Isothermal Amplification and Hypersensitive Fluorescence Dual-Enhancement Nucleic Acid Lateral Flow Assay for Rapid Detection of Acinetobacter baumannii and Its Drug Resistance
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
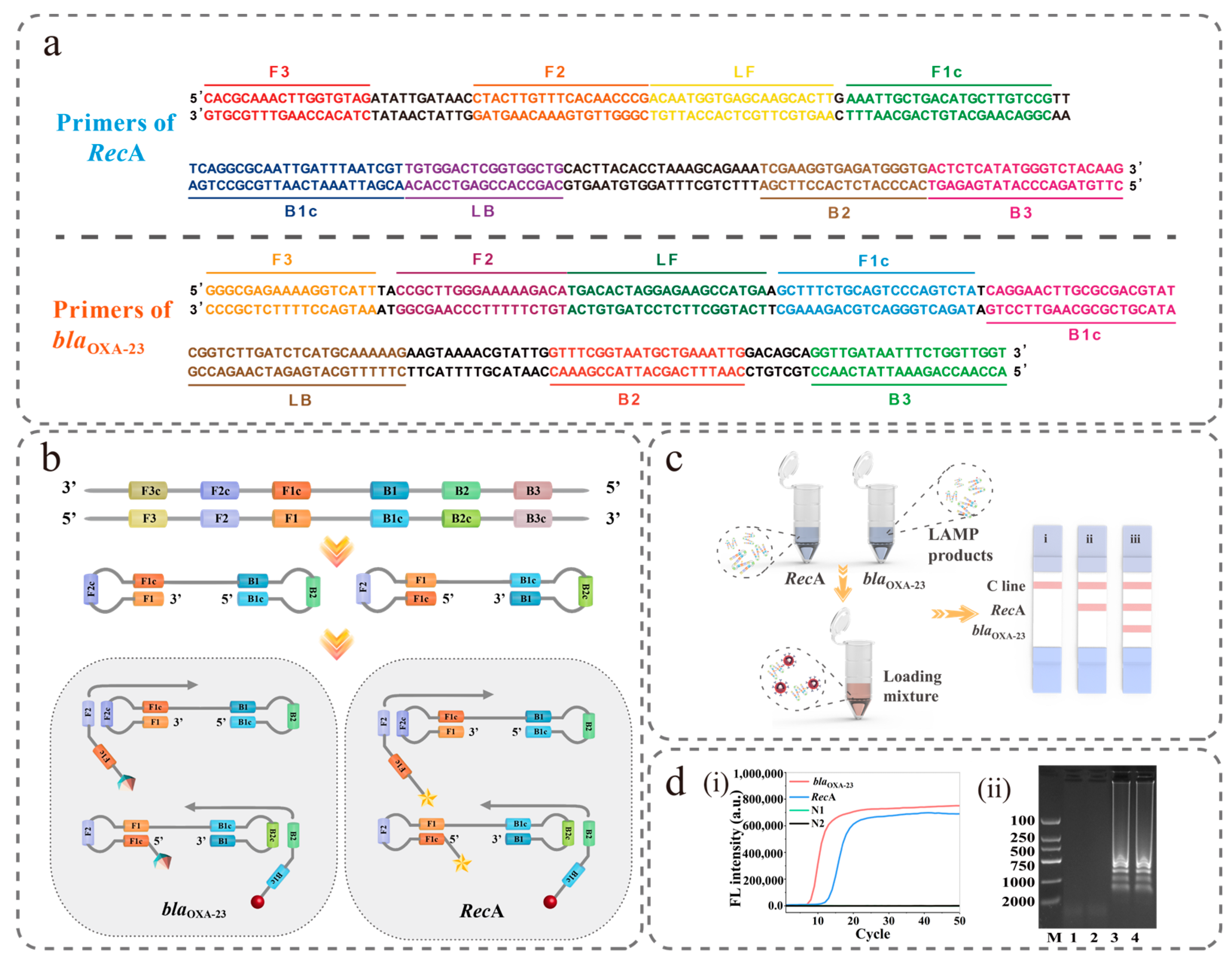
2.2. Primer Design
2.3. LAMP Reaction
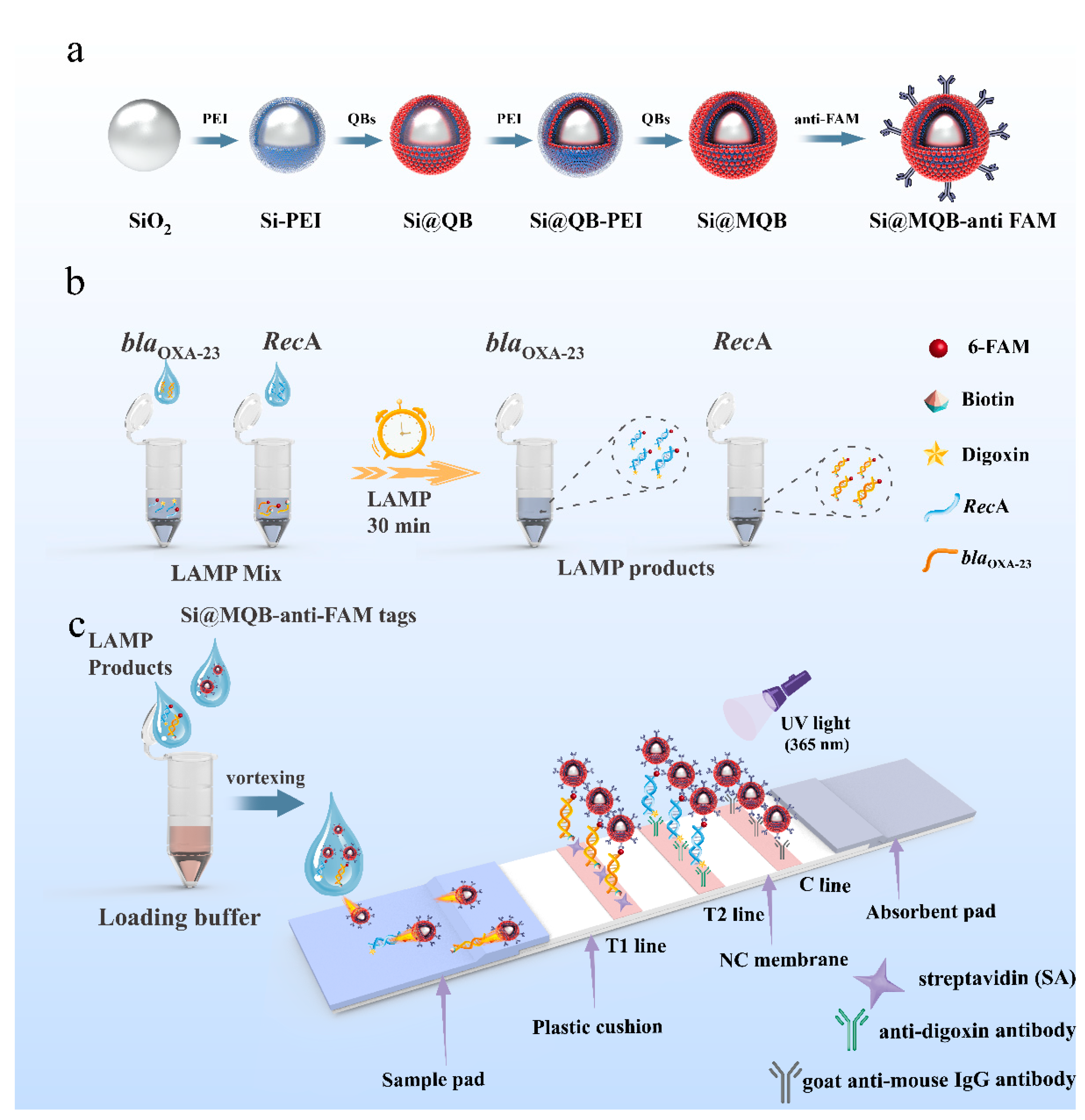
2.4. Preparation of Si@MQB
2.5. Preparation of Si@MQB-Anti-FAM Tags
2.6. Preparation of Samples
2.7. Fabrication of NFLFA
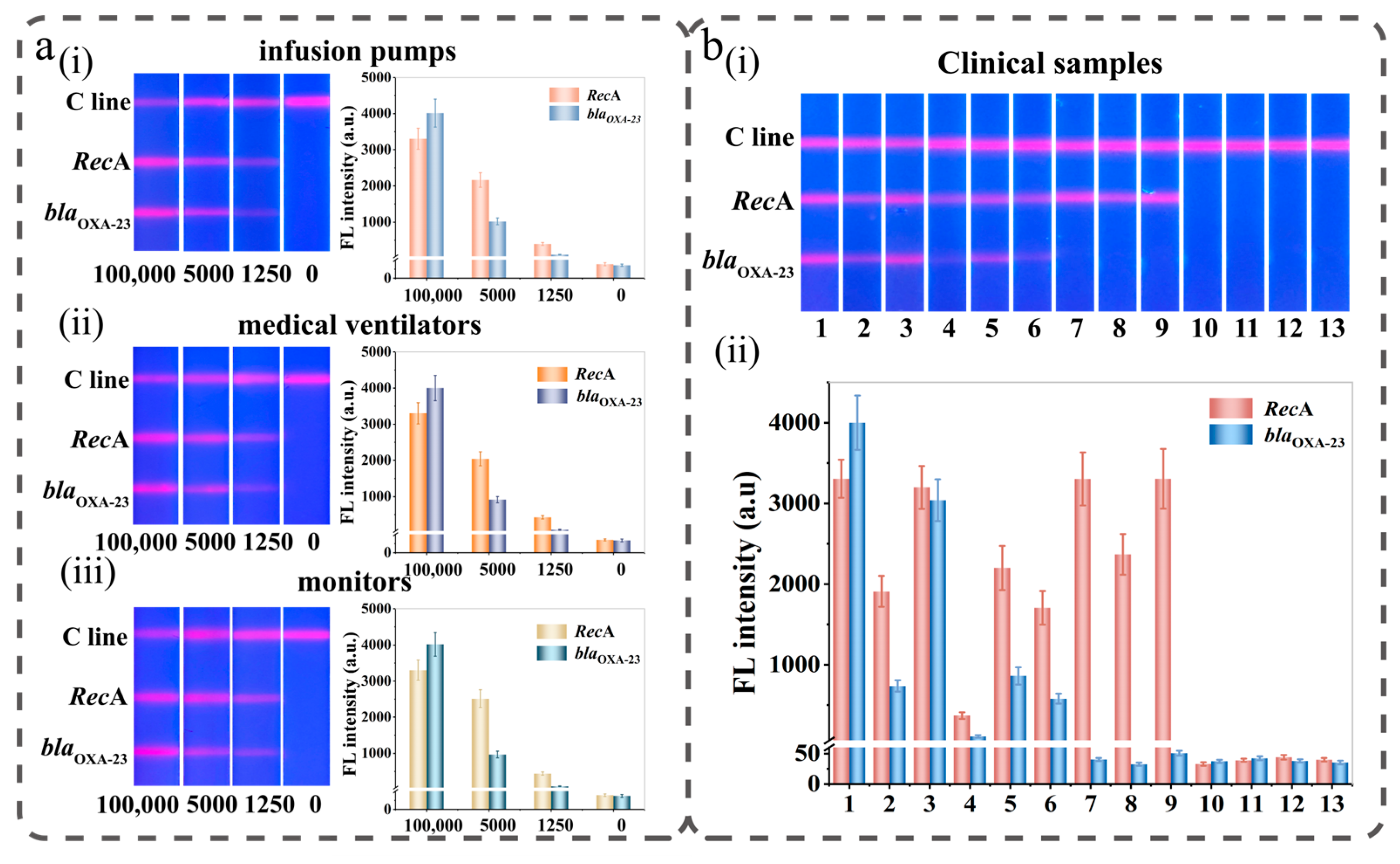
2.8. Actual Sample Testing
3. Results and Discussion
3.1. Principle of LAMP-NFLFA
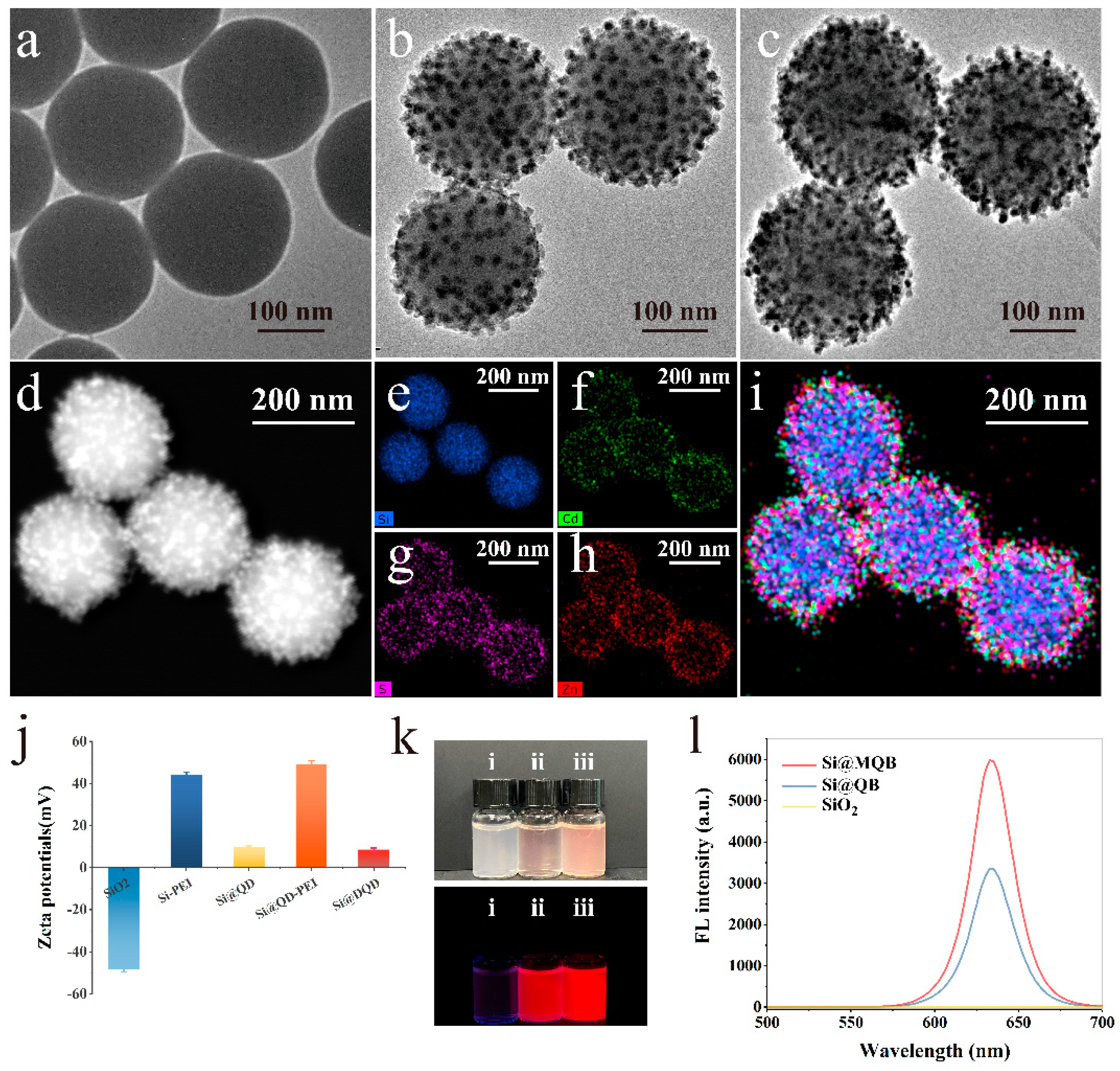
3.2. Characterisation of Si@MQB
3.3. Detection Feasibility of the LAMP-NFLFA System
3.4. Optimisation of the LAMP-NFLFA System
3.5. Construction of the LAMP-NFLFA Dual-Channel Method for the Detection of A. baumannii and CRAB
3.6. Performance of Si@MQB-Based LAMP-NFLFA in Bacteria Detection
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cisneros, J.M.; Rodriguez-Bano, J.; Fernandez-Cuenca, F.; Ribera, A.; Vila, J.; Pascual, A.; Martinez-Martinez, L.; Bou, G.; Pachon, J.; Clinical, M.; et al. Risk-factors for the acquisition of imipenem-resistant Acinetobacter baumannii in Spain: A nationwide study. Clin. Microbiol. Infect. 2005, 11, 874–879. [Google Scholar] [CrossRef] [PubMed]
- Liou, M.L.; Chen, K.H.; Yeh, H.L.; Lai, C.Y.; Chen, C.H. Persistent nasal carriers of Acinetobacter baumannii in long-term-care facilities. Am. J. Infect. Control. 2017, 45, 723–727. [Google Scholar] [CrossRef]
- Pakharukova, N.; Malmi, H.; Tuittila, M.; Dahlberg, T.; Ghosal, D.; Chang, Y.W.; Myint, S.L.; Paavilainen, S.; Knight, S.D.; Lamminmaki, U.; et al. Archaic chaperone-usher pili self-secrete into superelastic zigzag springs. Nature 2022, 609, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Perez, S.; Innes, G.K.; Walters, M.S.; Mehr, J.; Arias, J.; Greeley, R.; Chew, D. Increase in Hospital-Acquired Carbapenem-Resistant Acinetobacter baumannii Infection and Colonization in an Acute Care Hospital During a Surge in COVID-19 Admissions—New Jersey, February–July 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 1827–1831. [Google Scholar] [CrossRef]
- Cruz-Lopez, F.; Villarreal-Trevino, L.; Morfin-Otero, R.; Martinez-Melendez, A.; Camacho-Ortiz, A.; Rodriguez-Noriega, E.; Garza-Gonzalez, E. Dynamics of colonization in patients with health care-associated infections at step-down care units from a tertiary care hospital in Mexico. Am. J. Infect. Control 2020, 48, 1329–1335. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.Y.; Yiu, S.M.; Kuo, H.Y.; Tan, T.S.; Liao, K.H.; Liu, C.C.; Hon, W.K.; Liou, M.L. Application of 16S rRNA metagenomics to analyze bacterial communities at a respiratory care centre in Taiwan. Appl. Microbiol. Biotechnol. 2015, 99, 2871–2881. [Google Scholar] [CrossRef]
- Hong, K.B.; Oh, H.S.; Song, J.S.; Lim, J.H.; Kang, D.K.; Son, I.S.; Park, J.D.; Kim, E.C.; Lee, H.J.; Choi, E.H. Investigation and control of an outbreak of imipenem-resistant Acinetobacter baumannii Infection in a Pediatric Intensive Care Unit. Pediatr. Infect. Dis. J. 2012, 31, 685–690. [Google Scholar] [CrossRef]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Hu, D.; Liu, B.; Dijkshoorn, L.; Wang, L.; Reeves, P.R. Diversity in the major polysaccharide antigen of Acinetobacter baumannii assessed by DNA sequencing, and development of a molecular serotyping scheme. PLoS ONE 2013, 8, e70329. [Google Scholar] [CrossRef]
- Ecker, J.A.; Massire, C.; Hall, T.A.; Ranken, R.; Pennella, T.T.; Agasino Ivy, C.; Blyn, L.B.; Hofstadler, S.A.; Endy, T.P.; Scott, P.T.; et al. Identification of Acinetobacter species and genotyping of Acinetobacter baumannii by multilocus PCR and mass spectrometry. J. Clin. Microbiol. 2006, 44, 2921–2932. [Google Scholar] [CrossRef]
- Hornsey, M.; Loman, N.; Wareham, D.W.; Ellington, M.J.; Pallen, M.J.; Turton, J.F.; Underwood, A.; Gaulton, T.; Thomas, C.P.; Doumith, M.; et al. Whole-genome comparison of two Acinetobacter baumannii isolates from a single patient, where resistance developed during tigecycline therapy. J. Antimicrob. Chemother. 2011, 66, 1499–1503. [Google Scholar] [CrossRef] [PubMed]
- Parolo, C.; Sena-Torralba, A.; Bergua, J.F.; Calucho, E.; Fuentes-Chust, C.; Hu, L.; Rivas, L.; Alvarez-Diduk, R.; Nguyen, E.P.; Cinti, S.; et al. Tutorial: Design and fabrication of nanoparticle-based lateral-flow immunoassays. Nat. Protoc. 2020, 15, 3788–3816. [Google Scholar] [CrossRef]
- Wu, P.; Xue, F.; Zuo, W.; Yang, J.; Liu, X.; Jiang, H.; Dai, J.; Ju, Y. A Universal Bacterial Catcher Au-PMBA-Nanocrab-Based Lateral Flow Immunoassay for Rapid Pathogens Detection. Anal. Chem. 2022, 94, 4277–4285. [Google Scholar] [CrossRef]
- Orlov, A.V.; Malkerov, J.A.; Novichikhin, D.O.; Znoyko, S.L.; Nikitin, P.I. Express high-sensitive detection of ochratoxin A in food by a lateral flow immunoassay based on magnetic biolabels. Food Chem. 2022, 383, 132427. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, X.; Rong, Z.; Tu, Z.; Zhang, X.; Gu, B.; Wang, C.; Wang, S. Introduction of graphene oxide-supported multilayer-quantum dots nanofilm into multiplex lateral flow immunoassay: A rapid and ultrasensitive point-of-care testing technique for multiple respiratory viruses. Nano Res. 2023, 16, 3063–3073. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Li, J.; Tu, Z.; Gu, B.; Wang, S. Ultrasensitive and multiplex detection of four pathogenic bacteria on a bi-channel lateral flow immunoassay strip with three-dimensional membrane-like SERS nanostickers. Biosens. Bioelectron. 2022, 214, 114525. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.; Wang, K.; Zheng, W.; Cheng, Y.; Li, T.; Cao, B.; Jin, Q.; Cui, D. Rapid developments in lateral flow immunoassay for nucleic acid detection. Analyst 2021, 146, 1514–1528. [Google Scholar] [CrossRef]
- Kim, J.M.; Park, J.S.; Yoon, T.H.; Park, J.; Park, K.S. Nucleic acid lateral flow assay for simultaneous detection of hygiene indicator bacteria. Anal. Bioanal. Chem. 2021, 413, 5003–5011. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Safenkova, I.V.; Zherdev, A.V.; Dzantiev, B.B. Nucleic acid lateral flow assay with recombinase polymerase amplification: Solutions for highly sensitive detection of RNA virus. Talanta 2020, 210, 120616. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.H.; Kim, S.; Park, J.S.; Cha, B.S.; Lee, E.S.; Han, J.; Shin, J.; Jang, Y.; Park, K.S. Loop-mediated isothermal amplification-based nucleic acid lateral flow assay for the specific and multiplex detection of genetic markers. Anal. Chim. Acta 2022, 1205, 339781. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Cao, J.; Zhu, M.; Xu, M.; Shi, F. Loop-mediated isothermal amplification-lateral-flow dipstick (LAMP-LFD) to detect Mycoplasma ovipneumoniae. World J. Microbiol. Biotechnol. 2019, 35, 31. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Hu, X.; Wang, W.; Yang, Y.; Zhang, X.; Fang, W.; Zhang, L.; Li, S.; Gu, B. RT-LAMP assay for rapid detection of the R203M mutation in SARS-CoV-2 Delta variant. Emerg. Microbes Infect. 2022, 11, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Kitao, M.; Tomita, N.; Notomi, T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 2004, 59, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Komura, R.; Kawakami, T.; Nakajima, K.; Suzuki, H.; Nakashima, C. Simultaneous detection of benzimidazole-resistant strains of Fusarium head blight using the loop-mediated isothermal amplification-fluorescent loop primer method. J. Gen. Plant Pathol. 2018, 84, 247–253. [Google Scholar] [CrossRef]
- Chen, X.; Ma, K.; Yi, X.; Xiong, L.; Wang, Y.; Li, S. The rapid and visual detection of methicillin-susceptible and methicillin-resistant Staphylococcus aureus using multiplex loop-mediated isothermal amplification linked to a nanoparticle-based lateral flow biosensor. Antimicrob. Resist. Infect. Control. 2020, 9, 111. [Google Scholar] [CrossRef]
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens. Actuators B 2021, 329, 129196. [Google Scholar] [CrossRef]
- Li, J.; Wang, C.; Shi, L.; Shao, L.; Fu, P.; Wang, K.; Xiao, R.; Wang, S.; Gu, B. Rapid identification and antibiotic susceptibility test of pathogens in blood based on magnetic separation and surface-enhanced Raman scattering. Microchim. Acta 2019, 186, 475. [Google Scholar] [CrossRef]
- Humphries, R.; Bobenchik, A.M.; Hindler, J.A.; Schuetz, A.N. Overview of Changes to the Clinical and Laboratory Standards Institute Performance Standards for Antimicrobial Susceptibility Testing, M100, 31st Edition. J. Clin. Microbiol. 2021, 59, e0021321. [Google Scholar] [CrossRef]
- Hombach, M.; Jetter, M.; Blochliger, N.; Kolesnik-Goldmann, N.; Keller, P.M.; Bottger, E.C. Rapid disc diffusion antibiotic susceptibility testing for Pseudomonas aeruginosa, Acinetobacter baumannii and Enterococcus spp. J. Antimicrob. Chemother. 2018, 73, 385–391. [Google Scholar] [CrossRef]
- Gao, F.; Lei, C.; Liu, Y.; Song, H.; Kong, Y.; Wan, J.; Yu, C. Rational Design of Dendritic Mesoporous Silica Nanoparticles’ Surface Chemistry for Quantum Dot Enrichment and an Ultrasensitive Lateral Flow Immunoassay. ACS Appl. Mater. Interfaces 2021, 13, 21507–21515. [Google Scholar] [CrossRef]
- Song, S.; Liu, N.; Zhao, Z.; Njumbe Ediage, E.; Wu, S.; Sun, C.; De Saeger, S.; Wu, A. Multiplex lateral flow immunoassay for mycotoxin determination. Anal. Chem. 2014, 86, 4995–5001. [Google Scholar] [CrossRef]
- Virgilio, A.; Silva, A.B.S.; Nogueira, A.R.A.; Nóbrega, J.A.; Donati, G.L. Calculating limits of detection and defining working ranges for multi-signal calibration methods. J. Anal. At. Spectrom. 2020, 35, 1614–1620. [Google Scholar] [CrossRef]
- Wang, C.; Xiao, R.; Wang, S.; Yang, X.; Bai, Z.; Li, X.; Rong, Z.; Shen, B.; Wang, S. Magnetic quantum dot based lateral flow assay biosensor for multiplex and sensitive detection of protein toxins in food samples. Biosens. Bioelectron. 2019, 146, 111754. [Google Scholar] [CrossRef] [PubMed]
- Varona, M.; Eitzmann, D.R.; Anderson, J.L. Sequence-specific detection of ORF1a, BRAF, and ompW DNA sequences with loop mediated isothermal amplification on lateral flow immunoassay strips enabled by molecular beacons. Anal. Chem. 2021, 93, 4149–4153. [Google Scholar] [CrossRef] [PubMed]
- Gadsby, N.J.; McHugh, M.P.; Russell, C.D.; Mark, H.; Conway Morris, A.; Laurenson, I.F.; Hill, A.T.; Templeton, K.E. Development of two real-time multiplex PCR assays for the detection and quantification of eight key bacterial pathogens in lower respiratory tract infections. Clin. Microbiol. Infect. 2015, 21, 788.e1–788.e3. [Google Scholar] [CrossRef]
- Zheng, Y.; Wang, X.; Jiang, H. Label-free detection of Acinetobacter baumannii through the induced fluorescence quenching of thiolated AuAg nanoclusters. Sens. Actuators B. 2018, 277, 388–393. [Google Scholar] [CrossRef]
- Liu, T.-H.; Cheng, S.-S.; You, H.-L.; Lee, M.S.; Lee, G.-B. Bacterial detection and identification from human synovial fluids on an integrated microfluidic system. Analyst 2019, 144, 1210–1222. [Google Scholar] [CrossRef]
- Su, C.-H.; Tsai, M.-H.; Lin, C.-Y.; Ma, Y.-D.; Wang, C.-H.; Chung, Y.-D.; Lee, G.-B. Dual aptamer assay for detection of Acinetobacter baumannii on an electromagnetically-driven microfluidic platform. Biosens. Bioelectron. 2020, 159, 112148. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, H.S.; Lee, J.M.; Yoon, S.S.; Yong, D. Rapid Detection of Pseudomonas aeruginosa and Acinetobacter baumannii Harboring blaVIM-2, blaIMP-1 and blaOXA-23 Genes by Using Loop-Mediated Isothermal Amplification Methods. Ann. Lab. Med. 2016, 36, 15–22. [Google Scholar] [CrossRef]
- Sharma, A.; Gaind, R. Development of Loop-Mediated Isothermal Amplification Assay for Detection of Clinically Significant Members of Acinetobacter calcoaceticus–baumannii Complex and Associated Carbapenem Resistance. Front. Mol. Biosci. 2021, 8, 659256. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Gaind, R. Standardization of LAMP Assay for early detection of Acinetobacter baumannii and its resistant variants from patients with sepsis. Int. J. Infect. Dis. 2020, 101, 195. [Google Scholar] [CrossRef]
- Kanapathy, S.; Obande, G.A.; Chuah, C.; Shueb, R.H.; Yean, C.Y.; Banga Singh, K.K. Sequence-Specific Electrochemical Genosensor for Rapid Detection of bla(OXA-51-like) Gene in Acinetobacter baumannii. Microorganisms 2022, 10, 1413. [Google Scholar] [CrossRef] [PubMed]


| Detection Method | Differentiation of Drug Resistance | Detection Limit | Assay Time (min) | Reference |
|---|---|---|---|---|
| RT multiplex PCR | No | 8333 CFU/mL | >60 | [36] |
| Fluorescence | No | 103CFU/mL | 40 | [37] |
| Microfluidics | No | 102 CFU/mL | 90 | [38] |
| Microfluidics | No | 104 CFU/mL | 30 | [39] |
| LAMP | Yes | 10 pg | 60 | [40] |
| LAMP | No | 104 CFU/mL | 15 | [41] |
| LAMP | Yes | 104 CFU/mL | − | [42] |
| Electrochemical Genosensor | No | 103 CFU/mL | − | [43] |
| LAMP-NFLFA | Yes | 199 CFU/mL | 43 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zheng, S.; Liu, Y.; Wang, C.; Gu, B.; Zhang, L.; Wang, S. Isothermal Amplification and Hypersensitive Fluorescence Dual-Enhancement Nucleic Acid Lateral Flow Assay for Rapid Detection of Acinetobacter baumannii and Its Drug Resistance. Biosensors 2023, 13, 945. https://doi.org/10.3390/bios13100945
Wang Q, Zheng S, Liu Y, Wang C, Gu B, Zhang L, Wang S. Isothermal Amplification and Hypersensitive Fluorescence Dual-Enhancement Nucleic Acid Lateral Flow Assay for Rapid Detection of Acinetobacter baumannii and Its Drug Resistance. Biosensors. 2023; 13(10):945. https://doi.org/10.3390/bios13100945
Chicago/Turabian StyleWang, Qian, Shuai Zheng, Yong Liu, Chongwen Wang, Bing Gu, Long Zhang, and Shu Wang. 2023. "Isothermal Amplification and Hypersensitive Fluorescence Dual-Enhancement Nucleic Acid Lateral Flow Assay for Rapid Detection of Acinetobacter baumannii and Its Drug Resistance" Biosensors 13, no. 10: 945. https://doi.org/10.3390/bios13100945
APA StyleWang, Q., Zheng, S., Liu, Y., Wang, C., Gu, B., Zhang, L., & Wang, S. (2023). Isothermal Amplification and Hypersensitive Fluorescence Dual-Enhancement Nucleic Acid Lateral Flow Assay for Rapid Detection of Acinetobacter baumannii and Its Drug Resistance. Biosensors, 13(10), 945. https://doi.org/10.3390/bios13100945







