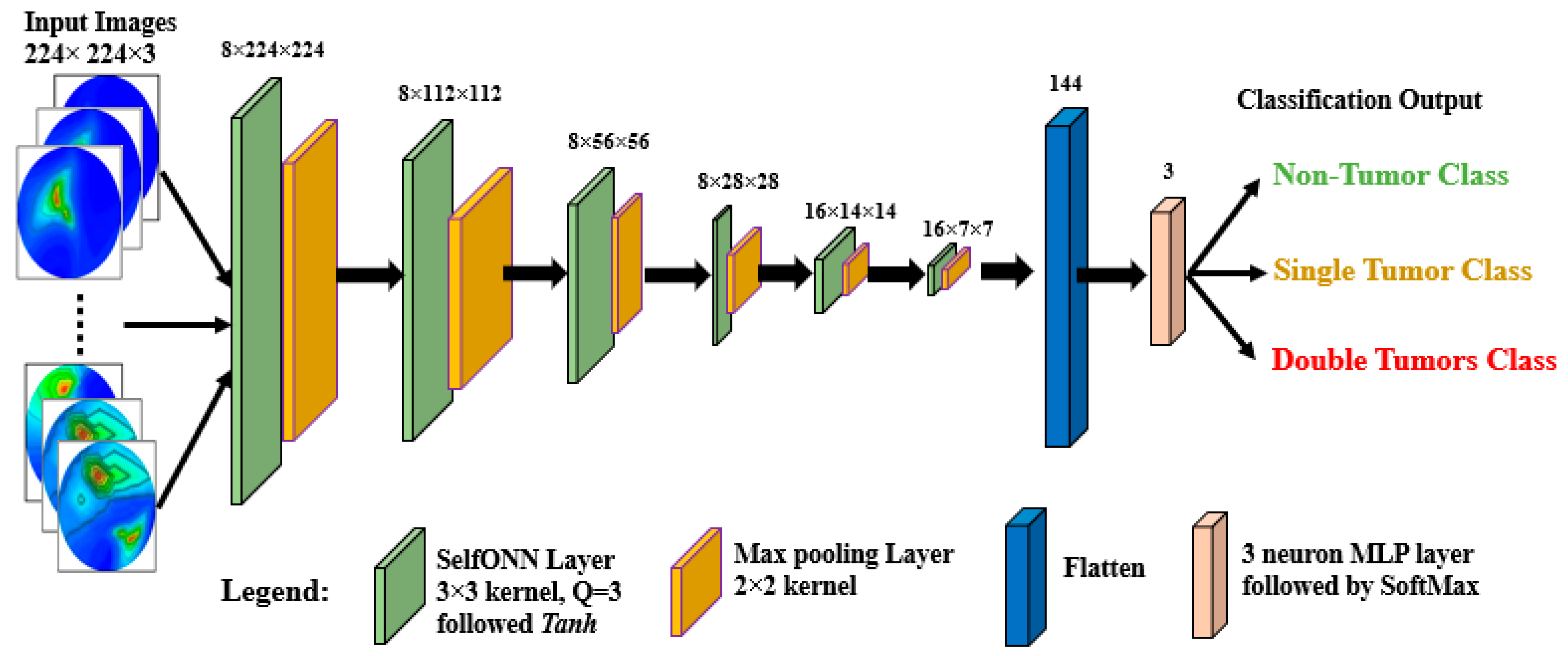
- A lightweight classification model called BrainImageNet (BINet) is proposed to classify the raw and segmented RMW brain images using a new machine learning paradigm, the self-organized operational neural network (Self-ONN) architecture.
- To segment both large and small brain tumors, the proposed MSegNet model is developed and tested on RMW brain tumor images.
- We formulated a tissue-mimicking head phantom model to investigate the imaging system for generating the RMW brain image dataset.
- A new Self-ONN model, BINet, three other Self-ONN models, and two conventional CNN classification models are investigated on the raw and segmented RMW brain tumor images to classify non-tumor, single tumor, and double tumor classes to show the efficacy of the proposed BINet classification model.
2. Experimental Setup of a Sensor-Based Microwave Brain Imaging System and Sample Image Collection Process
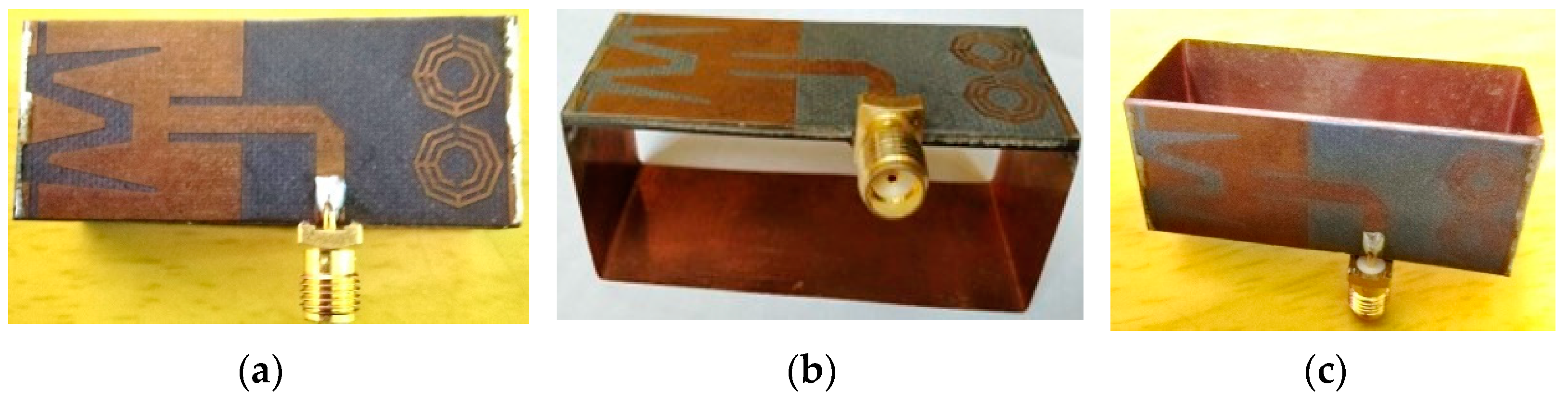
2.1. Antenna Sensor Design and Measurement
2.2. Phantom’s Composition Process and RMW Image Sample Collection
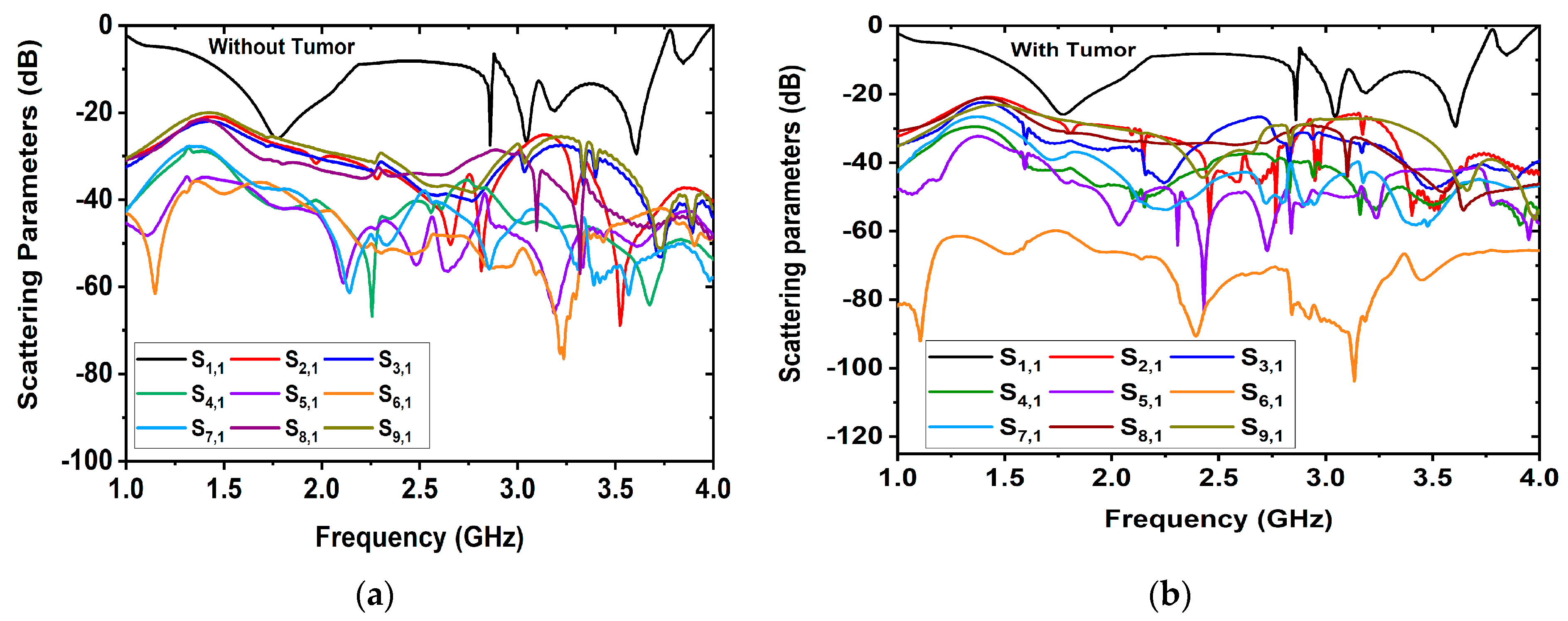
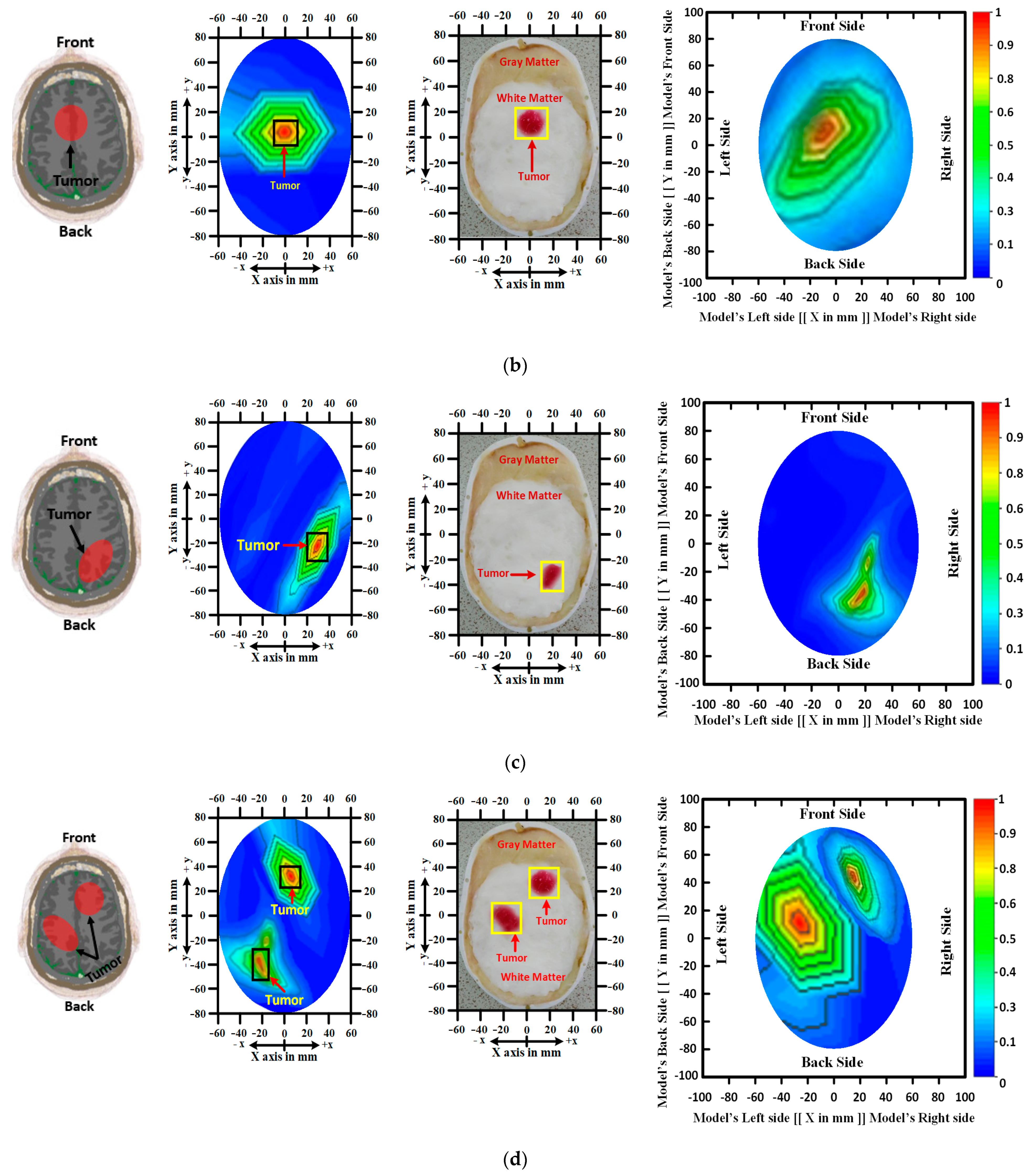
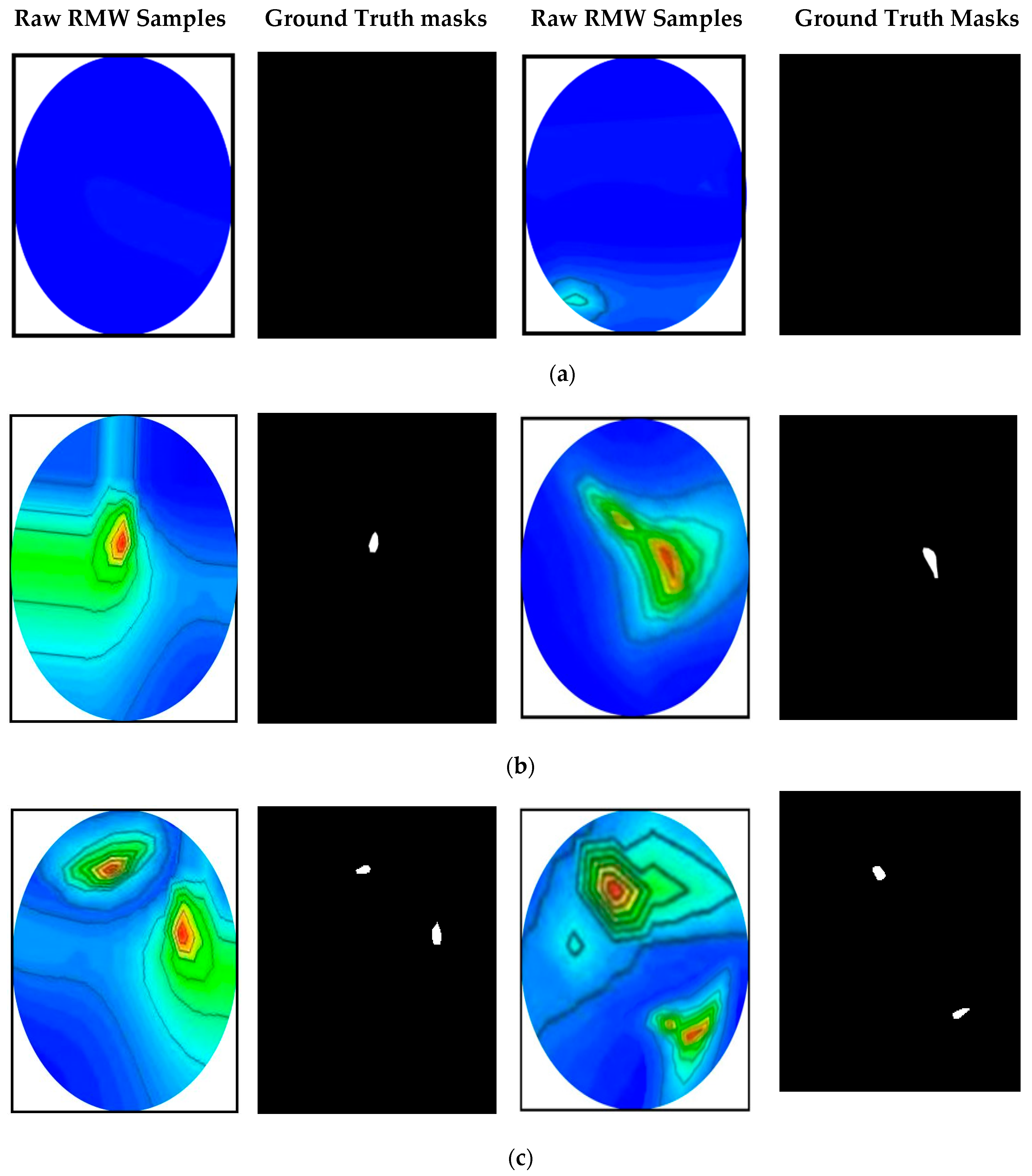
RMW Brain Tumor Image Sample Collection
3. Methodology and Materials
3.1. Dataset Preparation
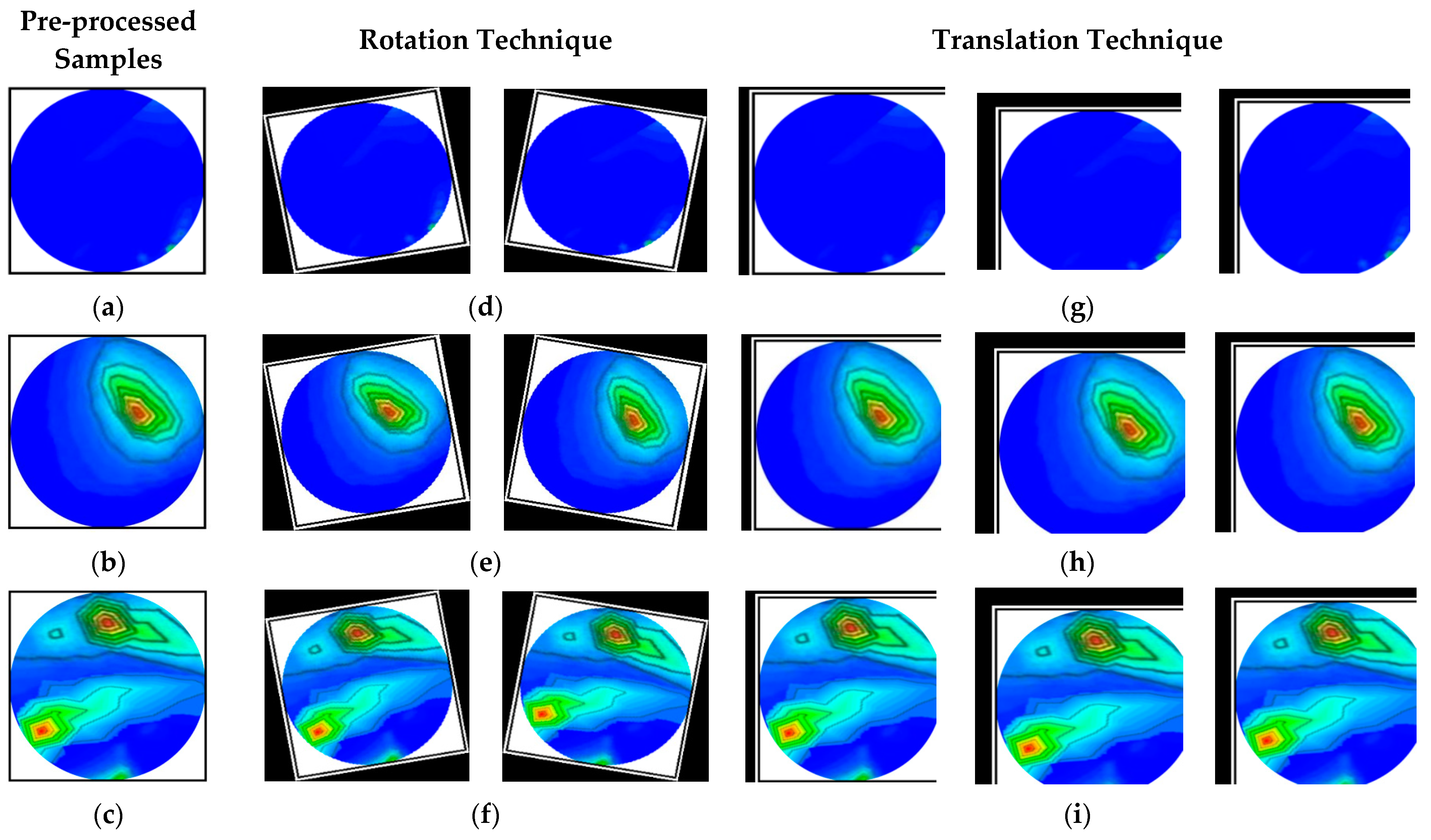
3.2. Image Pre-Processing and Method of Augmentation
3.3. Dataset Splitting and Ratio Consideration for Training and Testing Dataset
3.4. Experiments
3.4.1. Proposed MicrowaveSegNet (MSegNet)—Brain Tumor Segmentation Model
3.4.2. Experimental Analysis of the Segmentation Models
3.4.3. Proposed BrainImageNet (BINet)—Brain Image Classification Model
3.4.4. Experimental Analysis of the Classification Models
3.5. Performance Evaluation Matrices
3.5.1. Assessment Matrix for the Segmentation Model
3.5.2. Assessment Matrix for the Classification Model
4. Results and Discussion
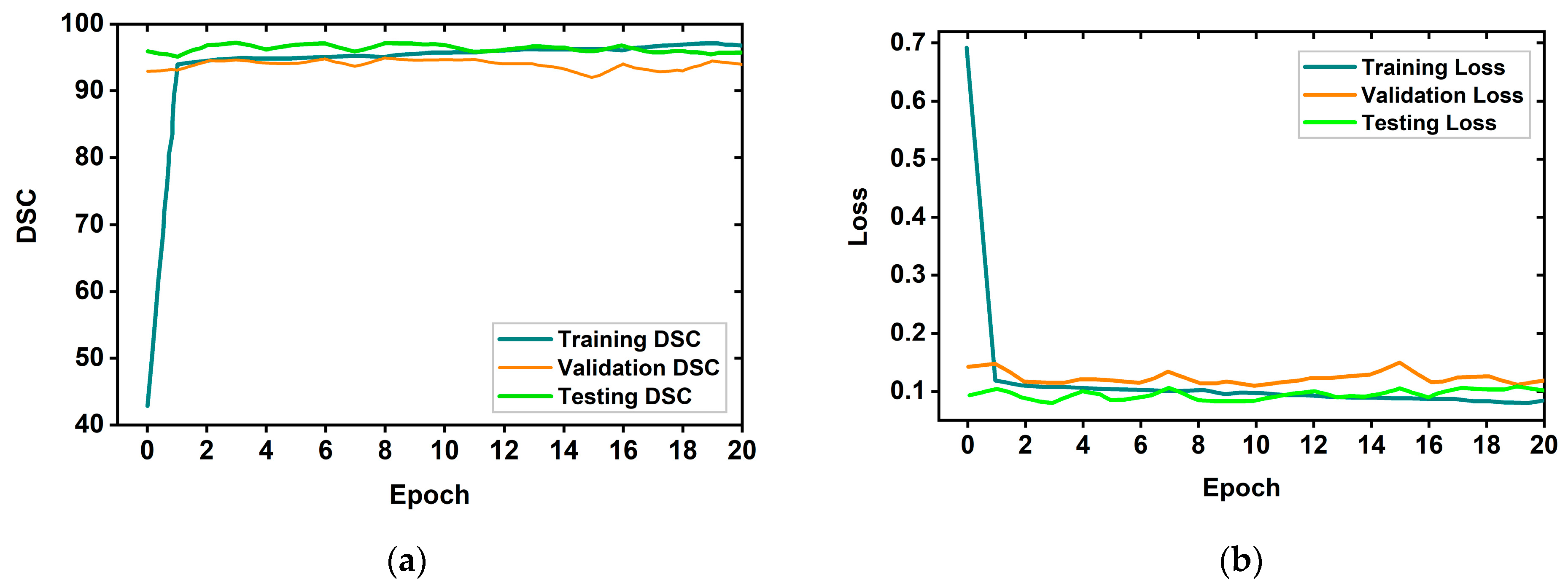
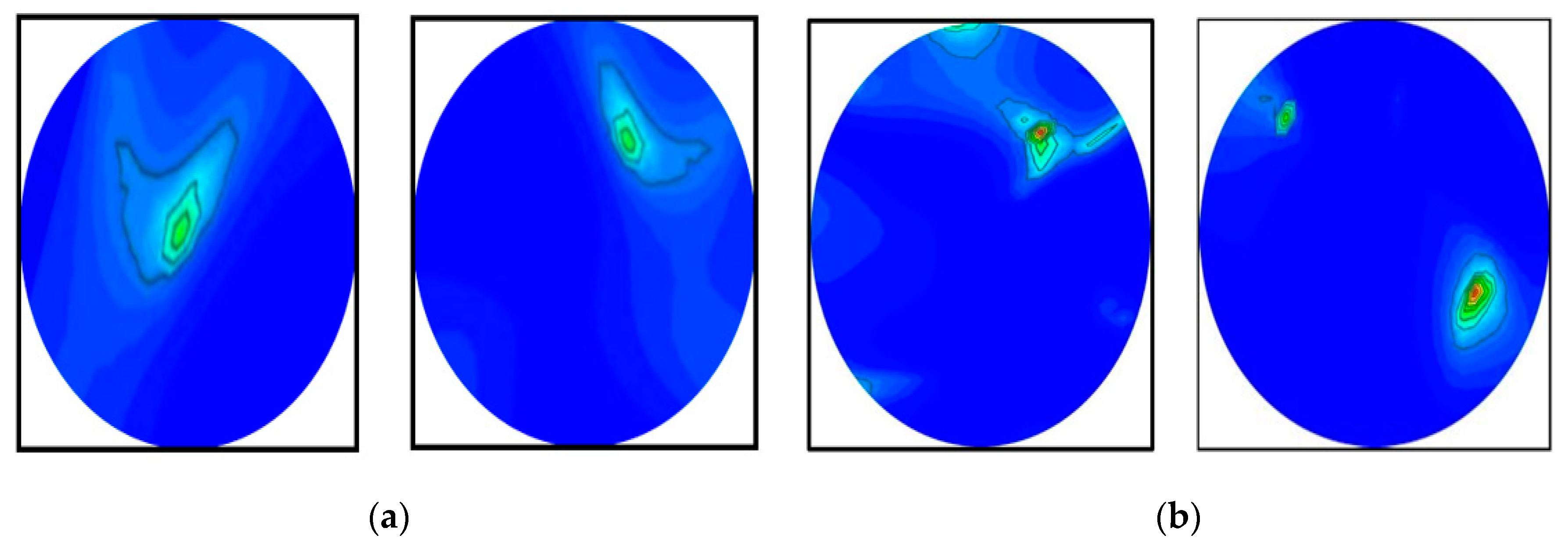
4.1. Brain Tumor Segmentation Performances
4.2. Raw and Segmented RMW Brain Images Classification Performances
4.3. Performance Analysis
5. Discussion about Classification Classes
Future Improvement and Future Directions to Microwave Biomedical Community
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tracy Wyant, R.A. Cynthia Ogoro. In Key Statistics for Brain and Spinal Cord Tumors, 12-01-2021 ed.; American Cancer Society: Atlanta, GA, USA, 2021. [Google Scholar]
- Ahmad, H.A.; Yu, H.J.; Miller, C.G. Medical imaging modalities. In Medical Imaging in Clinical Trials; Springer: Berlin/Heidelberg, Germany, 2014; pp. 3–26. [Google Scholar]
- Frangi, A.F.; Tsaftaris, S.A.; Prince, J.L. Simulation and synthesis in medical imaging. IEEE Trans. Med. Imaging 2018, 37, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Siddiqi, A.A.; Narejo, G.B.; Andleeb, S. A cross sectional study of tumors using bio-medical imaging modalities. Curr. Med. Imaging 2019, 15, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Adamson, E.B.; Ludwig, K.D.; Mummy, D.G.; Fain, S.B. Magnetic resonance imaging with hyperpolarized agents: Methods and applications. Phys. Med. Biol. 2017, 62, R81. [Google Scholar] [CrossRef] [PubMed]
- Cazzato, R.L.; Garnon, J.; Shaygi, B.; Koch, G.; Tsoumakidou, G.; Caudrelier, J.; Addeo, P.; Bachellier, P.; Namer, I.J.; Gangi, A. PET/CT-guided interventions: Indications, advantages, disadvantages and the state of the art. Minim. Invasive Ther. Allied Technol. 2018, 27, 27–32. [Google Scholar] [CrossRef]
- Chakraborty, S.; Chatterjee, S.; Ashour, A.S.; Mali, K.; Dey, N. Intelligent computing in medical imaging: A study. In Advancements in Applied Metaheuristic Computing; IGI Global: Hershey, PA, USA, 2018; pp. 143–163. [Google Scholar]
- Dougeni, E.; Faulkner, K.; Panayiotakis, G. A review of patient dose and optimisation methods in adult and paediatric CT scanning. Eur. J. Radiol. 2012, 81, e665–e683. [Google Scholar] [CrossRef]
- Jacobs, M.A.; Ibrahim, T.S.; Ouwerkerk, R. MR imaging: Brief overview and emerging applications. Radiographics 2007, 27, 1213–1229. [Google Scholar] [CrossRef]
- Jones, K.M.; Michel, K.A.; Bankson, J.A.; Fuller, C.D.; Klopp, A.H.; Venkatesan, A.M. Emerging magnetic resonance imaging technologies for radiation therapy planning and response assessment. Int. J. Radiat. Oncol. Biol. Phys. 2018, 101, 1046–1056. [Google Scholar] [CrossRef]
- Alqadami, A.S.; Bialkowski, K.S.; Mobashsher, A.T.; Abbosh, A.M. Wearable electromagnetic head imaging system using flexible wideband antenna array based on polymer technology for brain stroke diagnosis. IEEE Trans. Biomed. Circuits Syst. 2018, 13, 124–134. [Google Scholar] [CrossRef]
- Stancombe, A.E.; Bialkowski, K.S.; Abbosh, A.M. Portable microwave head imaging system using software-defined radio and switching network. IEEE J. Electromagn. RF Microw. Med. Biol. 2019, 3, 284–291. [Google Scholar] [CrossRef]
- Tobon Vasquez, J.A.; Scapaticci, R.; Turvani, G.; Bellizzi, G.; Joachimowicz, N.; Duchêne, B.; Tedeschi, E.; Casu, M.R.; Crocco, L.; Vipiana, F. Design and experimental assessment of a 2D microwave imaging system for brain stroke monitoring. Int. J. Antennas Propag. 2019, 2019, 8065036. [Google Scholar] [CrossRef]
- Hossain, A.; Islam, M.T.; Almutairi, A.F.; Singh, M.S.J.; Mat, K.; Samsuzzaman, M. An octagonal ring-shaped parasitic resonator based compact ultrawideband antenna for microwave imaging applications. Sensors 2020, 20, 1354. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Islam, M.T.; Chowdhury, M.E.; Samsuzzaman, M. A grounded coplanar waveguide-based slotted inverted delta-shaped wideband antenna for microwave head imaging. IEEE Access 2020, 8, 185698–185724. [Google Scholar] [CrossRef]
- Hossain, A.; Islam, M.T.; Islam, M.; Chowdhury, M.E.; Rmili, H.; Samsuzzaman, M. A Planar Ultrawideband Patch Antenna Array for Microwave Breast Tumor Detection. Materials 2020, 13, 4918. [Google Scholar] [CrossRef] [PubMed]
- Mobashsher, A.; Bialkowski, K.; Abbosh, A.; Crozier, S. Design and experimental evaluation of a non-invasive microwave head imaging system for intracranial haemorrhage detection. PLoS ONE 2016, 11, e0152351. [Google Scholar] [CrossRef] [PubMed]
- Mobashsher, A.T.; Abbosh, A.M.; Wang, Y. Microwave system to detect traumatic brain injuries using compact unidirectional antenna and wideband transceiver with verification on realistic head phantom. IEEE Trans. Microw. Theory Tech. 2014, 62, 1826–1836. [Google Scholar] [CrossRef]
- Mohammed, B.J.; Abbosh, A.M.; Mustafa, S.; Ireland, D. Microwave system for head imaging. IEEE Trans. Instrum. Meas. 2013, 63, 117–123. [Google Scholar] [CrossRef]
- Salleh, A.; Yang, C.; Alam, T.; Singh, M.; Samsuzzaman, M.; Islam, M. Development of microwave brain stroke imaging system using multiple antipodal vivaldi antennas based on raspberry Pi technology. J. Kejuruterran 2020, 32, 1–6. [Google Scholar]
- Fedeli, A.; Estatico, C.; Pastorino, M.; Randazzo, A. Microwave detection of brain injuries by means of a hybrid imaging method. IEEE Open J. Antennas Propag. 2020, 1, 513–523. [Google Scholar] [CrossRef]
- Inum, R.; Rana, M.; Shushama, K.N.; Quader, M. EBG based microstrip patch antenna for brain tumor detection via scattering parameters in microwave imaging system. Int. J. Biomed. Imaging 2018, 2018, 8241438. [Google Scholar] [CrossRef]
- Islam, M.S.; Islam, M.T.; Hoque, A.; Islam, M.T.; Amin, N.; Chowdhury, M.E. A portable electromagnetic head imaging system using metamaterial loaded compact directional 3D antenna. IEEE Access 2021, 9, 50893–50906. [Google Scholar] [CrossRef]
- Rezaeieh, S.A.; Zamani, A.; Abbosh, A. 3-D wideband antenna for head-imaging system with performance verification in brain tumor detection. IEEE Antennas Wirel. Propag. Lett. 2014, 14, 910–914. [Google Scholar] [CrossRef]
- Rokunuzzaman, M.; Ahmed, A.; Baum, T.C.; Rowe, W.S. Compact 3-D antenna for medical diagnosis of the human head. IEEE Trans. Antennas Propag. 2019, 67, 5093–5103. [Google Scholar] [CrossRef]
- Gerazov, B.; Conceicao, R.C. Deep learning for tumour classification in homogeneous breast tissue in medical microwave imaging. In Proceedings of IEEE EUROCON 2017-17th International Conference on Smart Technologies; IEEE: Piscataway, NJ, USA, 2017; pp. 564–569. [Google Scholar]
- Khoshdel, V.; Asefi, M.; Ashraf, A.; LoVetri, J. Full 3D microwave breast imaging using a deep-learning technique. J. Imaging 2020, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.P.; Dey, M.; Tiberi, G.; Sani, L.; Vispa, A.; Raspa, G.; Duranti, M.; Ghavami, M.; Dudley, S. Machine learning approaches for automated lesion detection in microwave breast imaging clinical data. Sci. Rep. 2019, 9, 10510. [Google Scholar] [CrossRef]
- Salucci, M.; Polo, A.; Vrba, J. Multi-step learning-by-examples strategy for real-time brain stroke microwave scattering data inversion. Electronics 2021, 10, 95. [Google Scholar] [CrossRef]
- Shah, P.; Moghaddam, M. Super resolution for microwave imaging: A deep learning approach. In Proceedings of 2017 IEEE International Symposium on Antennas and Propagation & USNC/URSI National Radio Science Meeting; IEEE: Piscataway, NJ, USA, 2017; pp. 849–850. [Google Scholar]
- Shao, W.; Du, Y. Microwave imaging by deep learning network: Feasibility and training method. IEEE Trans. Antennas Propag. 2020, 68, 5626–5635. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.; Berger, C.; Ha, S.; Rozycki, M. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv 2018, arXiv:1811.02629. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Cheng, G.; Ji, H. Adversarial Perturbation on MRI Modalities in Brain Tumor Segmentation. IEEE Access 2020, 8, 206009–206015. [Google Scholar] [CrossRef]
- Ding, Y.; Chen, F.; Zhao, Y.; Wu, Z.; Zhang, C.; Wu, D. A stacked multi-connection simple reducing net for brain tumor segmentation. IEEE Access 2019, 7, 104011–104024. [Google Scholar] [CrossRef]
- Aboelenein, N.M.; Songhao, P.; Koubaa, A.; Noor, A.; Afifi, A. HTTU-Net: Hybrid two track U-net for automatic brain tumor segmentation. IEEE Access 2020, 8, 101406–101415. [Google Scholar] [CrossRef]
- Hu, K.; Gan, Q.; Zhang, Y.; Deng, S.; Xiao, F.; Huang, W.; Cao, C.; Gao, X. Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 2019, 7, 92615–92629. [Google Scholar] [CrossRef]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. S3D-UNet: Separable 3D U-Net for brain tumor segmentation. In Proceedings of International MICCAI Brainlesion Workshop; Springer: Berlin/Heidelberg, Germany, 2019; pp. 358–368. [Google Scholar]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Imran, M.; Shoaib, M. A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access 2020, 8, 55135–55144. [Google Scholar] [CrossRef]
- Ding, Y.; Li, C.; Yang, Q.; Qin, Z.; Qin, Z. How to improve the deep residual network to segment multi-modal brain tumor images. IEEE Access 2019, 7, 152821–152831. [Google Scholar] [CrossRef]
- Hao, J.; Li, X.; Hou, Y. Magnetic resonance image segmentation based on multi-scale convolutional neural network. IEEE Access 2020, 8, 65758–65768. [Google Scholar] [CrossRef]
- Kumar, R.L.; Kakarla, J.; Isunuri, B.V.; Singh, M. Multi-class brain tumor classification using residual network and global average pooling. Multimed. Tools Appl. 2021, 80, 13429–13438. [Google Scholar] [CrossRef]
- Devecioglu, O.C.; Malik, J.; Ince, T.; Kiranyaz, S.; Atalay, E.; Gabbouj, M. Real-time glaucoma detection from digital fundus images using Self-ONNs. IEEE Access 2021, 9, 140031–140041. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Malik, J.; Abdallah, H.B.; Ince, T.; Iosifidis, A.; Gabbouj, M. Self-organized operational neural networks with generative neurons. Neural Netw. 2021, 140, 294–308. [Google Scholar] [CrossRef]
- Hossain, A.; Islam, M.T.; Islam, M.S.; Chowdhury, M.E.; Almutairi, A.F.; Razouqi, Q.A.; Misran, N. A YOLOv3 Deep Neural Network Model to Detect Brain Tumor in Portable Electromagnetic Imaging System. IEEE Access 2021, 9, 82647–82660. [Google Scholar] [CrossRef]
- Mobashsher, A.; Abbosh, A. Three-dimensional human head phantom with realistic electrical properties and anatomy. IEEE Antennas Wirel. Propag. Lett. 2014, 13, 1401–1404. [Google Scholar] [CrossRef]
- Hossain, A.; Islam, M.T.; Almutairi, A.F. A deep learning model to classify and detect brain abnormalities in portable microwave based imaging system. Sci. Rep. 2022, 12, 6319. [Google Scholar] [CrossRef]
- Karadima, O.; Rahman, M.; Sotiriou, I.; Ghavami, N.; Lu, P.; Ahsan, S.; Kosmas, P. Experimental validation of microwave tomography with the DBIM-TwIST algorithm for brain stroke detection and classification. Sensors 2020, 20, 840. [Google Scholar] [CrossRef] [PubMed]
- Joachimowicz, N.; Duchêne, B.; Conessa, C.; Meyer, O. Anthropomorphic breast and head phantoms for microwave imaging. Diagnostics 2018, 8, 85. [Google Scholar] [CrossRef] [PubMed]
- Wood, S.; Krishnamurthy, N.; Santini, T.; Raval, S.B.; Farhat, N.; Holmes, J.A.; Ibrahim, T.S. Correction: Design and fabrication of a realistic anthropomorphic heterogeneous head phantom for MR purposes. PLoS ONE 2018, 13, e0192794. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, B.; Li, H.; Fu, F.; Shi, X.; Dong, X.; Dai, M. A novel 3D-printed head phantom with anatomically realistic geometry and continuously varying skull resistivity distribution for electrical impedance tomography. Sci. Rep. 2017, 7, 4608. [Google Scholar] [CrossRef]
- Pokorny, T.; Vrba, D.; Tesarik, J.; Rodrigues, D.B.; Vrba, J. Anatomically and dielectrically realistic 2.5 D 5-layer reconfigurable head phantom for testing microwave stroke detection and classification. Int. J. Antennas Propag. 2019, 2019, 5459391. [Google Scholar] [CrossRef]
- Li, C.-W.; Hsu, A.-L.; Huang, C.-W.C.; Yang, S.-H.; Lin, C.-Y.; Shieh, C.-C.; Chan, W.P. Reliability of synthetic brain MRI for assessment of ischemic stroke with phantom validation of a relaxation time determination method. J. Clin. Med. 2020, 9, 1857. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.W.; Liao, S.W. KUnet: Microscopy image segmentation with deep unet based convolutional networks. In Proceedings of 2019 IEEE International Conference on Systems, Man and Cybernetics (SMC); IEEE: Piscataway, NJ, USA, 2019; pp. 3561–3566. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. arXiv 2019, arXiv:1505.04597. [Google Scholar]
- Kiranyaz, S.; Ince, T.; Iosifidis, A.; Gabbouj, M. Operational neural networks. Neural Comput. Appl. 2020, 32, 6645–6668. [Google Scholar] [CrossRef]
- Kiranyaz, S.; Malik, J.; Abdallah, H.B.; Ince, T.; Iosifidis, A.; Gabbouj, M. Exploiting heterogeneity in operational neural networks by synaptic plasticity. Neural Comput. Appl. 2021, 33, 7997–8015. [Google Scholar] [CrossRef]
- Malik, J.; Kiranyaz, S.; Gabbouj, M. Operational vs. convolutional neural networks for image denoising. arXiv 2020, arXiv:2009.00612. [Google Scholar]
- Malik, J.; Kiranyaz, S.; Gabbouj, M. Self-organized operational neural networks for severe image restoration problems. Neural Netw. 2021, 135, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Rahman, T.; Khandakar, A.; Qiblawey, Y.; Tahir, A.; Kiranyaz, S.; Kashem, S.B.A.; Islam, M.T.; Al Maadeed, S.; Zughaier, S.M.; Khan, M.S. Exploring the effect of image enhancement techniques on COVID-19 detection using chest X-ray images. Comput. Biol. Med. 2021, 132, 104319. [Google Scholar] [CrossRef] [PubMed]
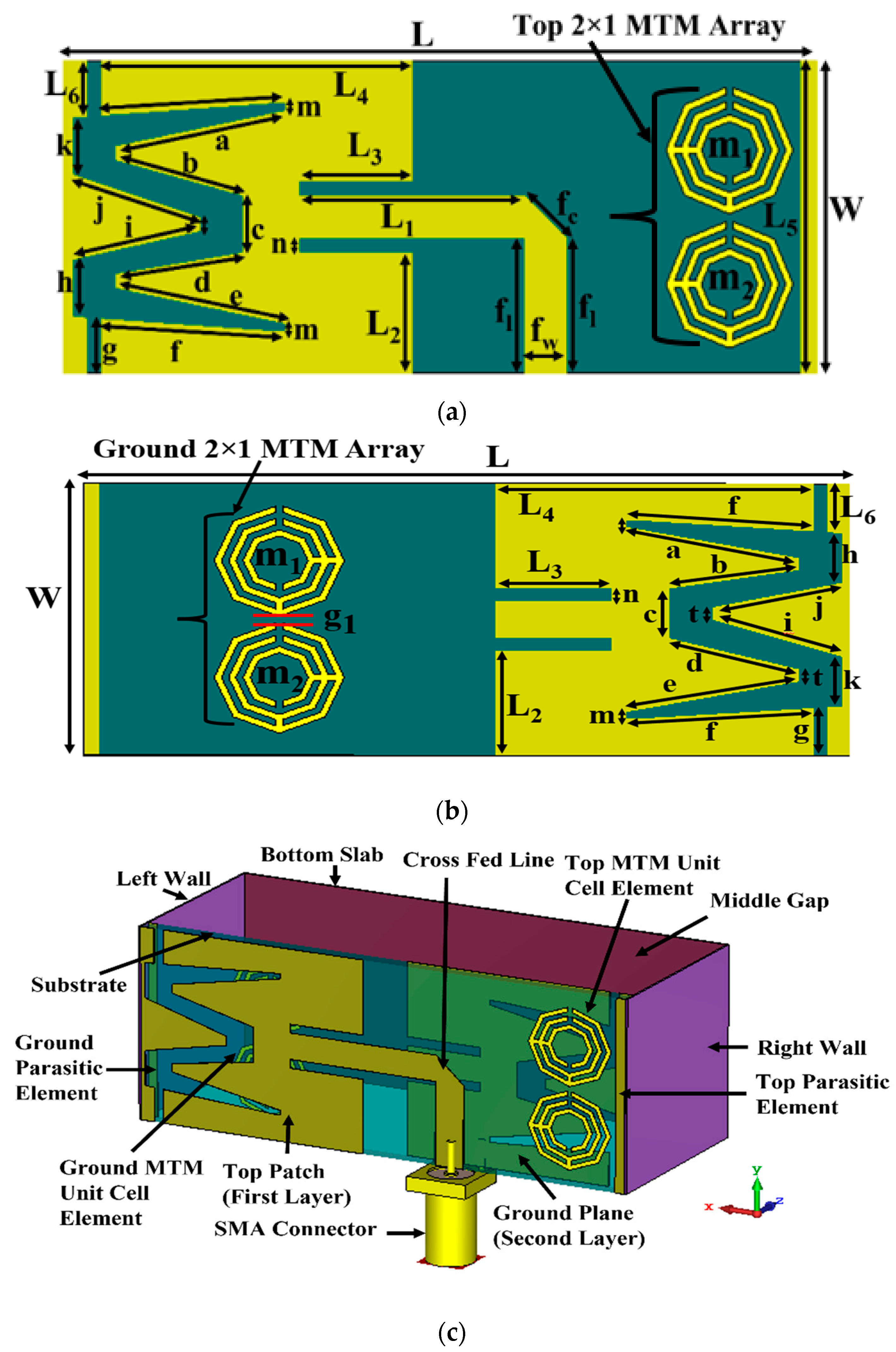








| Parameters | Value (mm) | Parameters | Value (mm) | Parameters | Value (mm) |
|---|---|---|---|---|---|
| L | 53.00 | b | 9.34 | k | 4.00 |
| W | 22.00 | c | 4.00 | t | 1.00 |
| L1 | 16.00 | d | 9.12 | fl | 9.50 |
| L2 | 8.50 | e | 12.26 | fw | 3.00 |
| L3 | 8.00 | f | 12.26 | fc | 4.24 |
| L4 | 22.00 | g | 3.86 | g1 | 0.50 |
| L5 | 22.00 | h | 4.00 | m | 0.50 |
| L6 | 3.93 | i | 9.22 | n | 1.00 |
| a | 12.26 | j | 9.49 | .. | .. |
| Ref. | Types of Phantom | Fabricated Tissues | Imaging System | Image Reconstruction Algorithm | No. of Detection | Application |
|---|---|---|---|---|---|---|
| [23] | Semi-solid heterogeneous | DURA, CSF, WM, GM | Nine-antenna-based experimental system | IC-CF-DMAS | Only one object | Microwave stroke imaging |
| [49] | Liquid, homogeneous | Only brain tissue | Eight-antenna-based experimental system | DBIM-TwIST | Single tumor with noisy image | Microwave tomography imaging |
| [22] | Semi-solid heterogeneous | Brain CSF, DURA | Single-antenna-based simulated system | Radar-based confocal | Single tumor with noisy image | Microwave brain imaging |
| [50] | Solid, acrylonitrile butadiene styrene (ABS) | CSF, WM, and GM | Single-antenna-based simulated system | Not stated | Single tumor with noisy image | Microwave brain imaging |
| [51] | Liquid, heterogeneous | Brain, CSF, fat, and muscle | Simulated imaging System | Segmentation slice-based | Single tumor with noisy image | Magnetic resonance imaging and electromagnetic imaging |
| [52] | Solid, acrylonitrile butadiene styrene (ABS) | Skull, CSF, brain | Two-antenna-based experimental system | EIT-based | Single tumor with blurry images | Microwave tomography imaging |
| [53] | Semi-solid heterogeneous | Scalp, skull, CSF | Single-antenna-based simulated system | Multi-layer time stable confocal | Single object with noisy image | Microwave brain imaging |
| [54] | Liquid, heterogeneous | CSF, WM, GM | Single-antenna-based experimental system | Not stated | Only one object | Microwave brain imaging |
| Used Phantom | Semi-solid heterogeneous | DURA, CSF, GM, WM, fat, skin | Nine-antenna-based experimental imaging system | M-DMAS | Two tumors with clear image | Sensor-based Microwave brain tumor imaging system (SMBIS) |
| Dataset | Number of Original Images | Image Classes | Training Dataset | |||
|---|---|---|---|---|---|---|
| Number of Images per Class | Augmented Train Images per Fold | Testing Images per Fold | Validation Image per Fold | |||
| Raw RMW brain image samples | 300 | Non-tumor | 100 | 1980 | 20 | 16 |
| Single tumor | 100 | 2008 | 20 | 16 | ||
| Double tumors | 100 | 2012 | 20 | 16 | ||
| Total | 300 | 6000 | 60 | 48 | ||
| Parameter’s Name | Assigned Value | Parameter’s Name | Assigned Value |
|---|---|---|---|
| Input channels | 3 | Output channels | 1 |
| Batch size | 8 | Optimizer | Adam |
| Learning rate (LR) | 0.0005 | Loss type | Dice loss |
| Maximum number of epochs | 30 | Epochs patience | 10 |
| Maximum epochs stop | 15 | Learning factor | 0.2 |
| Initial feature | 32 | Number of folds | 5 |
| Parameter’s Name | Assigned Value | Parameter’s Name | Assigned Value |
|---|---|---|---|
| Input channels | 3 | Q order | 1 for CNN, 3 for Self-ONNs |
| Batch size | 16 | Optimizer | Adam |
| Learning rate (LR) | 0.0005 | Stop criteria | Loss |
| Maximum number of epochs | 30 | Epochs patience | 5 |
| Maximum epochs stop | 10 | Learning factor | 0.2 |
| Image size | 224 | Number of folds | 5 |
| Network Model Name | Accuracy (%) | IoU (%) | Dice Score (%) | Loss |
|---|---|---|---|---|
| U-net | 99.96 | 85.72 | 91.58 | 0.1127 |
| Modified Unet (M-Unet) | 99.96 | 86.47 | 92.20 | 0.1086 |
| Keras Unet (K-Unet) | 99.96 | 86.01 | 91.91 | 0.1156 |
| MultiResUnet | 99.96 | 86.55 | 92.20 | 0.1064 |
| ResNet50 | 99.95 | 86.43 | 92.13 | 0.1121 |
| DenseNet161 | 99.95 | 85.62 | 91.59 | 0.1145 |
| ResNet152 FPN | 99.94 | 82.86 | 89.58 | 0.1312 |
| DenseNet121 FPN | 99.95 | 83.30 | 89.91 | 0.1318 |
| nnU-net | 99.96 | 84.95 | 92.85 | 0.1112 |
| Proposed MSegNet | 99.97 | 86.92 | 93.10 | 0.1010 |
| Network Model Name | Parameters (M) | Training Time (Second/Fold) | Inference Time (Second/Image) |
|---|---|---|---|
| U-net | 30 | 480 | 0.025 |
| Modified Unet (M-Unet) | 28 | 440 | 0.023 |
| Keras Unet (K-Unet) | 30 | 490 | 0.026 |
| MultiResUnet | 25 | 425 | 0.02 |
| ResNet50 | 25 | 420 | 0.023 |
| DenseNet161 | 28.5 | 450 | 0.033 |
| ResNet152 FPN | 40 | 720 | 0.05 |
| DenseNet121 FPN | 20 | 410 | 0.021 |
| nnU-net | 18 | 340 | 0.015 |
| Proposed MSegNet | 8 | 305 | 0.007 |
| Image Type | Network Model Name | Overall | Weighted | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (A) | Precession (P) | Recall (R) | Specificity (S) | F1 Score (Fs) | ||||||||
| Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | |||
| Raw RMW Brain Images | Vanilla CNN6L | 84.33 | 4.11 | 84.17 | 4.13 | 84.33 | 4.11 | 92.17 | 3.04 | 84.06 | 4.14 | <0.05 |
| Vanilla CNN8L | 85.33 | 4.00 | 85.62 | 3.97 | 85.33 | 4.00 | 92.67 | 2.95 | 85.14 | 4.03 | <0.05 | |
| Self-ONN4L | 85.00 | 4.04 | 84.91 | 4.05 | 85.00 | 4.04 | 92.50 | 2.98 | 84.87 | 4.06 | <0.05 | |
| Self-ONN4L1DN | 87.00 | 3.81 | 87.05 | 3.80 | 87.00 | 3.81 | 93.50 | 2.79 | 86.95 | 3.81 | <0.05 | |
| Self-ONN6L | 87.00 | 3.81 | 86.85 | 3.82 | 87.00 | 3.81 | 93.50 | 2.79 | 86.82 | 3.83 | <0.05 | |
| Proposed BINet | 89.33 | 3.49 | 88.74 | 3.58 | 88.67 | 3.59 | 94.33 | 2.62 | 88.61 | 3.59 | <0.05 | |
| Image Type | Network Model Name | Overall | Weighted | p-Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy (A) | Precession (P) | Recall (R) | Specificity (S) | F1 Score (Fs) | ||||||||
| Mean | STD | Mean | STD | Mean | STD | Mean | STD | Mean | STD | |||
| Segmented RMW Brain Images | Vanilla CNN6L | 95.00 | 2.47 | 94.98 | 2.47 | 95.00 | 2.47 | 97.50 | 1.77 | 94.96 | 2.48 | <0.05 |
| Vanilla CNN8L | 95.67 | 2.30 | 95.77 | 2.28 | 95.67 | 2.30 | 97.83 | 1.65 | 95.65 | 2.31 | <0.05 | |
| Self-ONN4L | 94.00 | 2.69 | 93.96 | 2.70 | 94.00 | 2.69 | 97.00 | 1.93 | 93.96 | 2.70 | <0.05 | |
| Self-ONN4L1DN | 96.33 | 2.13 | 96.41 | 2.11 | 97.00 | 1.93 | 98.17 | 1.52 | 97.00 | 1.93 | <0.05 | |
| Self-ONN6L | 96.67 | 2.03 | 96.79 | 1.99 | 96.67 | 2.03 | 98.33 | 1.45 | 96.66 | 2.03 | <0.05 | |
| Proposed BINet | 98.33 | 1.45 | 98.35 | 1.44 | 98.33 | 1.45 | 99.17 | 1.03 | 98.33 | 1.45 | <0.05 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hossain, A.; Islam, M.T.; Rahman, T.; Chowdhury, M.E.H.; Tahir, A.; Kiranyaz, S.; Mat, K.; Beng, G.K.; Soliman, M.S. Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models. Biosensors 2023, 13, 302. https://doi.org/10.3390/bios13030302
Hossain A, Islam MT, Rahman T, Chowdhury MEH, Tahir A, Kiranyaz S, Mat K, Beng GK, Soliman MS. Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models. Biosensors. 2023; 13(3):302. https://doi.org/10.3390/bios13030302
Chicago/Turabian StyleHossain, Amran, Mohammad Tariqul Islam, Tawsifur Rahman, Muhammad E. H. Chowdhury, Anas Tahir, Serkan Kiranyaz, Kamarulzaman Mat, Gan Kok Beng, and Mohamed S. Soliman. 2023. "Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models" Biosensors 13, no. 3: 302. https://doi.org/10.3390/bios13030302
APA StyleHossain, A., Islam, M. T., Rahman, T., Chowdhury, M. E. H., Tahir, A., Kiranyaz, S., Mat, K., Beng, G. K., & Soliman, M. S. (2023). Brain Tumor Segmentation and Classification from Sensor-Based Portable Microwave Brain Imaging System Using Lightweight Deep Learning Models. Biosensors, 13(3), 302. https://doi.org/10.3390/bios13030302








