Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review
Abstract
:1. Introduction
2. Cell Lysis
2.1. Chemical Cell Lysis
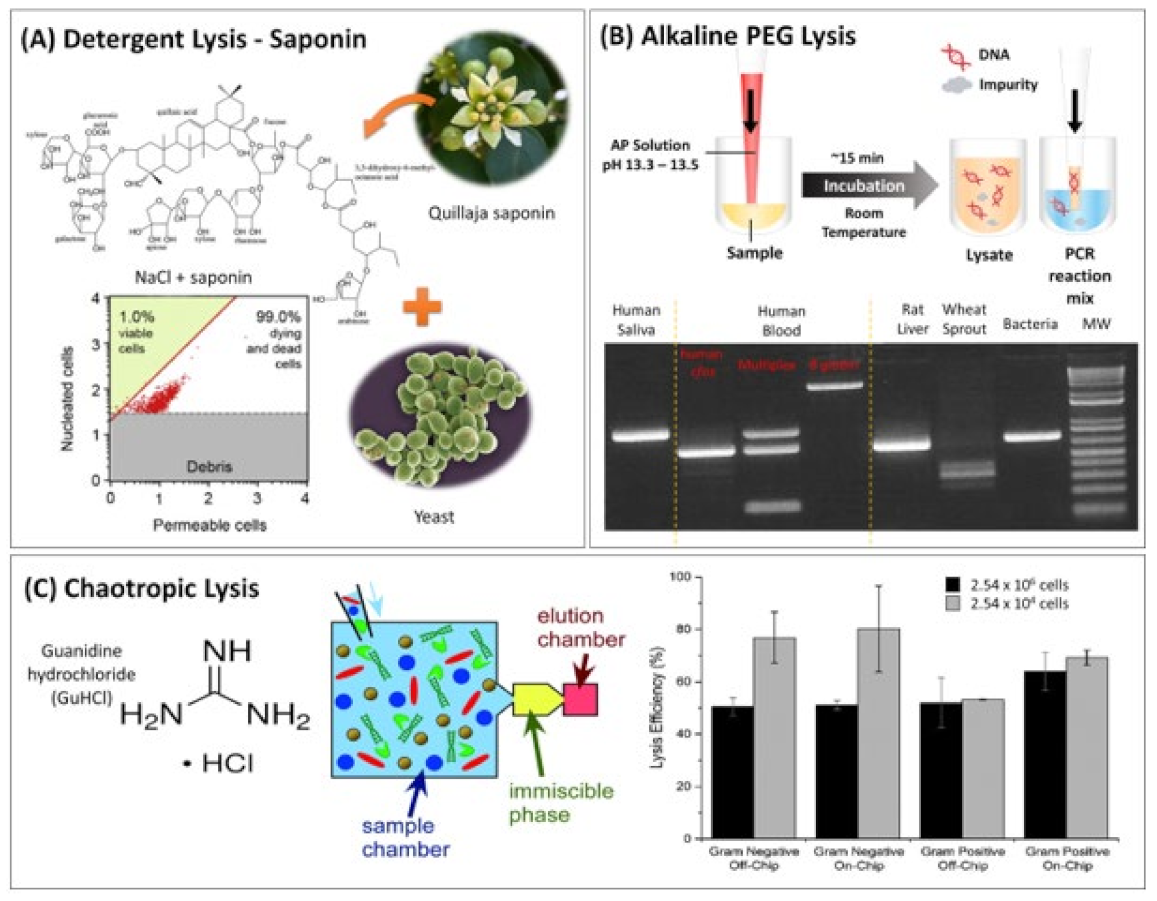
2.1.1. Detergents
2.1.2. Enzymatic Lysis
2.1.3. Alkaline Lysis
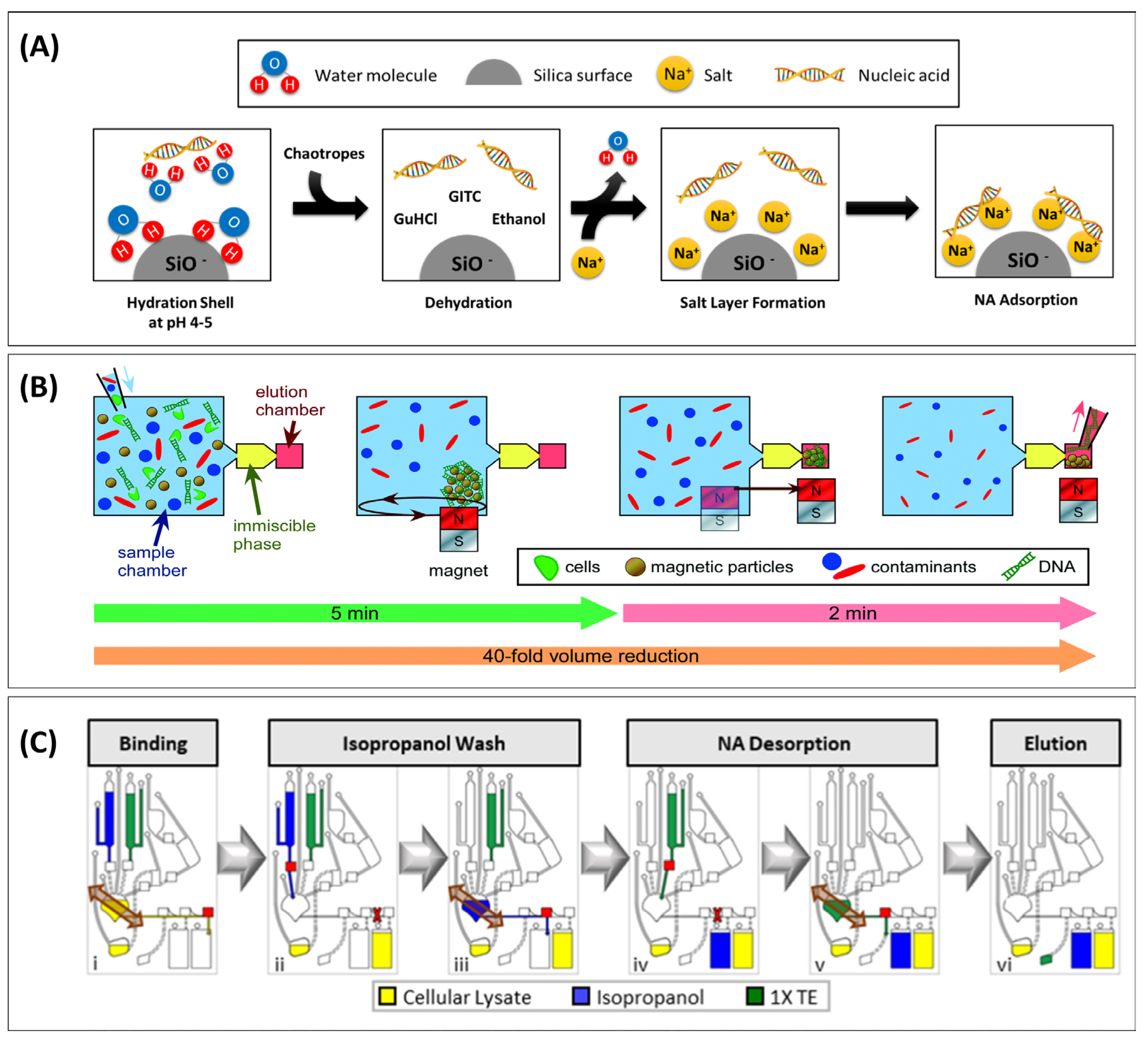
2.1.4. Chaotropic Lysis
2.1.5. New Reagents for Cell Lysis
2.2. Other Cell Lysis Methods
3. Nucleic Acid Extraction
3.1. Solid-Phase Extraction (SPE)
3.1.1. Reagents
Anionic Supports under Chaotropic Conditions
| Solid Phase | Surface | Binding Buffer | Washing Buffer | Elution Buffer | Elution Volume | Target | Sample Matrix | Amplification | LOD | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| MB | Silica | Ethanol | Kit | Kit | 10 µL | HPV virus | Synthetic DNA | qPCR | - | [19] |
| MB | Silica | GuHCl | Mineral oil + GuHCl | Water | 10 µL | Bacteria | Liquid stool (clinical) | PCR | - | [41] |
| Glass membrane | Whatman glass pad | 20 mM Tris-HCl, 4 M GuSCN, 1 mM DTT, pH 7.7 (lysis buffer) | Isopropanol, 15% v/v | Water | 5 µL | Animal | Mixed meat (minced) | qPCR | [42] | |
| Paper/Disc | Cellulose | 1.5 M GuHCl, 50 mM Tris [pH 8], 100 mM NaCl, 5 mM EDTA, 1% Tween-20 | 10 mM Tris pH 8.0, 0.1% Tween-20 | Water | 10 µL | Viral gene in fish | Blood (fish) | PCR | 104 cells | [49] |
| MB | Silica | GuHCl, TRIS, EDTA, NaCl in ethanol (50%) | Wash buffer I (GuHCl + 68% v/v ethanol) Wash buffer II (70% v/v ethanol) | 10 mM Tris, 0.1 mM EDTA pH 8 | 200 µL | Bacteria | Serum and saliva | RPA | - | [48] |
| MB | Silica | GuSCN + Triton X-100 (pH 6.8) | Organogel (12-HAS) GuSCN (pH 6.8) Ethanol NaCl2 (pH 7.6) | 10 mM Tris, 0.1 mM EDTA pH 8 (TE buffer) | 100 µL | Virus | Spiked blood | qPCR | 5 particles | [50] |
| MB | Silica | 4 M GuSCN, 10 mM MES (2-ethanesulfonic acid), 1% Triton X-100, with 1% ß-mercaptoethanol | oil immersed, Ethanol, 50% v/v, then water Water | None (on-bead amplification) | Virus | Nasopharyngeal swab | LAMP | 1–10 copies/µL | [51] | |
| MB | Silica | 3 M GuHCl, protein kinase K, at elevated T | Isopropanol | 10 mM Tris, 0.1 mM EDTA pH 8 | 8 µL | Virus | Buccal swab | LAMP, qPCR | - | [45] |
| MB + Steel wool | Silica | GuSCN + EDTA + Tris-HCl + Triton X-100 | GTIC Ethanol Tris-HCl + EDTA | 30 µL | Synthetic sputum + residual urine sample | qPCR | - | [52] | ||
| MB | Silica | Isopropanol | Washing buffer 1 and 2 | Elution buffer | Virus | Cervical swab | PCR | 103 copies/mL | [112] | |
| MB | Silica | 3.5 M GuSCN, isopropanol, 45% v/v, 2.5% Tween 20, 10 mM Tris pH 8.0, 1 mM EDTA | 3 M GuSCN isopropanol 30% v/v, 5% Tween 20, 40 mM Bis-Tris pH 6.0, 2 mM EDTA then 50 mM Tris pH 8.0 0.5 mM EDTA ethanol 80% v/v then ethanol, 100% v/v | human | Plasma | PCR | - | [113] | ||
| GB | Glass for DNA, oligo(dT) functionalized for RNA | Ethanol | Fluorinated oil Buffer AW1/2 (DNA) or Tris-HCl (pH 7.5) + LiCl + EDTA (RNA) | DNA: Tris HCl, 10 mM, EDTA, 0.5 mM, pH 9/RNA, Tris-HCl (pH 7.5) | 100 µL | mRNA | THP-1 cell | qPCR/RT-qPCR | 10 cells | [110] |
| MB | Selective recognition using NA probe | 100 mM phosphate, 150 mM NaCl, pH 6.0 | 10 mM Tris, 50 mM NaCl, pH 8.0 | 10 µL | Virus | Lab culture | Amplification-free | 0.021 pfu/mL | [114] | |
| Membrane (kit, ground) | Silica | Buffer AW | Buffer AW1/2 | Kit | Virus | Lab culture | RT-LAMP | 25 copies | [115] | |
| Porous silicon | Silica | 6 M GuSCN in 10 mM Tris, 1 mM EDTA (pH 8) with 1% Triton-X 100 (adjusted to pH 6.4) | Ethanol, 70% v/v in 10 mM NaCl | 10 mM Tris, 0.1 mM EDTA pH 8 (TE buffer) | 10 µL, 25 µL | [116] | ||||
| Silica filter | Silica | Binding buffer | GuSCN,3 M 25% v/v, Ethanol, 75% v/v then Ethanol 96% v/v | Water | 50 µL | HPV 16 | Cervical specimens | NASBA | [117] | |
| Paper | Polyether sulfone (PES) | GuSCN, NaCl, 1-butanol, glycoblue (coprecipitant) | Ethanol 70% v/v, then 100% v/v | LAMP mix | 12.5 µL | HPV 16 | Cervical specimens | LAMP | 1.2.105 copies | [118] |
| Paper | Paper also polymer monolith | 2.6 M GuSCN, 300 mM NaCl, 35% v/v 1-butanol 45 μg glycoblue (co-precipitant) | Ethanol 70% v/v, then 100% v/v | Tris EDTA | 200 µL | Bacteria | Synthetic urine | Isothermal helicase-dependent amplification (tHDA) | [119] |
Non-Chaotropic Binding

| Solid Phase | Surface Group | Binding Buffer | Washing Buffer | Washing Steps, Volume | Elution Buffer | Elution Volume | Target | Sample Matrix | Amplification | LOD | Kit vs. Assay | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MB | Fe3O4 nanoparticles | PEG + NaCl | Ethanol, 75% v/v | Water | 50 µL | Human saliva | PCR | n/a | Assay | [43] | ||
| Glass filter | Glass | 200 mM NaOH with 1% SDS (=lysis buffer) | Isopropanol, 15% v/v | 1, 75 µL | Water | 2 µL | Bacteria (aerosol spiked) | Cultured | qPCR | 10 CFU | Kit | [34] |
| MB | Silica | Lysis buffer (SDS and Protein Kinase K) | Washing buffer 1 and 2 | 2 | Low ionic strength elution buffer | 100 µL | K562, CHO-K1 cells | Culture media | PCR | 18 cells | Comparable but faster | [97] |
| MB | Carboxyl (to compare with silanol) | PEG 8000 (18 wt%), NaCl, 1 M | Ethanol, 80% v/v | Water | Animal | Faecal swab | PCR | 35.53 ± 15.03 ng (faecal) 261.12 ± 390.08 ng (cloacal swab) 233.52 ± 142.83 (oral swab) 87.3 ± 7.2% N/A | [139] | |||
| MB | Silica | 5% PEG8000, 0.5 M NaCl, and 3.5 mM KOH | Wash can be eliminated | On-bead amplification | - | Bacteria | Artificial saliva, sweat, urine | qPCR | 0.15 CFU/50 μL | Assay | [140] |
Cationic Supports
| Solid Phase | Surface | Binding Buffer | Washing Buffer | Elution Buffer | Elution Volume | Target | Sample Matrix | Amplification | LOD | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Acrylonitrile butadiene styrene (ABS) device | (3-Aminopropyl)triethoxysilane (APTES) | Dimethyl suberimidate in TE-based lysis buffer | PBS | NaHCO3 10 mM, pH > 10 | Virus | Clinical | RPA | 10 copies | [31] | |
| MB | Imidazole | Tris-HCl (pH3) | PBS | NaHCO3 (pH 10.6) | Bacteria | Human urine/milk | qPCR | 5 CFU/10 mL | [44] | |
| PMMA | Histidine or polyhistidine | 0.5 M KAc, pH 5.0 | 0.5 M potassium acetate, pH 5.0 | NaHCO3 (pH 10.6) | 10 µL | Bacteria | Culture | PCR | <5000 cells | [141] |
| Amine-functionalized diatomaceous earth | 3-aminopropyl(diethoxy)methyl silane | Dimethyl suberimidate in lysis buffer (Proteinase K, Tris-HCl [pH 8.0], EDTA, SDS, Triton X-100, lysozyme solution RNase-Free Dnase) | PBS | NaHCO3 10 mM, pH > 10 | 100 µL | PCR qPCR | [131] | |||
| Fe3O4 | PDA | PEG, 20% v/v, 4 M NaCl | Ethanol, 70% v/v | 10 mM Tris-HCl, 1 mM EDTA, pH 8 | 50 µL | PCR | [142] | |||
| Glass slide | APTES | Dimethyl adipimidate (DMA) | PBS | NaHCO3 (pH 10.6) | 150 µL | HRAS gene | Urine | PCR | - | [145] |
| ABS chamber wall | APTES | Dimethyl pimelimidate (DMP) in 100 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% SDS, and 10% Triton X-100) with either proteinase K (for DNA) or proteinase K and DNase I (for RNA) | PBS | NaHCO3 pH < 10.6 | 50 µL/min | Viral/bacterial, cancer | Plasma | PCR | 1 CFU/mL (10 cells/100 µL for cancerous cells) | [53] |
| MB | ChargeSwitch magnetic beads (commercial, + charge) | ChargeSwitch binding buffer, pH.5 | ChargeSwitch wash solution AP001 + Tween20 pH 7; silicone oil | Bacteria | Cultured | dPCR | [147] | |||
| Glass beads | TEOS or APTES or GO | Acetate pH5 | Tris-HCl pH7 | Tris-HCl pH 9 | 200 µL | Bacteria + virus | Toilet seat | qPCR | 0.007 CFU/cm2 | [148] |
| Membrane Polyvinylidene Fluoride (PVDF) | Amine-functionalized diatomaceous earth | Dimethyl pimelimidate dihydrochloride (DMP) | PBS | Elution buffer | 100 µL | Bacteria | Lab cultured | PCR qPCR | [149] | |
| SOI wafer | APTES | Dimethyl adipimidate (DMA), 25 mg/mL | PBS | NaHCO3 10 mM, pH 10.6 | 50 µL | Methylated DNA | Blood, urine | PCR | [150] | |
| Capillary | Poly-diallyl dimethylammonium chloride (PDDA) | None (thermal lysate) | no wash | No elution, in capillary amplification | - | Bacteria | Lab cultured | qPCR | 10 ng/µL | [151] |
| MB | Chitosan | Tris, 10 mM; Triton X-100, 0.1% v/v, pH 8.5 | Tris, 10 mM; Triton X-100, 0.1% v/v, pH 8.5 | On-bead amplification | Virus | Whole blood | PCR | 5 copes/µg of particles | ||
| Membrane | Tertiary amine | None | Direct amplification | Bacteria | LAMP |
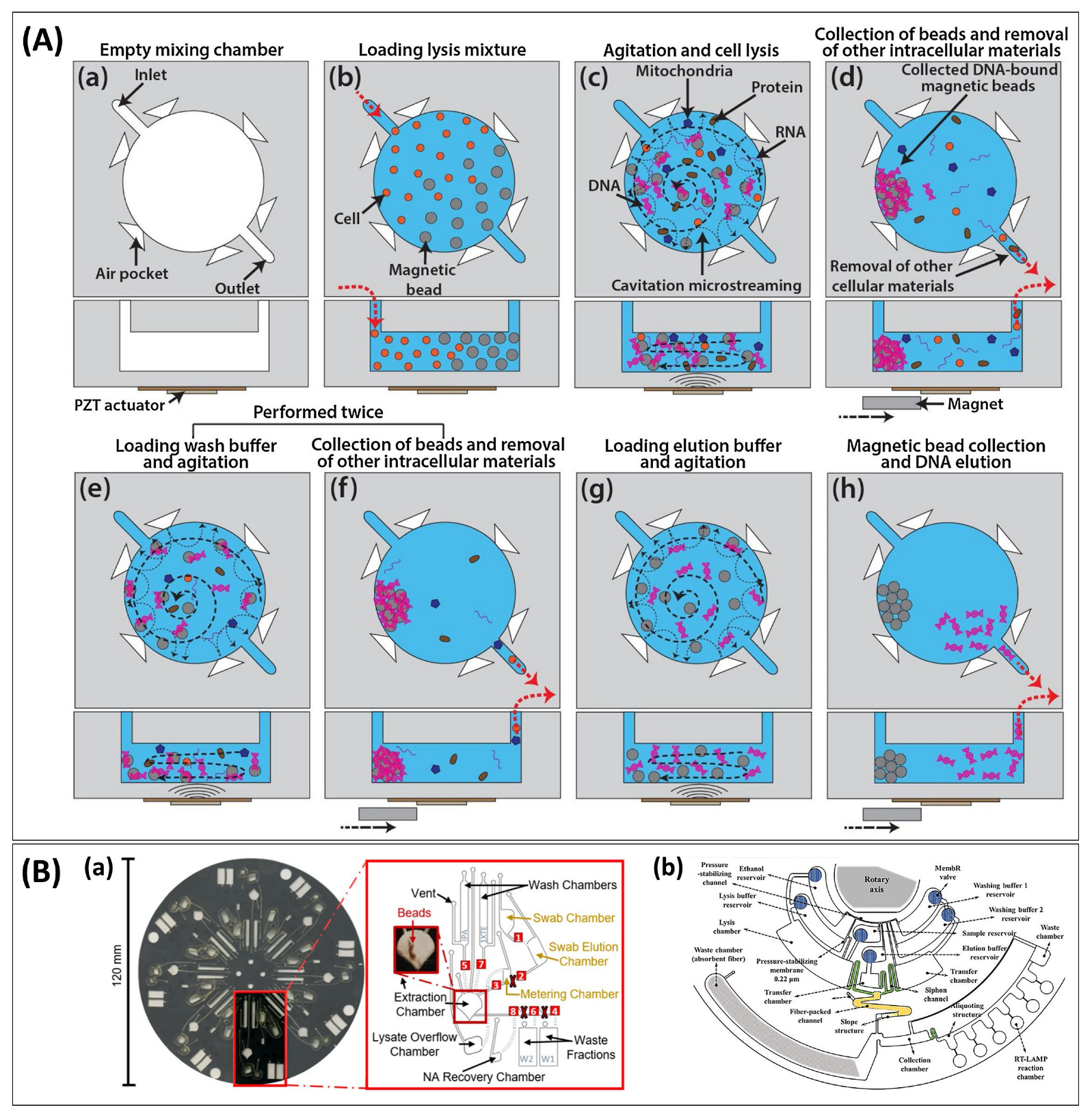
3.1.2. Fluidics
Liquid Handling

| Immiscible Phase | Bead Surface | Mechanism | Binding | Washing | Elution | Target | Sample Matrix | Amplification | LOD | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| FC-40, silicone oil, mineral oil | Silica | Chaotropic | Kit | Kit | Kit | HIV virus | Plasma | qPCR | - | [164] |
| FC40 | silica | Chaotropic lysis | 5 m GuHCl, pH 4.1 (citrate) with triton X OR Sarkoosyl OR Tween 20 | Carryover study | Carryover study | Carryover study | Carryover study | Carryover study | [159] | |
| Silicone oil | Silica | Chaotropic | Kit | Kit | HBV virus | Spiked blood | qPCR | n/r | [121] | |
| Silicone oil | Silica | Chaotropic lysis | 4 M GuSCN, 10 mM MES, 1% Triton X-100, with 1% ß-mercapto-ethanol | Oil immersed, Ethanol, 50% v/v, then water Water | None (on-bead amplification) | Virus | Nasopharyngeal swab | LAMP | 1–10 copies/μL | [51] |
| Mineral oil | Silica | Chaotropic | GuHCl | GuHCl, 5 M | Water | Bacteria | Liquid stool (clinical) | PCR | - | [41] |
| Mineral oil | Silica | Chaotropic | 5 M GuHCl, 0.005% TWEEN-20 | LAMP-CRISPR | [163] | |||||
| Mineral oil | Silica | Chaotropic | lysis buffer: Tris-HCl, lysozyme, protein kinase K, SDS, EDTA, RNase | GuHCl, 6 M | Magnesium acetate | Bacteria | Spiked milk | dRPA | 10 cells | [29] |
| Mineral oil | Silica | Chaotropic | 5 M GuHCl in 10 mM Tris-HCl 1 mM EDTA pH 8 | Ethanol, 70% v/v | Water | Animal identification | Dung | qPCR, LAMP | [162] | |
| Mineral oil | Silica | Chaotropic | 3 M GuHCl | Plasmid | Cultured | dRPA | 1.7e5 CFU/mL | [161] | ||
| Olive oil | Silica | Chaotropic | GuSCN, isopropanol and carrier RNA to facilitate nucleic acid precipitation | Aqueous low-salt solution | Virus | nasopharyngeal swab | RT-qPCR | 12.7 ± 4.6 ng/μL | [158] | |
| Liquid wax | Silica | Chaotropic | Alcohol | HIV | Whole blood | qPCR | 1200 copies/mL (RNA) | [155] | ||
| Organogel (12-HAS) | Silica | Chaotropic | GuSCN + Triton X-100 (pH 6.8) | GITC (pH 6.8) Ethanol NaCl2 (pH 7.6) | 10 mM Tris, 0.1 mM EDTA pH 8 (TE buffer) | HBV virus | Spiked blood | qPCR | 5 particles | [50] |
| Olive oil (silicone and mineral also evaluated) | ChargeSwitch | Ionic (cationic surface) | ChargeSwitch binding buffer, pH 5 | [165] | ||||||
| Silicone oil | ChargeSwitch | Ionic (cationic surface) | ChargeSwitch binding buffer, pH 5 | ChargeSwitch wash solution AP001 + Tween20 pH 7 | Direct amplification in LD Amplitaq Gold pH 8.3 | Bacteria | Cultured | dPCR | Only proof of concept provided | [147] |
| Olive oil | ChargeSwitch | Ionic (cationic surface) | ChargeSwitch binding buffer, pH 5 | ChargeSwitch washing buffer with SDS and TWEEN-20 | [157] | |||||
| FC-40 | ZrO | Zr−O−P coordination bond and hydrogen bond | Lysis buffer: 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA·1% Triton X-100, 0.1% Sodium Deoxycholate, 0.1% SDS, 140 mM NaCl | 10 mM PBS | Target DNA | Plasma | LAMP | [166] | ||
| Castor oil | Cellulose | Chaotropic | kit | Kit | HPV plasmids | Transport medium | 10 copies/100 μL | [167] | ||
| FC-40 | Silica | Chaotropic | Kit | Kit | HBV virus | Spiked plasma | dLAMP | 104 copies/ml | [120] | |
| Mineral oil | (dT)coated | Chaotropic | 6 M GuHCl | 0.005% Tween-20 | On-bead amplification | Virus | Artificial sputum | LAMP | 470 copies/mL | [156] |
| Liquid wax olive oil | Oligo-dT PMPs | Recognition | kit | Kit | Tris-HCl | Breast cancer cells | - | RT-qPCR | - | [160] |
| FC-40 | Biotinylated oligo | Recognition | 20 mM Tris pH 7.5, 100 mM KCl, 5 mM MgCl2, and 0.3% Nonidet P-40/Igepal, 17U RNAseOUT™, and 2.5 μL of 100X Halt™ protease inhibitor cocktail | Diethyl pyrocarbonate in PBS | Diethyl pyrocarbonate in PBS | microRNA | Culture medium | RT-qPCR, dPCR, array | [168] |
Process Integration
| Solid Phase | Surface | Primary Binding Principle | Elution = Amplification Mix | Volume | Target | Sample | Amplification | LOD | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|
| Elution in amplification mix | MB | Silica | Chaotrope | LAMP mix | 10 μL | Bacteria | Food | LAMP | 50 cells per test, or 10 cells/μL | [172] |
| Paper | Whatmann FTA | Ionic interaction | LAMP mix | 15.5 | Lambda DNA | Saliva | LAMP | 100 copies/mL | [178] | |
| Paper | Polyether sulfone (PES) | Chaotropic | LAMP mix | 12.5 | HPV 16 | Cervical specimens | LAMP | 1.27.105 Copies | [118] | |
| MB | ChargeSwitch magnetic beads (commercial, + charge) | Ionic interaction | Amplitaq Gold | Bacteria | Cultured | dPCR | [147] | |||
| Elution-free | Paper | Whatmann FTA | Ionic interaction | On-paper amplification | Bacteria | Whole blood | LAMP | 10 CFU/mL | [179] | |
| MB | Silica | Alkaline crowding | On-bead amplification | Bacteria | Artificial saliva, sweat, urine | qPCR | 0.15 CFU/50 µL | [140] | ||
| MB | Chitosan | Ionic interaction | On-bead amplification | Virus | Whole blood | PCR | 5 copes/µg of particles | [180] | ||
3.2. Extraction Methods without Stationary Phase
| Substrate/Method | Primary Extraction Principle | Extraction | Comment | Target | Sample Matrix | Amplification | LOD | Ref. |
|---|---|---|---|---|---|---|---|---|
| LLE | Preferential solubility | Various ILs and MILs | Thermal elution | White blood cells | White blood cells | qPCR | 500 pg DNA from 50 µL blood | [181] |
| LLE | Preferential solubility | [P6,6,6,14+] [Ni(hfacac)3−] MIL, [P6,6,6,14+] [Co(hfacac)3−]+ Tris | Thermal elution | Plant | Plant | qPCR | 311.8 ng of A. thaliana DNA per mg sample | [54] |
| Hydrogel | No binding, just physical exclusion of debris | In-gel amplification | Bacteria | Artificially infected fruits and vegetables | dLAMP | Single cell | [130] | |
| Polyacrylamide gel | Electrophoresis | Free flow electrophoresis, followed by lysis | DNA migrates through gel after lysis | Phage | Culture medium | qPCR | 1 PFU/mL or 0.02 copies/µL | [186] |
| ITP | Electromigration | LE 50 mM Tris HCl pH 8.2 TE 50 mM Tris HEPES pH 7.8 | Lambda DNA | Blood | PCR | 10 cells | [187] | |
| ITP | Electromigration | LE 100 mM of Tris-HCl TE 100 mM Tris and 100 mM of HEPES | Hydrogel as immiscible interface | Cell-free DNA | Plasma | PCR | [188] | |
| ITP | Electromigration | LE: 200 mM HCl with 400 mM Bistris as the LE solution TE: 10 mM Tricine with 20 mM Bistris | Paper to generate EOF counterflow, not for binding | Morpholino NA probes | - | Amplification-free detection | 5 pM after 10 min | [189] |
| ITP | Electromigration | LE 250 mM HCl and 375 mM Tris pH 7.8; TE 25 mM serine and 25 mM Tris pH 8.7 | Virus | Whole blood (spiked DNA) | RPA | 1000 copies/mL | [190] | |
| ITP | Electromigration | LE Tris HCl MgCl2, PEG1450, PVP, Triton X-100, and tetramethyl ammonium chloride pH 8.1 TE β-alanine, Tris, PVP, Triton X-100 pH 8.9–9.1 | Paper as carrier, also focuses RPA reagents | Synthetic viral DNA | Whole blood | RPA | 104 copies/mL | [191] |
4. Evaluation of Sample Preparation Method for NA-PONT Assays Based on the REASSURED Criteria
| Environment Lysis Reagents | Environment Extraction and Device | Equipment-Free | Deliverable to End-Users | User-Friendly | Affordable | Equipment-Free | Ref. | |
|---|---|---|---|---|---|---|---|---|
| 1. Centrifugal | Guanidium for lysis and binding | Plastic | Needs spinning | Relies on advanced instrumentation | All integrated/automated | Instrument expensive | Requires instrument | [45] |
| 2. Organogel as immiscible barrier | Guanidium for lysis, binding, and wash | Plastic | Only needs magnet | Yes, low reliance on equipment and skill | Can be operated after limited training | No instrumentation, simple device | Yes, other than a magnet | [50] |
| 3. Self-powered switch-controlled system | AL | Plastic device and manifold | Yes, powered by syringe (vacuum) | Yes, simple | Yes, easy to activate with gear and syringe | Simple device and tool | Syringe-powered | [145] |
| 4. Abridged solid-phase extraction with AP lysis (ASAP) | AL | Plastic tubes | Just needs magnet (and pipette in current form) | Needs training for manual pipetting | Few reagents, no wash but in its current form relies on manual handling | Affordable reagents, but still needs packaging in device/instrument | Not in device | [140] |
| 5. Paper-based ITP with on-paper RPA | Surfactant + enzyme | Electrolytes, paper | Membrane on paper | Paper devices easy to operate | Few reagents, all happens in electric field | Economic device, simple operation | Needs electrical power | [191] |
5. Summary and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 12-HAS | 12-Hydroxystearic Acid |
| ADE | Amine-Functionalized Diatomaceous Earth |
| AMPs | Antimicrobial Peptides |
| AL | Alkaline Lysis |
| AP | Alkaline polyethylene glycol-based lysis method |
| APTES | 3-Aminopropyltriethoxysilane |
| ASAP | Abridged solid-phase extraction with alkaline Poly(ethylene) glycol lysis |
| BSA | Bovine Serum Albumin |
| BAW | Bulk Acoustic Wave |
| CAI | Centrifugation-Assisted Immiscible Fluid Filtration |
| cdPCR | Chamber-Based Digital PCR |
| cfDNA | Cell-Free DNA |
| CFU | Colony-Forming Unit |
| CMC | Critical Micellar Concentration |
| CMV | Cucumber Mosaic Virus |
| CIFF | Centrifugation-Assisted Immiscible Fluid Filtration |
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| CTAB | Cetyltrimethylammonium Bromide |
| cSPE | Conventional Solid-Phase Extraction |
| DMP | Dimethyl Pimelimidate |
| DNA | Deoxyribonucleic Acid |
| dNTP | Deoxynucleotide Triphosphate |
| dMDA | Digital Multiple Displacement Amplification |
| dPCR | Digital Polymerase Chain Reaction |
| DI | Deionized |
| DMS | Dimethyl Suberimidate |
| DMA | Dimethyl Adipimidate |
| DTT | Dithiothereitol |
| E. coli | Escherichia coli |
| EDTA | Ethylenediaminetetraacetic Acid |
| EL | Electrical Lysis |
| FC-40 | Fluorinert FC-40 |
| Fe3O4 | Iron (II, III) oxide |
| fg | Femtogram |
| gDNA | Genomic DNA |
| GFP | Green Fluorescent Protein |
| GUSCN | Guanidinium Thiocyanate |
| Gu | Guanidine |
| GuHCl | Guanidine Hydrochloride |
| HAdV | Human Adenovirus |
| HBV | Hepatitis B Virus |
| HCl | Hydrochloric acid |
| HCV | Hepatitis C Virus |
| HCN | Hydrogen Cyanide |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| HGMS | High-Gradient Magnetic Separation |
| HI | Homobifuctional Imidoester |
| HPV | Human Papillomavirus |
| hrs | Hours |
| IA | Immunoassay |
| ICP | Ion Concentration Polarisation |
| ISM | Ion-Selective Membrane |
| IL | Ionic Liquid |
| ITP | Isotachophoresis |
| IPA | Isopropyl Alcohol |
| KCl | Potassium Chloride |
| IFAST | Immiscible Phase Filtration Assisted by Surface Tension |
| kb | kilo basepair |
| LAMP | Loop-mediated Isothermal Amplification |
| LLE | Liquid–Liquid Extraction |
| LMOGs | Low-Molecular-Mass Organogelators |
| LOD | Limit of Detection |
| MB | Magnetic Bead |
| MB-SPE | Magnetic Bead Solid-Phase Extraction |
| MES | 2-ethanesulfonic Acid |
| mL | Millilitre |
| MILs | Magnetic Ionic Liquids |
| Min | minutes |
| MSIs | Magainin analogues (synthetic antimicrobial peptides) |
| MTB | Mycobacterium tuberculosis |
| mtDNA | Mitochondrial DNA |
| NA | Nucleic Acid |
| NAAT | Nucleic Acid Amplification Test |
| NA-PONT | Nucleic Acid Pont-Of-Need Testing |
| NA-SPE | Nucleic Acid Solid-Phase Extraction |
| NaCl | Sodium Chloride |
| NaHCO3 | Sodium Hydrogencarbonate |
| NaOH | Sodium Hydroxide |
| -NH2 | Amine group |
| PCR | Polymerase Chain Reaction |
| PDA | Polydopamine |
| PDMS | Polydimethylsiloxane |
| PEG | Polyethylene Glycol |
| pg | Picogram |
| PMPs | Paramagnetic Particles |
| PONT | Point-of-Need Testing |
| Q. saponaria | Quillaja saponaria |
| qPCR | Quantitative Polymerase Chain Reaction |
| RNA | Ribonucleic Acid |
| RBCs | Red Blood Cells |
| RT | Reverse Transcription |
| RT-RPA | Reverse Transcription Recombinase Polymerase Amplification |
| RPA | Recombinase Polymerase Amplification |
| SARS-CoV-2 | Severe Acute Respiratory Syndrome Coronavirus 2 |
| SAW | Surface Acoustic Wave |
| SDS | Sodium Dodecyl Sulfate |
| sec | Seconds |
| SIO− | Silanol |
| SPE | Solid-Phase Extraction |
| SPRI | Solid-Phase Reversible Immobilization |
| SSNES | Self-powered Switch-Controlled NA Extraction System |
| STT | Sodium dodecyl sulphate, Tween 20, and Triton X-100 |
| TE | Tris-EDTA |
| TPW | Two-Phase Wash |
| µL | Microlitre |
References
- Ke, R.; Sanche, S.; Romero-Severson, E.; Hengartner, N. Fast spread of COVID-19 in Europe and the US suggests the necessity of early, strong and comprehensive interventions. medRxiv 2020. [Google Scholar] [CrossRef]
- Ali, Z.; Wang, J.; Mou, X.; Tang, Y.; Li, T.; Liang, W.; Shah, M.A.A.; Ahmad, R.; Li, Z.; He, N. Integration of Nucleic Acid Extraction Protocol with Automated Extractor for Multiplex Viral Detection. J. Nanosci. Nanotechnol. 2017, 17, 862–870. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, H.; Majidi, M.R.; Fakhraei, M.; Jahanban-Esfahlan, A.; Hejazi, M.; Oroojalian, F.; Baradaran, B.; Tohidast, M.; Guardia, M.d.l.; Mokhtarzadeh, A. Lateral flow assays (LFA) for detection of pathogenic bacteria: A small point-of-care platform for diagnosis of human infectious diseases. Talanta 2022, 243, 123330. [Google Scholar] [CrossRef] [PubMed]
- Ince, B.; Sezgintürk, M.K. Lateral flow assays for viruses diagnosis: Up-to-date technology and future prospects. TrAC Trends Anal. Chem. 2022, 157, 116725. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Lee, L.P. Toward Rapid and Accurate Molecular Diagnostics at Home. Adv. Mater. 2022, 35, e2206525. [Google Scholar] [CrossRef]
- Boonbanjong, P.; Treerattrakoon, K.; Waiwinya, W.; Pitikultham, P.; Japrung, D. Isothermal Amplification Technology for Disease Diagnosis. Biosensors 2022, 12, 677. [Google Scholar] [CrossRef]
- Peeling, R.W.; Holmes, K.K.; Mabey, D.; Ronald, A. Rapid tests for sexually transmitted infections (STIs): The way forward. Sex. Transm. Infect. 2006, 82, v1–v6. [Google Scholar] [CrossRef]
- Manz, A.; Graber, N.; Widmer, H.M. Miniaturized total chemical analysis systems: A novel concept for chemical sensing. Sens. Actuators B Chem. 1990, 1, 244–248. [Google Scholar] [CrossRef]
- Otoo, J.A.; Schlappi, T.S. REASSURED Multiplex Diagnostics: A Critical Review and Forecast. Biosensors 2022, 12, 124. [Google Scholar] [CrossRef]
- Lau, H.Y.; Botella, J.R. Advanced DNA-Based Point-of-Care Diagnostic Methods for Plant Diseases Detection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Song, Q.; Sun, X.; Dai, Z.; Gao, Y.; Gong, X.; Zhou, B.; Wu, J.; Wen, W. Point-of-care testing detection methods for COVID-19. Lab. A Chip. 2021, 21, 1634–1660. [Google Scholar] [CrossRef]
- Emaus, M.N.; Varona, M.; Eitzmann, D.R.; Hsieh, S.-A.; Zeger, V.R.; Anderson, J.L. Nucleic acid extraction: Fundamentals of sample preparation methodologies, current advancements, and future endeavors. TrAC Trends Anal. Chem. 2020, 130, 115985. [Google Scholar] [CrossRef]
- Seo, M.-J.; Yoo, J.-C. Fully Automated Lab-On-A-Disc Platform for Loop-Mediated Isothermal Amplification Using Micro-Carbon-Activated Cell Lysis. Sensors 2020, 20, 4746. [Google Scholar] [CrossRef] [PubMed]
- Dong, T.; Ma, X.; Sheng, N.; Qi, X.; Chu, Y.; Song, Q.; Zou, B.; Zhou, G. Point-of-care DNA testing by automatically and sequentially performing extraction, amplification and identification in a closed-type cassette. Sens. Actuators B Chem. 2021, 327, 128919. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhu, Y.; Yang, Y.; Li, J.; Hoffmann, M.R. Electrochemical cell lysis of gram-positive and gram-negative bacteria: DNA extraction from environmental water samples. Electrochim. Acta 2020, 338, 135864. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Luan, Z.; Liu, Y.; Yang, C.; Wang, Y.; Ma, C.; Shi, C. Ultrafast bacterial cell lysis using a handheld corona treater and loop-mediated isothermal amplification for rapid detection of foodborne pathogens. Food Control 2021, 128, 108178. [Google Scholar] [CrossRef]
- Xin, Y.; Xie, J.; Nan, B.; Tang, C.; Xiao, Y.; Wu, Q.; Lin, Y.; Zhang, X.; Shen, H. Freeze-Thaw Pretreatment Can Improve Efficiency of Bacterial DNA Extraction From Meconium. Front. Microbiol. 2021, 12, 753688. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Chen, Y.; Farooq, U.; Xuan, W.; Jin, H.; Dong, S.; Luo, J. Ultrafast chemical-free cell lysis by high speed stream collision induced by surface acoustic waves. Appl. Phys. Lett. 2017, 110, 143504. [Google Scholar] [CrossRef]
- Deraney, R.N.; Schneider, L.; Tripathi, A. Synergistic use of electroosmotic flow and magnetic forces for nucleic acid extraction. Analyst 2020, 145, 2412–2419. [Google Scholar] [CrossRef]
- Nan, L.; Jiang, Z.; Wei, X. Emerging microfluidic devices for cell lysis: A review. Lab. A Chip. 2014, 14, 1060–1073. [Google Scholar] [CrossRef]
- Shehadul Islam, M.; Aryasomayajula, A.; Selvaganapathy, P.R. A Review on Macroscale and Microscale Cell Lysis Methods. Micromachines 2017, 8, 83. [Google Scholar] [CrossRef]
- Jue, E.; Witters, D.; Ismagilov, R.F. Two-phase wash to solve the ubiquitous contaminant-carryover problem in commercial nucleic-acid extraction kits. Sci. Rep. 2020, 10, 1940. [Google Scholar] [CrossRef] [PubMed]
- Schrader, C.; Schielke, A.; Ellerbroek, L.; Johne, R. PCR inhibitors—Occurrence, properties and removal. J. Appl. Microbiol. 2012, 113, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Paul, R.; Ostermann, E.; Wei, Q. Advances in point-of-care nucleic acid extraction technologies for rapid diagnosis of human and plant diseases. Biosens. Bioelectron. 2020, 169, 112592. [Google Scholar] [CrossRef] [PubMed]
- Bolsover, S.R.; Shephard, E.A.; White, H.A.; Hyams, J.S. Cell Biology: A Short Course; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Viet-Phuong Le, A.; Huang, D.; Blick, T.; Thompson, E.W.; Dobrovic, A. An optimised direct lysis method for gene expression studies on low cell numbers. Sci. Rep. 2015, 5, 12859. [Google Scholar] [CrossRef] [PubMed]
- Sewlikar, S.; D’Souza, D.H. Antimicrobial Effects of Quillaja saponaria Extract Against Escherichia coli O157:H7 and the Emerging Non-O157 Shiga Toxin-Producing E. coli. J. Food Sci. 2017, 82, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Sakai, Y. Improvements in Extraction Methods of High-molecular-weight DNA from Soils by Modifying Cell Lysis Conditions and Reducing Adsorption of DNA onto Soil Particles. Microbes Environ. 2021, 36, ME21017. [Google Scholar] [CrossRef]
- Yin, J.; Zou, Z.; Hu, Z.; Zhang, S.; Zhang, F.; Wang, B.; Lv, S.; Mu, Y. A “sample-in-multiplex-digital-answer-out” chip for fast detection of pathogens. Lab. A Chip. 2020, 20, 979–986. [Google Scholar] [CrossRef]
- Bender, A.T.; Sullivan, B.P.; Zhang, J.Y.; Juergens, D.C.; Lillis, L.; Boyle, D.S.; Posner, J.D. HIV detection from human serum with paper-based isotachophoretic RNA extraction and reverse transcription recombinase polymerase amplification. Analyst 2021, 146, 2851–2861. [Google Scholar] [CrossRef]
- Jin, C.E.; Lee, T.Y.; Koo, B.; Sung, H.; Kim, S.-H.; Shin, Y. Rapid virus diagnostic system using bio-optical sensor and microfluidic sample processing. Sens. Actuators B Chem. 2018, 255, 2399–2406. [Google Scholar] [CrossRef]
- Gan, W.; Zhuang, B.; Zhang, P.; Han, J.; Li, C.-X.; Liu, P. A filter paper-based microdevice for low-cost, rapid, and automated DNA extraction and amplification from diverse sample types. Lab. A Chip. 2014, 14, 3719–3728. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xie, S.; Chen, X.; Peng, C.; Xu, X.; Wei, W.; Ma, T.; Cai, J.; Xu, J. A rapid and convenient method for on-site detection of MON863 maize through real-time fluorescence recombinase polymerase amplification. Food Chem. 2020, 324, 126821. [Google Scholar] [CrossRef] [PubMed]
- Seok, Y.; Lee, J.; Kim, M.-G. Paper-Based Airborne Bacteria Collection and DNA Extraction Kit. Biosensors 2021, 11, 375. [Google Scholar] [CrossRef] [PubMed]
- Chomczynski, P.; Rymaszewski, M. Alkaline polyethylene glycol-based method for direct PCR from bacteria, eukaryotic tissue samples, and whole blood. BioTechniques 2006, 40, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.J.; Baek, C.; Lee, M.-H.; Min, J. Integrated microsystems for the in situ genetic detection of dengue virus in whole blood using direct sample preparation and isothermal amplification. Analyst 2020, 145, 2405–2411. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Xu, H.; Song, W.; Yang, Z.; Yu, J.; Tian, Y.; Jiang, M.; Shen, D.; Dou, D. Rapid and simple detection of Phytophthora cactorum in strawberry using a coupled recombinase polymerase amplification–lateral flow strip assay. Phytopathol. Res. 2021, 3, 12. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zheng, Y.; Zhang, F.; Yu, J.; Dai, T.; Wang, R.; Tian, Y.; Xu, H.; Shen, D.; Dou, D. A Rapid, Equipment-Free Method for Detecting Phytophthora infestans in the Field Using a Lateral Flow Strip-Based Recombinase Polymerase Amplification Assay. Plant Dis. 2020, 104, 2774–2778. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, C.; Hua, K.; Liang, J.; Li, H.; Ma, T.; Zhu, J.; Cui, Y. Direct genotyping from whole blood using alkaline polyethylene glycol. Anal. Biochem. 2019, 582, 113351. [Google Scholar] [CrossRef]
- Sillo, F.; Giordano, L.; Gonthier, P. Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a Loop-mediated isothermal AMPlification (LAMP) assay. For. Pathol. 2018, 48, e12396. [Google Scholar] [CrossRef]
- Mosley, O.; Melling, L.; Tarn, M.D.; Kemp, C.; Esfahani, M.M.N.; Pamme, N.; Shaw, K.J. Sample introduction interface for on-chip nucleic acid-based analysis of Helicobacter pylori from stool samples. Lab. A Chip. 2016, 16, 2108–2115. [Google Scholar] [CrossRef]
- Batule, B.S.; Seok, Y.; Kim, M.-G. An innovative paper-based device for DNA extraction from processed meat products. Food Chem. 2020, 321, 126708. [Google Scholar] [CrossRef] [PubMed]
- Bhati, A.; Varghese, A.; Rajan, G.; Sridhar, V.; Mohan, Y.; Pradeep, S.; Babu, S.; Kaikkolante, N.; Sarma, M.; Arun, S.; et al. An effective method for saliva stabilization and magnetic nanoparticles based DNA extraction for genomic applications. Anal. Biochem. 2021, 624, 114182. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Kim, S.; Na, J.-H.; Han, K.; Lee, T.Y. A single-tube sample preparation method based on a dual-electrostatic interaction strategy for molecular diagnosis of gram-negative bacteria. Microchim. Acta 2020, 187, 558. [Google Scholar] [CrossRef] [PubMed]
- Dignan, L.M.; Woolf, M.S.; Tomley, C.J.; Nauman, A.Q.; Landers, J.P. Multiplexed Centrifugal Microfluidic System for Dynamic Solid-Phase Purification of Polynucleic Acids Direct from Buccal Swabs. Anal. Chem. 2021, 93, 7300–7309. [Google Scholar] [CrossRef] [PubMed]
- Krõlov, K.; Uusna, J.; Grellier, T.; Andresen, L.; Jevtuševskaja, J.; Tulp, I.; Langel, Ü. Implementation of antimicrobial peptides for sample preparation prior to nucleic acid amplification in point-of-care settings. Expert Rev. Mol. Diagn. 2017, 17, 1117–1125. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Yang, H.; Chen, Z.; Cao, X.; Li, Z.; Stavrakis, S.; Choo, J.; deMello, A.J.; Howes, P.D.; He, N. A sample-in-digital-answer-out system for rapid detection and quantitation of infectious pathogens in bodily fluids. Anal. Bioanal. Chem. 2018, 410, 7019–7030. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Li, X.; Lin, S.; Zhao, Y.; Lin, P.; Wang, B.; Tang, R.; Guo, J.; Zu, Y.; Zhou, Y.; et al. Low-Cost and Rapid Method of DNA Extraction from Scaled Fish Blood and Skin Mucus. Viruses 2022, 14, 840. [Google Scholar] [CrossRef]
- Ohashi, T.; Kuyama, H. Magnetic particle transport through organogel—An application to DNA extraction. Anal. Biochem. 2020, 611, 113932. [Google Scholar] [CrossRef]
- Juang, D.S.; Juang, T.D.; Dudley, D.M.; Newman, C.M.; Accola, M.A.; Rehrauer, W.M.; Friedrich, T.C.; O’Connor, D.H.; Beebe, D.J. Oil immersed lossless total analysis system for integrated RNA extraction and detection of SARS-CoV-2. Nat. Commun. 2021, 12, 4317. [Google Scholar] [CrossRef]
- Pearlman, S.I.; Leelawong, M.; Richardson, K.A.; Adams, N.M.; Russ, P.K.; Pask, M.E.; Wolfe, A.E.; Wessely, C.; Haselton, F.R. Low-Resource Nucleic Acid Extraction Method Enabled by High-Gradient Magnetic Separation. ACS Appl. Mater. Interfaces 2020, 12, 12457–12467. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.E.; Lee, T.Y.; Koo, B.; Choi, K.-C.; Chang, S.; Park, S.Y.; Kim, J.Y.; Kim, S.-H.; Shin, Y. Use of Dimethyl Pimelimidate with Microfluidic System for Nucleic Acids Extraction without Electricity. Anal. Chem. 2017, 89, 7502–7510. [Google Scholar] [CrossRef] [PubMed]
- Emaus, M.N.; Cagliero, C.; Gostel, M.R.; Johnson, G.; Anderson, J.L. Simple and efficient isolation of plant genomic DNA using magnetic ionic liquids. Plant Methods 2022, 18, 37. [Google Scholar] [CrossRef] [PubMed]
- Helenius, A.; Simons, K. Solubilization of membranes by detergents. Biochim. Biophys. Acta (BBA) Rev. Biomembr. 1975, 415, 29–79. [Google Scholar] [CrossRef]
- Lichtenberg, D.; Ahyayauch, H.; Goñi, F.M. The mechanism of detergent solubilization of lipid bilayers. Biophys. J. 2013, 105, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Ahyayauch, H.; Bennouna, M.; Alonso, A.; Goñi, F.M. Detergent Effects on Membranes at Subsolubilizing Concentrations: Transmembrane Lipid Motion, Bilayer Permeabilization, and Vesicle Lysis/Reassembly Are Independent Phenomena. Langmuir 2010, 26, 7307–7313. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Mak, T.W.; Krylov, S.N. Cell lysis inside the capillary facilitated by transverse diffusion of laminar flow profiles (TDLFP). Anal. Bioanal. Chem. 2007, 387, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Syn, C.K.; Teo, W.L.; Swarup, S. Three-detergent method for the extraction of RNA from several bacteria. BioTechniques 1999, 27, 1140–1141. [Google Scholar] [CrossRef]
- Berlowska, J.; Dudkiewicz, M.; Kregiel, D.; Czyzowska, A.; Witonska, I. Cell lysis induced by membrane-damaging detergent saponins from Quillaja saponaria. Enzym. Microb. Technol. 2015, 75–76, 44–48. [Google Scholar] [CrossRef]
- Partearroyo, M.A.; Ostolaza, H.; Goñi, F.M.; Barberá-Guillem, E. Surfactant-induced cell toxicity and cell lysis: A study using B16 melanoma cells. Biochem. Pharmacol. 1990, 40, 1323–1328. [Google Scholar] [CrossRef]
- Jones, S.A.; Laskaris, G.; Vincent-Bonnieu, S.; Farajzadeh, R.; Rossen, W.R. Effect of surfactant concentration on foam: From coreflood experiments to implicit-texture foam-model parameters. J. Ind. Eng. Chem. 2016, 37, 268–276. [Google Scholar] [CrossRef]
- Pereiro, I.; Fomitcheva Khartchenko, A.; Petrini, L.; Kaigala, G.V. Nip the bubble in the bud: A guide to avoid gas nucleation in microfluidics. Lab. A Chip. 2019, 19, 2296–2314. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, R. Simple and rapid preparation of samples for PCR. In PCR Technology; Springer: Berlin/Heidelberg, Germany, 1989; pp. 31–38. [Google Scholar]
- Villarreal, J.V.; Jungfer, C.; Obst, U.; Schwartz, T. DNase I and Proteinase K eliminate DNA from injured or dead bacteria but not from living bacteria in microbial reference systems and natural drinking water biofilms for subsequent molecular biology analyses. J. Microbiol. Methods 2013, 94, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Genoud, V.; Stortz, M.; Waisman, A.; Berardino, B.G.; Verneri, P.; Dansey, V.; Salvatori, M.; Remes Lenicov, F.; Levi, V. Extraction-free protocol combining proteinase K and heat inactivation for detection of SARS-CoV-2 by RT-qPCR. PLoS ONE 2021, 16, e0247792. [Google Scholar] [CrossRef] [PubMed]
- Bera, A.; Herbert, S.; Jakob, A.; Vollmer, W.; Götz, F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol. Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Norioka, S.; Sakiyama, F. Purification, staphylolytic activity, and cleavage sites of α-lytic protease from Achromobacter lyticus. J. Biochem. 1997, 122, 772–778. [Google Scholar] [CrossRef] [PubMed]
- Heiniger, E.K.; Buser, J.R.; Mireles, L.; Zhang, X.; Ladd, P.D.; Lutz, B.R.; Yager, P. Comparison of point-of-care-compatible lysis methods for bacteria and viruses. J. Microbiol. Methods 2016, 128, 80–87. [Google Scholar] [CrossRef]
- Shah, K.G.; Roller, M.; Kumar, S.; Bennett, S.; Heiniger, E.; Looney, K.; Buser, J.; Bishop, J.D.; Yager, P. Disposable platform for bacterial lysis and nucleic acid amplification based on a single USB-powered printed circuit board. PLoS ONE 2023, 18, e0284424. [Google Scholar] [CrossRef]
- Buser, J.R.; Zhang, X.; Byrnes, S.A.; Ladd, P.D.; Heiniger, E.K.; Wheeler, M.D.; Bishop, J.D.; Englund, J.A.; Lutz, B.; Weigl, B.H.; et al. A disposable chemical heater and dry enzyme preparation for lysis and extraction of DNA and RNA from microorganisms. Anal. Methods 2016, 8, 2880–2886. [Google Scholar] [CrossRef]
- Chondrogiannis, G.; Réu, P.; Hamedi, M.M. Paper-Based Bacterial Lysis Enables Sample-to-Answer Home-based DNA Testing. Adv. Mater. Technol. 2023, 8, 2201004. [Google Scholar] [CrossRef]
- Birnboim, H.C.; Doly, J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979, 7, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Preparation of Plasmid DNA by Alkaline Lysis with Sodium Dodecyl Sulfate: Minipreps. Cold Spring Harb. Protoc. 2016, 2016, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Panda, S.; Gekara, N.O. Chapter Eighteen—Comet and micronucleus assays for analyzing DNA damage and genome integrity. Methods Enzymol. 2019, 625, 299–307. [Google Scholar] [PubMed]
- Ickenstein, L.M.; Sandström, M.C.; Mayer, L.D.; Edwards, K. Effects of phospholipid hydrolysis on the aggregate structure in DPPC/DSPE-PEG2000 liposome preparations after gel to liquid crystalline phase transition. Biochim. Biophys. Acta Biomembr. 2006, 1758, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Girish, P.S.; Barbuddhe, S.B.; Kumari, A.; Rawool, D.B.; Karabasanavar, N.S.; Muthukumar, M.; Vaithiyanathan, S. Rapid detection of pork using alkaline lysis- Loop Mediated Isothermal Amplification (AL-LAMP) technique. Food Control 2020, 110, 107015. [Google Scholar] [CrossRef]
- Zhao, G.; Wang, J.; Yao, C.; Xie, P.; Li, X.; Xu, Z.; Xian, Y.; Lei, H.; Shen, X. Alkaline lysis-recombinase polymerase amplification combined with CRISPR/Cas12a assay for the ultrafast visual identification of pork in meat products. Food Chem. 2022, 383, 132318. [Google Scholar] [CrossRef] [PubMed]
- Gautam, A. Isolation of Plasmid DNA by Alkaline Lysis. In DNA and RNA Isolation Techniques for Non-Experts; Gautam, A., Ed.; Springer International Publishing: Cham, Swizerland, 2022; pp. 55–61. [Google Scholar] [CrossRef]
- Salvi, G.; De Los Rios, P.; Vendruscolo, M. Effective interactions between chaotropic agents and proteins. Proteins Struct. Funct. Bioinform. 2005, 61, 492–499. [Google Scholar] [CrossRef]
- Melzak, K.A.; Sherwood, C.S.; Turner, R.F.B.; Haynes, C.A. Driving Forces for DNA Adsorption to Silica in Perchlorate Solutions. J. Colloid Interface Sci. 1996, 181, 635–644. [Google Scholar] [CrossRef]
- Yang, W. Nucleases: Diversity of structure, function and mechanism. Q. Rev. Biophys. 2011, 44, 1–93. [Google Scholar] [CrossRef]
- Lee, S.; Kim, S.; Kim, S. A novel paper-based lysis strip for SARS-CoV-2 RNA detection at low resource settings. Anal. Biochem. 2023, 664, 115037. [Google Scholar] [CrossRef]
- Zhang, Y.; Ren, G.; Buss, J.; Barry, A.J.; Patton, G.C.; Tanner, N.A. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. BioTechniques 2020, 69, 178–185. [Google Scholar] [CrossRef]
- Cho, H.-S.; Choi, M.; Lee, Y.; Jeon, H.; Ahn, B.; Soundrarajan, N.; Hong, K.; Kim, J.-H.; Park, C. High-Quality Nucleic Acid Isolation from Hard-to-Lyse Bacterial Strains Using PMAP-36, a Broad-Spectrum Antimicrobial Peptide. Int. J. Mol. Sci. 2021, 22, 4149. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Chen, B.; Koo, Y.-M.; MacFarlane, D.R. Introduction: Ionic Liquids. Chem. Rev. 2017, 117, 6633–6635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Liu, L.; Liu, X.; Zhang, X.; Zhang, S. Insight into the Relationship between Viscosity and Hydrogen Bond of a Series of Imidazolium Ionic Liquids: A Molecular Dynamics and Density Functional Theory Study. Ind. Eng. Chem. Res. 2019, 58, 18848–18854. [Google Scholar] [CrossRef]
- George, A.; Brandt, A.; Tran, K.; Zahari, S.M.S.N.S.; Klein-Marcuschamer, D.; Sun, N.; Sathitsuksanoh, N.; Shi, J.; Stavila, V.; Parthasarathi, R.; et al. Design of low-cost ionic liquids for lignocellulosic biomass pretreatment. Green Chem. 2015, 17, 1728–1734. [Google Scholar] [CrossRef]
- Vandeventer, P.E.; Weigel, K.M.; Salazar, J.; Erwin, B.; Irvine, B.; Doebler, R.; Nadim, A.; Cangelosi, G.A.; Niemz, A. Mechanical disruption of lysis-resistant bacterial cells by use of a miniature, low-power, disposable device. J. Clin. Microbiol. 2011, 49, 2533–2539. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.-W.; Sakamuri, R.; Kumar, P.; Ferguson, T.M.; Doebler, R.W.; Herrington, K.D.; Talbot, R.P.; Weigel, K.M.; Nguyen, F.K.; Cangelosi, G.A.; et al. Integrated nucleic acid testing system to enable TB diagnosis in peripheral settings. Lab. A Chip. 2020, 20, 4071–4081. [Google Scholar] [CrossRef] [PubMed]
- Shin, D.J.; Athamanolap, P.; Chen, L.; Hardick, J.; Lewis, M.; Hsieh, Y.H.; Rothman, R.E.; Gaydos, C.A.; Wang, T.H. Mobile nucleic acid amplification testing (mobiNAAT) for Chlamydia trachomatis screening in hospital emergency department settings. Sci. Rep. 2017, 7, 4495. [Google Scholar] [CrossRef]
- Grigorov, E.; Kirov, B.; Marinov, M.B.; Galabov, V. Review of Microfluidic Methods for Cellular Lysis. Micromachines 2021, 12, 498. [Google Scholar] [CrossRef]
- Danaeifar, M. New horizons in developing cell lysis methods: A review. Biotechnol. Bioeng. 2022, 119, 3007–3021. [Google Scholar] [CrossRef]
- Marentis, T.C.; Kusler, B.; Yaralioglu, G.G.; Liu, S.; Haeggström, E.O.; Khuri-Yakub, B.T. Microfluidic sonicator for real-time disruption of eukaryotic cells and bacterial spores for DNA analysis. Ultrasound Med. Biol. 2005, 31, 1265–1277. [Google Scholar] [CrossRef]
- Branch, D.W.; Vreeland, E.C.; McClain, J.L.; Murton, J.K.; James, C.D.; Achyuthan, K.E. Rapid Nucleic Acid Extraction and Purification Using a Miniature Ultrasonic Technique. Micromachines 2017, 8, 228. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Mutafopulos, K.; Heyman, J.A.; Spink, P.; Shen, L.; Wang, C.; Franke, T.; Weitz, D.A. Rapid additive-free bacteria lysis using traveling surface acoustic waves in microfluidic channels. Lab. A Chip. 2019, 19, 4064–4070. [Google Scholar] [CrossRef] [PubMed]
- Kaba, A.M.; Jeon, H.; Park, A.; Yi, K.; Baek, S.; Park, A.; Kim, D. Cavitation-microstreaming-based lysis and DNA extraction using a laser-machined polycarbonate microfluidic chip. Sens. Actuators B Chem. 2021, 346, 130511. [Google Scholar] [CrossRef]
- Zevnik, J.; Dular, M. Cavitation bubble interaction with compliant structures on a microscale: A contribution to the understanding of bacterial cell lysis by cavitation treatment. Ultrason. Sonochem. 2022, 87, 106053. [Google Scholar] [CrossRef] [PubMed]
- Zupanc, M.; Zevnik, J.; Filipić, A.; Gutierrez-Aguirre, I.; Ješelnik, M.; Košir, T.; Ortar, J.; Dular, M.; Petkovšek, M. Inactivation of the enveloped virus phi6 with hydrodynamic cavitation. Ultrason. Sonochem. 2023, 95, 106400. [Google Scholar] [CrossRef] [PubMed]
- Nittala, P.V.K.; Hohreiter, A.; Rosas Linhard, E.; Dohn, R.; Mishra, S.; Konda, A.; Divan, R.; Guha, S.; Basu, A. Integration of silicon chip microstructures for in-line microbial cell lysis in soft microfluidics. Lab. A Chip. 2023, 23, 2327–2340. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, S. An electro-conductive plane heating element for rapid thermal lysis of bacterial cells. J. Microbiol. Methods 2018, 153, 99–103. [Google Scholar] [CrossRef]
- Shetty, P.; Ghosh, D.; Paul, D. Thermal lysis and isothermal amplification of Mycobacterium tuberculosis H37Rv in one tube. J. Microbiol. Methods 2017, 143, 1–5. [Google Scholar] [CrossRef]
- Kim, M.; Wu, L.; Kim, B.; Hung, D.T.; Han, J. Continuous and High-Throughput Electromechanical Lysis of Bacterial Pathogens Using Ion Concentration Polarization. Anal. Chem. 2018, 90, 872–880. [Google Scholar] [CrossRef]
- Zhao, J.; Li, N.; Zhou, X.; Yu, Z.; Lan, M.; Chen, S.; Miao, J.; Li, Y.; Li, G.; Yang, F. Electrolysis of Bacteria Based on Microfluidic Technology. Micromachines 2023, 14, 144. [Google Scholar] [CrossRef]
- Ma, S.; Bryson, B.D.; Sun, C.; Fortune, S.M.; Lu, C. RNA Extraction from a Mycobacterium under Ultrahigh Electric Field Intensity in a Microfluidic Device. Anal. Chem. 2016, 88, 5053–5057. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.S.; Shahid, A.; Kuryllo, K.; Li, Y.; Deen, M.J.; Selvaganapathy, P.R. Electrophoretic Concentration and Electrical Lysis of Bacteria in a Microfluidic Device Using a Nanoporous Membrane. Micromachines 2017, 8, 45. [Google Scholar] [CrossRef]
- Hong, S.; Park, K.S.; Weissleder, R.; Castro, C.M.; Lee, H. Facile silicification of plastic surface for bioassays. Chem. Commun. 2017, 53, 2134–2137. [Google Scholar] [CrossRef] [PubMed]
- Boom, R.; Sol, C.J.; Salimans, M.M.; Jansen, C.L.; Wertheim-van Dillen, P.M.; van der Noordaa, J. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 1990, 28, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Wang, H.; Tong, R.; Du, Q.; Zhong, W. Preparation and morphology of SiO2/PMMA nanohybrids by microemulsion polymerization. Colloid Polym. Sci. 2006, 284, 755–762. [Google Scholar] [CrossRef]
- Juang, D.S.; Berry, S.M.; Li, C.; Lang, J.M.; Beebe, D.J. Centrifugation-Assisted Immiscible Fluid Filtration for Dual-Bioanalyte Extraction. Anal. Chem. 2019, 91, 11848–11855. [Google Scholar] [CrossRef]
- Ngo, D.; Liu, H.; Chen, Z.; Kaya, H.; Zimudzi, T.J.; Gin, S.; Mahadevan, T.; Du, J.; Kim, S.H. Hydrogen bonding interactions of H2O and SiOH on a boroaluminosilicate glass corroded in aqueous solution. NPJ Mater. Degrad. 2020, 4, 1. [Google Scholar] [CrossRef]
- Wang, R.; Wu, J.; He, X.; Zhou, P.; Shen, Z. A Sample-In-Answer-Out Microfluidic System for the Molecular Diagnostics of 24 HPV Genotypes Using Palm-Sized Cartridge. Micromachines 2021, 12, 263. [Google Scholar] [CrossRef]
- Raymond, C.K.; Raymond, F.C.; Hill, K. UltraPrep is a scalable, cost-effective, bead-based method for purifying cell-free DNA. PLoS ONE 2020, 15, e0231854. [Google Scholar] [CrossRef]
- Du, K.; Cai, H.; Park, M.; Wall, T.A.; Stott, M.A.; Alfson, K.J.; Griffiths, A.; Carrion, R.; Patterson, J.L.; Hawkins, A.R.; et al. Multiplexed efficient on-chip sample preparation and sensitive amplification-free detection of Ebola virus. Biosens. Bioelectron. 2017, 91, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhang, X.; Chen, L.; Yao, Y.; Ke, S.; Zhao, W.; Yang, Z.; Sui, G. A sample-to-answer labdisc platform integrated novel membrane-resistance valves for detection of highly pathogenic avian influenza viruses. Sens. Actuators B Chem. 2018, 270, 371–381. [Google Scholar] [CrossRef]
- Chen, X.; Cui, D.; Liu, C.; Li, H.; Chen, J. Continuous flow microfluidic device for cell separation, cell lysis and DNA purification. Anal. Chim. Acta 2007, 584, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Gulliksen, A.; Keegan, H.; Martin, C.; O’Leary, J.; Solli, L.A.; Falang, I.M.; Grønn, P.; Karlgård, A.; Mielnik, M.M.; Johansen, I.-R.; et al. Towards a “Sample-In, Answer-Out” Point-of-Care Platform for Nucleic Acid Extraction and Amplification: Using an HPV E6/E7 mRNA Model System. J. Oncol. 2012, 2012, 905024. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.M.; Wong, W.S.; Liu, L.; Dewar, R.; Klapperich, C.M. A fully integrated paperfluidic molecular diagnostic chip for the extraction, amplification, and detection of nucleic acids from clinical samples. Lab. A Chip. 2016, 16, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Linnes, J.C.; Fan, A.; Rodriguez, N.M.; Lemieux, B.; Kong, H.; Klapperich, C.M. Paper-based molecular diagnostic for Chlamydia trachomatis. RSC Adv. 2014, 4, 42245–42251. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Li, J.; Zhang, Z.; Li, M.; Zhao, S.; Li, Z.; Peng, N. Smartphone-Based Droplet Digital LAMP Device with Rapid Nucleic Acid Isolation for Highly Sensitive Point-of-Care Detection. Anal. Chem. 2020, 92, 2258–2265. [Google Scholar] [CrossRef]
- Hu, F.; Li, J.; Peng, N.; Li, Z.; Zhang, Z.; Zhao, S.; Duan, M.; Tian, H.; Li, L.; Zhang, P. Rapid isolation of cfDNA from large-volume whole blood on a centrifugal microfluidic chip based on immiscible phase filtration. Analyst 2019, 144, 4162–4174. [Google Scholar] [CrossRef]
- Hallsworth, J.E. Ethanol-induced water stress in yeast. J. Ferment. Bioeng. 1998, 85, 125–137. [Google Scholar] [CrossRef]
- Page, R.; Scourfield, E.; Ficarelli, M.; McKellar, S.W.; Lee, K.L.; Maguire, T.J.A.; Bouton, C.; Lista, M.J.; Neil, S.J.D.; Malim, M.H.; et al. Homebrew: An economical and sensitive glassmilk-based nucleic-acid extraction method for SARS-CoV-2 diagnostics. Cell Rep. Methods 2022, 2, 100186. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Lin, L.; Wu, T.; Zhao, Z.; Ying, B.; Chang, L. A finger-driven disposable micro-platform based on isothermal amplification for the application of multiplexed and point-of-care diagnosis of tuberculosis. Biosens. Bioelectron. 2022, 195, 113663. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef] [PubMed]
- Mohsen-Nia, M.; Amiri, H.; Jazi, B. Dielectric Constants of Water, Methanol, Ethanol, Butanol and Acetone: Measurement and Computational Study. J. Solut. Chem. 2010, 39, 701–708. [Google Scholar] [CrossRef]
- Park, J.-G.; Lee, S.-H.; Ryu, J.-S.; Hong, Y.-K.; Kim, T.-G.; Busnaina, A.A. Interfacial and Electrokinetic Characterization of IPA Solutions Related to Semiconductor Wafer Drying and Cleaning. J. Electrochem. Soc. 2006, 153, G811. [Google Scholar] [CrossRef]
- Lee, W.D.; Gawri, R.; Shiba, T.; Ji, A.-R.; Stanford, W.L.; Kandel, R.A. Simple Silica Column–Based Method to Quantify Inorganic Polyphosphates in Cartilage and Other Tissues. Cartilage 2017, 9, 417–427. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Kharb, A.; Vazirani, A.; Chauhan, R.S.; Pramanik, G.; Sengupta, M.; Ghosh, S. Nucleic acid extraction from complex biofluid using toothpick-actuated over-the-counter medical-grade cotton. Bioorganic Med. Chem. 2022, 73, 117009. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Fang, M.; Yi, C.; Jiang, Y.; Zhang, C.; Pan, X.; Luo, Z. Functional hydrogel for fast, precise and inhibition-free point-of-care bacteria analysis in crude food samples. Biomaterials 2022, 280, 121278. [Google Scholar] [CrossRef]
- Zhao, F.; Lee, E.Y.; Noh, G.S.; Shin, J.; Liu, H.; Qiao, Z.; Shin, Y. A robust, hand-powered, instrument-free sample preparation system for point-of-care pathogen detection. Sci. Rep. 2019, 9, 16374. [Google Scholar] [CrossRef]
- Akabayov, B.; Akabayov, S.R.; Lee, S.-J.; Wagner, G.; Richardson, C.C. Impact of macromolecular crowding on DNA replication. Nat. Commun. 2013, 4, 1615. [Google Scholar] [CrossRef]
- Phillip, Y.; Sherman, E.; Haran, G.; Schreiber, G. Common crowding agents have only a small effect on protein-protein interactions. Biophys. J. 2009, 97, 875–885. [Google Scholar] [CrossRef]
- Miyoshi, D.; Sugimoto, N. Molecular crowding effects on structure and stability of DNA. Biochimie 2008, 90, 1040–1051. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, T.L.; O’Connor-Morin, T.; Roy, A.; Santillan, C. DNA purification and isolation using a solid-phase. Nucleic Acids Res. 1994, 22, 4543–4544. [Google Scholar] [CrossRef] [PubMed]
- Rohland, N.; Reich, D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res. 2012, 22, 939–946. [Google Scholar] [CrossRef] [PubMed]
- Maghini, D.G.; Moss, E.L.; Vance, S.E.; Bhatt, A.S. Improved high-molecular-weight DNA extraction, nanopore sequencing and metagenomic assembly from the human gut microbiome. Nat. Protoc. 2021, 16, 458–471. [Google Scholar] [CrossRef] [PubMed]
- Stortchevoi, A.; Kamelamela, N.; Levine, S.S. SPRI Beads-based Size Selection in the Range of 2–10 kb. J. Biomol. Tech. 2020, 31, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Vo, A.T.E.; Jedlicka, J.A. Protocols for metagenomic DNA extraction and Illumina amplicon library preparation for faecal and swab samples. Mol. Ecol. Resour. 2014, 14, 1183–1197. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Nai, Y.H.; Doeven, E.H.; Balakrishnan, H.K.; Yuan, D.; Guijt, R.M. Abridged solid-phase extraction with alkaline Poly(ethylene) glycol lysis (ASAP) for direct DNA amplification. Talanta 2023, 266, 125006. [Google Scholar] [CrossRef] [PubMed]
- Kastania, A.S.; Petrou, P.S.; Loukas, C.-M.; Gogolides, E. Poly-L-histidine coated microfluidic devices for bacterial DNA purification without chaotropic solutions. Biomed. Microdevices 2020, 22, 44. [Google Scholar] [CrossRef]
- Zhang, J.; Su, X.; Xu, J.; Wang, J.; Zeng, J.; Li, C.; Chen, W.; Li, T.; Min, X.; Zhang, D.; et al. A point of care platform based on microfluidic chip for nucleic acid extraction in less than 1 minute. Biomicrofluidics 2019, 13, 034102. [Google Scholar] [CrossRef]
- Zandieh, M.; Liu, J. Spherical Nucleic Acid Mediated Functionalization of Polydopamine-Coated Nanoparticles for Selective DNA Extraction and Detection. Bioconjugate Chem. 2021, 32, 801–809. [Google Scholar] [CrossRef]
- Seong, H.; Park, J.; Bae, M.; Shin, S. Rapid and Efficient Extraction of Cell-Free DNA Using Homobifunctional Crosslinkers. Biomedicines 2022, 10, 1883. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Yoon, Y.-J.; Shin, Y.; Park, M.K. Self-powered switch-controlled nucleic acid extraction system. Lab. A Chip. 2016, 16, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hermanson, G.T. Chapter 5—Homobifunctional Crosslinkers. In Bioconjugate Techniques, 3rd ed.; Hermanson, G.T., Ed.; Academic Press: Boston, MA, USA, 2013; pp. 275–298. [Google Scholar] [CrossRef]
- Gaddes, D.E.; Lee, P.-W.; Trick, A.Y.; Athamanolap, P.; O’Keefe, C.M.; Puleo, C.; Hsieh, K.; Wang, T.-H. Facile Coupling of Droplet Magnetofluidic-Enabled Automated Sample Preparation for Digital Nucleic Acid Amplification Testing and Analysis. Anal. Chem. 2020, 92, 13254–13261. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-N.; Yoo, H.J.; Nguyen, K.H.; Baek, C.; Min, J. Semi-automatic instrumentation for nucleic acid extraction and purification to quantify pathogens on surfaces. Analyst 2019, 144, 6586–6594. [Google Scholar] [CrossRef] [PubMed]
- Noh, G.S.; Liu, H.; Kim, M.G.; Qiao, Z.; Jang, Y.O.; Shin, Y. Multi-Sample Preparation Assay for Isolation of Nucleic Acids Using Bio-Silica with Syringe Filters. Micromachines 2020, 11, 823. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.; Perera, A.P.; Wong, C.C.; Park, M.K. Solid phase nucleic acid extraction technique in a microfluidic chip using a novel non-chaotropic agent: Dimethyl adipimidate. Lab. A Chip. 2014, 14, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zhou, X.; Xing, D. Lab-on-capillary: A rapid, simple and quantitative genetic analysis platform integrating nucleic acid extraction, amplification and detection. Lab. A Chip. 2017, 17, 4334–4341. [Google Scholar] [CrossRef]
- O’Connell, K.C.; Landers, J.P. Integrated membranes within centrifugal microfluidic devices: A review. Lab. A Chip. 2023, 23, 3130–3159. [Google Scholar] [CrossRef]
- Ducrée, J. Systematic review of centrifugal valving based on digital twin modeling towards highly integrated lab-on-a-disc systems. Microsyst. Nanoeng. 2021, 7, 104. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Shi, Y.; Ping, J.; Wu, J.; Chen, H. Magnetic particles for integrated nucleic acid purification, amplification and detection without pipetting. TrAC Trends Anal. Chem. 2020, 127, 115912. [Google Scholar] [CrossRef]
- Sur, K.; McFall, S.M.; Yeh, E.T.; Jangam, S.R.; Hayden, M.A.; Stroupe, S.D.; Kelso, D.M. Immiscible Phase Nucleic Acid Purification Eliminates PCR Inhibitors with a Single Pass of Paramagnetic Particles through a Hydrophobic Liquid. J. Mol. Diagn. 2010, 12, 620–628. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Mateos, P.; Ngamsom, B.; Walter, C.; Dyer, C.E.; Gitaka, J.; Iles, A.; Pamme, N. A lab-on-a-chip platform for integrated extraction and detection of SARS-CoV-2 RNA in resource-limited settings. Anal. Chim. Acta 2021, 1177, 338758. [Google Scholar] [CrossRef] [PubMed]
- Troiano, D.; Deraney, R.N.; Tripathi, A. Effect of surfactants on carryover liquid volume in immiscible phase magnetic bead separation. Colloids Surf. A Physicochem. Eng. Asp. 2017, 513, 188–195. [Google Scholar] [CrossRef]
- Cui, F.R.; Wang, J.; Opal, S.M.; Tripathi, A. Isolating Influenza RNA from Clinical Samples Using Microfluidic Oil-Water Interfaces. PLoS ONE 2016, 11, e0149522. [Google Scholar] [CrossRef] [PubMed]
- Kistrup, K.; Skotte Sørensen, K.; Wolff, A.; Fougt Hansen, M. Liquid carry-over in an injection moulded all-polymer chip system for immiscible phase magnetic bead-based solid-phase extraction. J. Magn. Magn. Mater. 2015, 380, 191–196. [Google Scholar] [CrossRef]
- Berry, S.M.; Alarid, E.T.; Beebe, D.J. One-step purification of nucleic acid for gene expression analysis via Immiscible Filtration Assisted by Surface Tension (IFAST). Lab. A Chip. 2011, 11, 1747–1753. [Google Scholar] [CrossRef] [PubMed]
- Kalsi, S.; Valiadi, M.; Turner, C.; Sutton, M.; Morgan, H. Sample pre-concentration on a digital microfluidic platform for rapid AMR detection in urine. Lab. A Chip. 2019, 19, 168–177. [Google Scholar] [CrossRef]
- Wimbles, R.; Melling, L.M.; Cain, B.; Davies, N.; Doherty, J.; Johnson, B.; Shaw, K.J. On-site genetic analysis for species identification using lab-on-a-chip. Ecol. Evol. 2021, 11, 1535–1543. [Google Scholar] [CrossRef]
- Ngamsom, B.; Iles, A.; Kamita, M.; Kimani, R.; Wakaba, P.; Rodriguez-Mateos, P.; Mungai, M.; Dyer, C.E.; Walter, C.; Gitaka, J.; et al. A sample-to-answer COVID-19 diagnostic device based on immiscible filtration and CRISPR-Cas12a-assisted detection. Talanta Open 2022, 6, 100166. [Google Scholar] [CrossRef]
- Berry, S.M.; LaVanway, A.J.; Pezzi, H.M.; Guckenberger, D.J.; Anderson, M.A.; Loeb, J.M.; Beebe, D.J. HIV Viral RNA Extraction in Wax Immiscible Filtration Assisted by Surface Tension (IFAST) Devices. J. Mol. Diagn. 2014, 16, 297–304. [Google Scholar] [CrossRef]
- Strotman, L.N.; Lin, G.; Berry, S.M.; Johnson, E.A.; Beebe, D.J. Facile and rapid DNA extraction and purification from food matrices using IFAST (immiscible filtration assisted by surface tension). Analyst 2012, 137, 4023–4028. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Wang, X.; Ma, X.; Chu, Y.n.; Pang, S.; Chen, Y.; Guan, X.; Zou, B.; Wu, Y.; Zhou, G. Postsynthetic Modification of the Magnetic Zirconium–Organic Framework for Efficient and Rapid Solid-Phase Extraction of DNA. ACS Appl. Mater. Interfaces 2021, 13, 50309–50318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Deraney, R.N.; Tripathi, A. Adsorption and isolation of nucleic acids on cellulose magnetic beads using a three-dimensional printed microfluidic chip. Biomicrofluidics 2015, 9, 064118. [Google Scholar] [CrossRef] [PubMed]
- Poenitzsch Strong, A.M.; Berry, S.M.; Beebe, D.J.; Li, J.L.; Spiegelman, V.S. miFAST: A novel and rapid microRNA target capture method. Mol. Carcinog 2018, 57, 559–566. [Google Scholar] [CrossRef]
- Claveau, S.; Sasseville, M.; Beauregard, M. Alcohol-Mediated Error-Prone PCR. DNA Cell Biol. 2004, 23, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.; Loeffler, R.S.; Leigh, P.J.; Lopez, H.A.; Yoon, J.-Y. Recent Uses of Paper Microfluidics in Isothermal Nucleic Acid Amplification Tests. Biosensors 2023, 13, 885. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.-H.; Kim, Y.T.; Lee, K.; Kim, H.-M.; Lee, K.G.; Ahn, J.; Lee, J.; Lee, S.J.; Kim, K.-B. An electrophoretic DNA extraction device using a nanofilter for molecular diagnosis of pathogens. Nanoscale 2020, 12, 5048–5054. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Quyen, T.L.; Hung, T.Q.; Chin, W.H.; Wolff, A.; Bang, D.D. A lab-on-a-chip system with integrated sample preparation and loop-mediated isothermal amplification for rapid and quantitative detection of Salmonella spp. in food samples. Lab. A Chip. 2015, 15, 1898–1904. [Google Scholar] [CrossRef]
- Das, D.; Masetty, M.; Priye, A. Paper-Based Loop Mediated Isothermal Amplification (LAMP) Platforms: Integrating the Versatility of Paper Microfluidics with Accuracy of Nucleic Acid Amplification Tests. Chemosensors 2023, 11, 163. [Google Scholar] [CrossRef]
- Kaur, N.; Toley, B.J. Tuberculosis Diagnosis Using Isothermal Nucleic Acid Amplification in a Paper-and-Plastic Device. Methods Mol. Biol. 2023, 2621, 295–306. [Google Scholar] [CrossRef]
- Jawla, J.; Kumar, R.R.; Mendiratta, S.K.; Agarwal, R.K.; Singh, P.; Saxena, V.; Kumari, S.; Kumar, D. A novel paper based loop mediated isothermal amplification and lateral flow assay (LAMP-LFA) for point-of-care detection of buffalo tissue origin in diverse foods. J. Food Saf. 2023, 43, e13038. [Google Scholar] [CrossRef]
- Jawla, J.; Kumar, R.R.; Mendiratta, S.K.; Agarwal, R.K.; Kumari, S.; Saxena, V.; Kumar, D.; Singh, P.; Boby, N.; Rana, P. Paper-based loop-mediated isothermal amplification and lateral flow (LAMP-LF) assay for identification of tissues of cattle origin. Anal. Chim. Acta 2021, 1150, 338220. [Google Scholar] [CrossRef] [PubMed]
- Choopara, I.; Suea-Ngam, A.; Teethaisong, Y.; Howes, P.D.; Schmelcher, M.; Leelahavanichkul, A.; Thunyaharn, S.; Wongsawaeng, D.; deMello, A.J.; Dean, D.; et al. Fluorometric Paper-Based, Loop-Mediated Isothermal Amplification Devices for Quantitative Point-of-Care Detection of Methicillin-Resistant Staphylococcus aureus (MRSA). ACS Sens. 2021, 6, 742–751. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Yin, K.; Ding, X.; Li, Z.; Sun, X.; Li, B.; Lalla, R.V.; Gross, R.; Liu, C. An integrated E-Tube cap for sample preparation, isothermal amplification and label-free electrochemical detection of DNA. Biosens. Bioelectron. 2021, 186, 113306. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.R.; Hu, J.; Tang, R.; Gong, Y.; Feng, S.; Ren, H.; Wen, T.; Li, X.; Wan Abas, W.A.B.; Pingguan-Murphy, B.; et al. An integrated paper-based sample-to-answer biosensor for nucleic acid testing at the point of care. Lab. A Chip. 2016, 16, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Nanayakkara, I.A.; Cao, W.; White, I.M. Simplifying Nucleic Acid Amplification from Whole Blood with Direct Polymerase Chain Reaction on Chitosan Microparticles. Anal. Chem. 2017, 89, 3773–3779. [Google Scholar] [CrossRef] [PubMed]
- Emaus, M.N.; Anderson, J.L. Simultaneous cell lysis and DNA extraction from whole blood using magnetic ionic liquids. Anal. Bioanal. Chem. 2020, 412, 8039–8049. [Google Scholar] [CrossRef]
- Raeber, G.P.; Lutolf, M.P.; Hubbell, J.A. Molecularly Engineered PEG Hydrogels: A Novel Model System for Proteolytically Mediated Cell Migration. Biophys. J. 2005, 89, 1374–1388. [Google Scholar] [CrossRef]
- Phelps, E.A.; Enemchukwu, N.O.; Fiore, V.F.; Sy, J.C.; Murthy, N.; Sulchek, T.A.; Barker, T.H.; García, A.J. Maleimide Cross-Linked Bioactive PEG Hydrogel Exhibits Improved Reaction Kinetics and Cross-Linking for Cell Encapsulation and In Situ Delivery. Adv. Mater. 2012, 24, 64–70. [Google Scholar] [CrossRef]
- Xu, L.; Brito, I.L.; Alm, E.J.; Blainey, P.C. Virtual microfluidics for digital quantification and single-cell sequencing. Nat. Methods 2016, 13, 759–762. [Google Scholar] [CrossRef]
- Yi, C.; Luo, Z.; Lu, Y.; Belwal, T.; Pan, X.; Lin, X. Nanoporous hydrogel for direct digital nucleic acid amplification in untreated complex matrices for single bacteria counting. Biosens. Bioelectron. 2021, 184, 113199. [Google Scholar] [CrossRef] [PubMed]
- Hügle, M.; Behrmann, O.; Raum, M.; Hufert, F.T.; Urban, G.A.; Dame, G. A lab-on-a-chip for free-flow electrophoretic preconcentration of viruses and gel electrophoretic DNA extraction. Analyst 2020, 145, 2554–2561. [Google Scholar] [CrossRef] [PubMed]
- Persat, A.; Marshall, L.A.; Santiago, J.G. Purification of Nucleic Acids from Whole Blood Using Isotachophoresis. Anal. Chem. 2009, 81, 9507–9511. [Google Scholar] [CrossRef] [PubMed]
- Futai, N.; Fukazawa, Y.; Kashiwagi, T.; Tamaki, S.; Sakai, R.; Hogan, C.A.; Murugesan, K.; Ramachandran, A.; Banaei, N.; Santiago, J.G. A modular and reconfigurable open-channel gated device for the electrokinetic extraction of cell-free DNA assays. Anal. Chim. Acta 2022, 1200, 339435. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, T.; Bercovici, M. Amplification-free detection of DNA in a paper-based microfluidic device using electroosmotically balanced isotachophoresis. Lab. A Chip. 2018, 18, 861–868. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, B.P.; Bender, A.T.; Ngyuen, D.N.; Zhang, J.Y.; Posner, J.D. Nucleic acid sample preparation from whole blood in a paper microfluidic device using isotachophoresis. J. Chromatogr. B 2021, 1163, 122494. [Google Scholar] [CrossRef] [PubMed]
- Bender, A.T.; Borysiak, M.D.; Levenson, A.M.; Lillis, L.; Boyle, D.S.; Posner, J.D. Semiquantitative Nucleic Acid Test with Simultaneous Isotachophoretic Extraction and Amplification. Anal. Chem. 2018, 90, 7221–7229. [Google Scholar] [CrossRef]
| Main Lysis Method | Secondary Lysis Method | Reagents | Time/Temp (min or h/°C) | Target | Sample Matrix | Amplification | Lysis Efficiency | Yield Recovery Rate LOD | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Detergent | BSA | 0.3% IGEPAL CA-630 0.1% BSA | 5 min/on ice | Mammalian | N/A | RT-qPCR | N/A | N/A N/A 10 cells | [26] |
| Ethanol | 0.008% Q. saponaria 5% (w/v) NaCl 5% (v/v) ethanol | 48 h/55 °C | Yeast | N/A | N/A | 99.0% | N/A N/A N/A | [27] | |
| Enzymatic | Lysozyme Proteinase + SDS | 1. 1 h/45 °C 2. 5 h/50 °C | Soil microbiome | Soil | N/A | N/A | ~24 µg/g N/A N/A | [28] | |
| Enzymatic | 10 mg/mL Lysozyme 20 ng/mL Proteinase K 0.1% SDS 1 mM EDTA 10 mM Tris-HCl 1 µL RNase | 10 min/N/A | Bacteria | Milk (Spiked) | dRPA | N/A | ~20 ng/µL from 10 cells N/A 10 cells | [29] | |
| Enzymatic | 0.5% SDS 1 mg/mL Proteinase K 10 mM Dithiothreitol | 15 min/65 °C | Virus | Serum (Spiked) | RT-RPA | N/A | N/A N/A 500 copies/mL | [30] | |
| Enzymatic | 10 mM Tris-HCl (pH8) 10 mM EDTA 1% SDS 10% Triton X-100 Proteinase K DMS | 20 min/56 °C | Virus | Clinical | RPA | N/A | N/A 95% 10 copies | [31] | |
| Alkaline | N/A | 1.10 mM NaOH 2. 1 mM HCl | 5 min/N/A | Mammalian | Blood buccal swabs, saliva, cigarette butts | qPCR | N/A | 21.8 ng/µL N/A N/A | [32] |
| 0.5 M NaOH 10 mM Na2EDTA (pH 8) | 1 min/N/A | Plant | Plant | RT-RPA | N/A | N/A N/A 20 copies | [33] | ||
| Surfactant | 0.2 M NaOH 1% SDS (dried) | N/A | Bacteria | Aerosol (Spiked) | qPCR | N/A | N/A 10% 101 CFU | [34] | |
| PEG | 60% PEG200 20 mM KOH (pH 13.3–13.5) | 15 min/RT | Human Animal Bacteria Plant | Raw samples | PCR | N/A | N/A N/A 10 pg | [35] | |
| 1.25% PEG 200 10% PEG 8000 5% (v/v) NaOH | 1. 3 min/RT 2. 10 min/70 °C | Virus | Whole blood (Spiked) | LAMP | 100% | N/A N/A 102 PFU/mL | [36] | ||
| 6% PEG 200 0.08% NaOH | 3 min/RT | Fungal | Strawberry (Spiked) | RPA | N/A | N/A N/A 100 fg | [37] | ||
| 6% PEG 200 0.08% NaOH | 3 min/RT | Oomycete | Leaf | RPA | N/A | N/A N/A 500 fg | [38] | ||
| 60% PEG 400 100 mM KOH | N/A | Human | Whole blood | PCR | N/A | N/A N/A N/A | [39] | ||
| 50 g/L PEG 4600 20 mM KOH (pH 13.5) | 2 min/N/A | Fungal (mycelium) | Plant | LAMP | N/A | N/A N/A 19.9 pg/µL | [40] | ||
| Chaotropic | N/A | 5 M GuHCl | 5 min/RT | Bacteria | Liquid stool (clinical) | PCR | 50% (G−) 60% (G+) | Ave 109.5 ng/µL (3-chamber) Ave 59.3 ng/µL (5-chamber) ~60% N/A | [41] |
| 4 M GUSCN 20 mM Tris-HCl 1 mM DTT pH 7.7 | N/A | Animal | Mixed meat | qPCR | N/A | N/A N/A 0.1% | [42] | ||
| Enzymatic | GuHCl, Proteinase K | 30 min/56 °C | Bacteria | Human saliva | PCR | N/A | 157.2–165 ng/µL 7.86–8.25 µg N/A | [43] | |
| Proteinase K GUSCN | 10 min/56 °C | Bacteria | Human urine Milk | qPCR | N/A | N/A N/A 5 CFU/10 mL | [44] | ||
| 6 M GuHCl Proteinase K pH 6.1 | 10 min/56 °C | Virus | Buccal swab (spiked) | LAMP | N/A | N/A N/A N/A | [45] | ||
| Detergent | 6 M GuHCl 2% Triton X-100 13 mM EDTA 10 mM NaCl 51 mM Tris (pH 5.5) | 5 min/RT | Virus | Serum (Spiked) | RT-PCR | N/A | 1.3–2.0 µg/100 µL N/A N/A | [2] | |
| AMP (Melittin, Bombolitin III, MSI-78, or MSI-594) | 5 min/RT | Bacteria | N/A | qLAMP | 100% | N/A N/A N/A | [46] | ||
| 5 M GuSCN 100 mM EDTA 0.5% (v/v) Sarkosyl | 5–10 min/N/A | Bacteria | N/A | N/A | N/A | N/A N/A N/A | [47] | ||
| 6 M GuHCl 2% Triton X-100 13 mM EDTA 10 mM NaCl 51 mM Tris pH 5.5 | 5 min/RT | Bacteria | Serum Saliva (Spiked) | dRPA | N/A | 15–35 ng/µL 89.4%, 79.6% (saliva, serum) 1.1 × 108 copies/μL | [48] | ||
| 1.5 M GuHCl 50 mM Tris [pH 8] 100 mM NaCl 5 mM EDTA 1% Tween-20 | 10 s/RT | Fish | Blood (Fish) | PCR | N/A | N/A N/A 104 cells | [49] | ||
| 4.8% GuSCN 5% Triton X-100 (pH 6.8) | N/A | Virus | Spiked blood | PCR | N/A | N/A N/A 5 particles | [50] | ||
| 4 M GUSCN 1% Triton X-100 1% ß-mercaptoethanol 10 mM 2-Ethanesulfonic acid | 5 min/RT | Virus | Nasopharyngeal swab | LAMP | N/A | N/A N/A 1–10 copies/µL | [51] | ||
| 4 M GuSCN 10 mM Tris-HCl (pH 8) 1 mM EDTA (pH 8) 0.5% Triton X-100 300 µL Isopropanol 3 µL ß-mercaptoethanol 5.6 µg poly-A carrier RNA (For RNA) | 3 min/RT | Synthetic DNA | Synthetic sputum (Spiked) Residual urine sample | qPCR | N/A | N/A 10.2 ± 4.03%, 91.2 ± 7.46% (sputum, urine) N/A | [52] | ||
| 0.1 M Tris-HCl (pH8.0) 10 mM EDTA 1% SDS 10% Triton X-100 Proteinase K DNase I (RNA) | 10 min/RT (RNA) 20 min/56 °C (DNA) | Mammalian Bacteria | N/A | RT-qPCR qPCR | N/A | ~100 ng/µL N/A 103 CUF/mL, 101 cells/mL (DNA, RNA) | [53] | ||
| MIL | Peptide Enzymatic Detergent | 6 µL [P6,6,6,14+] [Ni(HfAcAc)3−] | 1 h/N/A | Plant | Plant | qPCR | N/A | ~8 ng N/A N/A | [54] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.M.; Balakrishnan, H.K.; Doeven, E.H.; Yuan, D.; Guijt, R.M. Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review. Biosensors 2023, 13, 980. https://doi.org/10.3390/bios13110980
Lee SM, Balakrishnan HK, Doeven EH, Yuan D, Guijt RM. Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review. Biosensors. 2023; 13(11):980. https://doi.org/10.3390/bios13110980
Chicago/Turabian StyleLee, Soo Min, Hari Kalathil Balakrishnan, Egan H. Doeven, Dan Yuan, and Rosanne M. Guijt. 2023. "Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review" Biosensors 13, no. 11: 980. https://doi.org/10.3390/bios13110980
APA StyleLee, S. M., Balakrishnan, H. K., Doeven, E. H., Yuan, D., & Guijt, R. M. (2023). Chemical Trends in Sample Preparation for Nucleic Acid Amplification Testing (NAAT): A Review. Biosensors, 13(11), 980. https://doi.org/10.3390/bios13110980






