Screening of Single-Stranded DNA Aptamer Specific for Florfenicol and Application in Detection of Food Safety
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Materials and Chemicals
2.2. Immobilization of the FFA by Using Magnetic Beads
2.3. Screening of FF Aptamers by MB-SELEX
2.4. Sequence Analysis and High-Throughput Sequencing
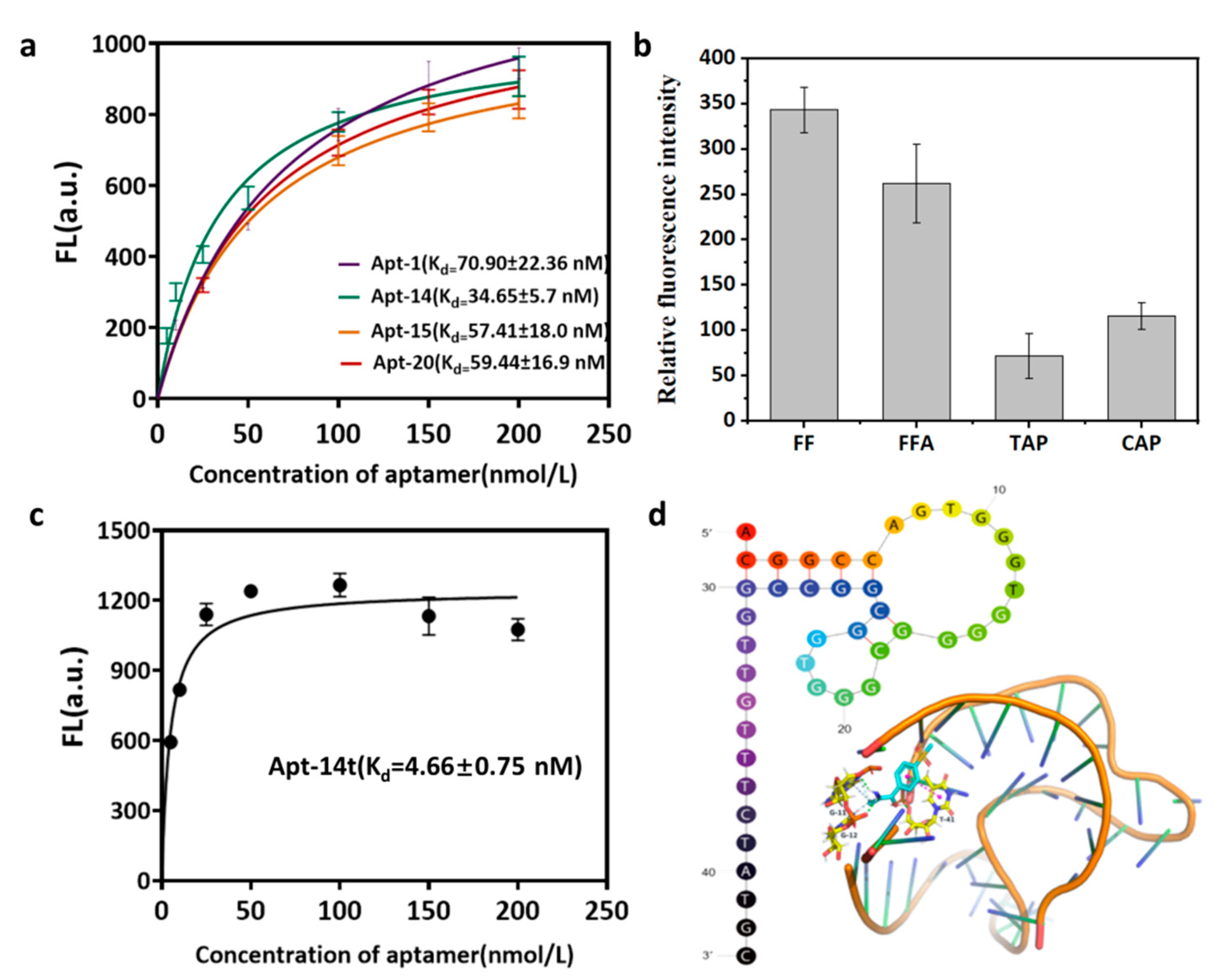
2.5. Dissociation Constants (Kd) Determination and Specificity Analysis of Candidate ssDNA
2.6. Molecular Docking Studies
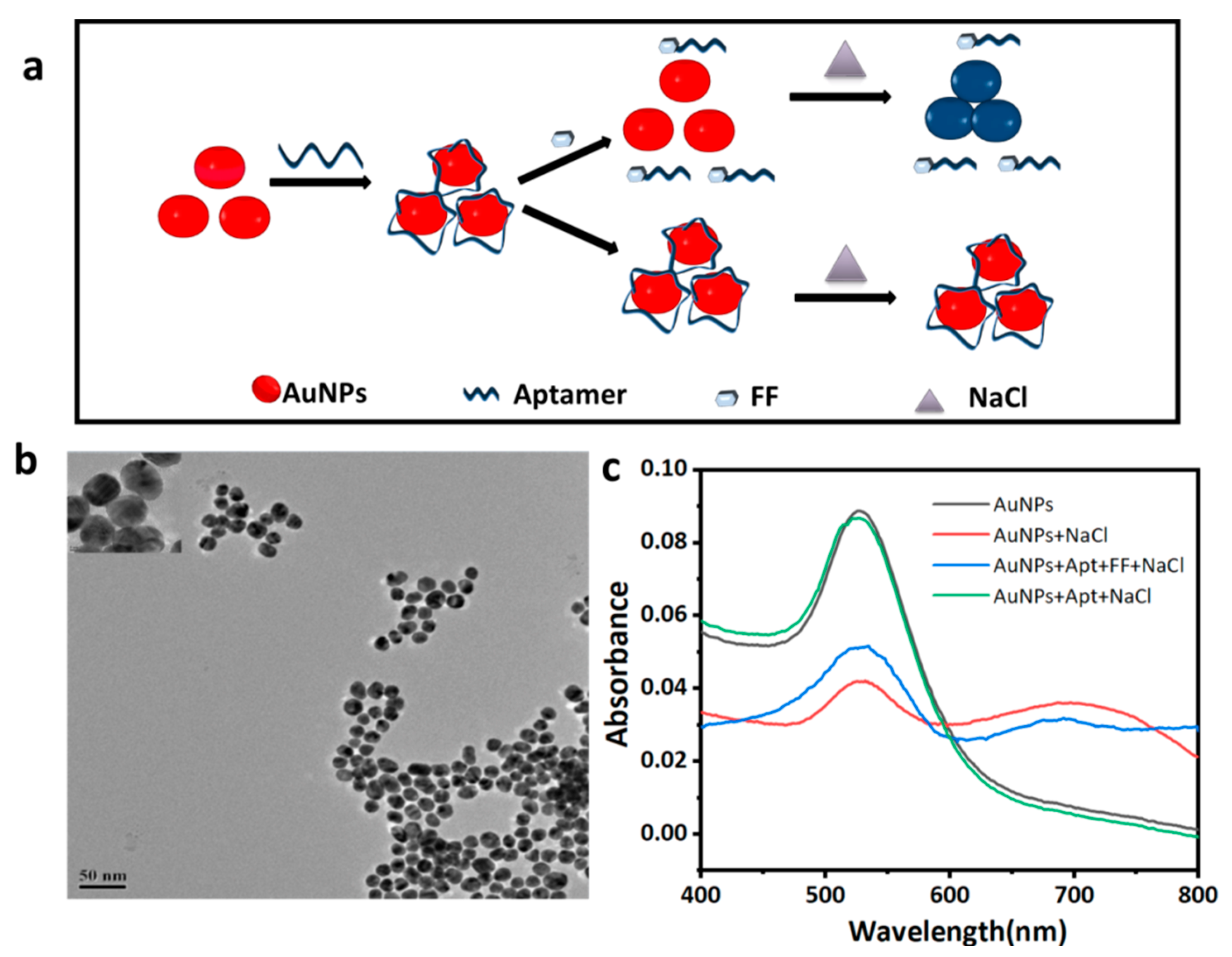
2.7. Detection of FF by AuNPs Colorimetry
3. Results and Discussion
3.1. Immobilization of Targets on Magnetic Beads
3.2. Selection of Aptamer In Vitro
3.3. Characterization of Specific Ligands
3.4. Molecular Docking
3.5. Detection of FF by AuNPs Colorimetry
3.5.1. AuNP Characterization
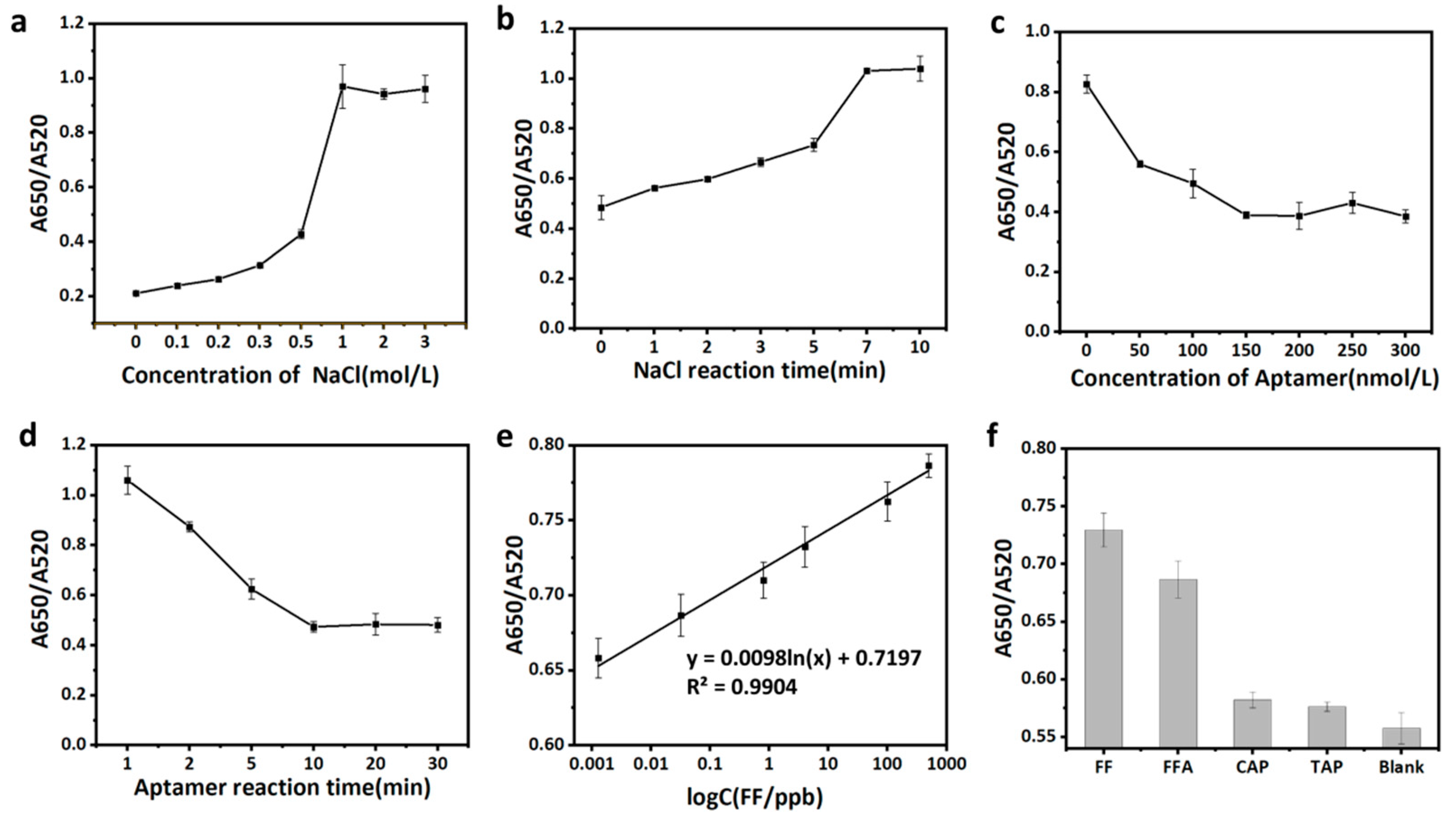
3.5.2. Condition Optimization
3.5.3. Colorimetric Method Performance Verification
3.6. Detection of FF in Milk and Egg Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ronquillo, M.G.; Hernandez, J.C.A. Antibiotic and synthetic growth promoters in animal diets: Review of impact and analytical methods. Food Control 2017, 72, 255–267. [Google Scholar] [CrossRef]
- Li, Z.; Yang, Y.-J.; Qin, Z.; Li, S.-H.; Bai, L.-X.; Li, J.-Y.; Liu, X.-W. Florfenicol-Polyarginine Conjugates Exhibit Promising Antibacterial Activity Against Resistant Strains. Front. Chem. 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhu, T.; Zhou, D.; Lu, W.; Liu, H.; Sun, Z.; Ying, J.; Lu, J.; Lin, X.; Li, K.; et al. Analysis of Resistance to Florfenicol and the Related Mechanism of Dissemination in Different Animal-Derived Bacteria. Front. Cell. Infect. Microbiol. 2020, 10, 11. [Google Scholar] [CrossRef]
- Lai, H.-T.; Hou, J.-H.; Su, C.-I.; Chen, C.-L. Effects of chloramphenicol, florfenicol, and thiamphenicol on growth of algae Chlorella pyrenoidosa, Isochrysis galbana, and Tetraselmis chui. Ecotoxicol. Environ. Saf. 2009, 72, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Jia, L.; Yao, Y.; Xu, D.; Chen, S.; Xie, X.; Pei, Y.; Bao, W.; Dai, G.; Wang, J.; et al. Simultaneous determination of thiamphenicol, florfenicol and florfenicol amine in eggs by reversed-phase high-performance liquid chromatography with fluorescence detection. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2011, 879, 2351–2354. [Google Scholar] [CrossRef] [PubMed]
- Samsonova, J.V.; Cannavan, A.; Elliott, C.T. A critical review of screening methods for the detection of chloramphenicol, thiamphenicol, and florfenicol residues in foodstuffs. Crit. Rev. Anal. Chem. 2012, 42, 50–78. [Google Scholar] [CrossRef]
- Pilehvar, S.; Gielkens, K.; Trashin, S.; Dardenne, F.; Blust, R.; De Wael, K. (Electro) Sensing of phenicol antibiotics—A review. Crit. Rev. Food Sci. Nutr. 2016, 56, 2416–2429. [Google Scholar] [CrossRef] [PubMed]
- Fodey, T.L.; George, S.E.; Traynor, I.M.; Delahaut, P.; Kennedy, D.G.; Elliott, C.T.; Crooks, S.R. Approaches for the simultaneous detection of thiamphenicol, florfenicol and florfenicol amine using immunochemical techniques. J. Immunol. Methods 2013, 393, 30–37. [Google Scholar] [CrossRef]
- Tao, X.; Jiang, H.; Yu, X.; Zhu, J.; Wang, X.; Wang, Z.; Niu, L.; Wu, X.; Shen, J. Simultaneous determination of chloramphenicol, florfenicol and florfenicol amine in ham sausage with a hybrid chemiluminescent immunoassay. Food Addit. Contam. Part A-Chem. Anal. Control. Expo. Risk Assess. 2013, 30, 804–812. [Google Scholar] [CrossRef]
- Van De Riet, J.M.; Potter, R.A.; Christie-Fougere, M.; Burns, B.G. Simultaneous determination of residues of chloramphenicol, thiamphenicol, florfenicol, and florfenicol amine in farmed aquatic species by liquid, chromatography/mass spectrometry. J. AOAC Int. 2003, 86, 510–514. [Google Scholar] [CrossRef]
- Ilgu, M.; Nilsen-Hamilton, M. Aptamers in analytics. Analyst 2016, 141, 1551–1568. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jo, M.; Kim, T.H.; Ahn, J.-Y.; Lee, D.-K.; Kim, S.; Hong, S. Aptamer sandwich-based carbon nanotube sensors for single-carbon-atomic-resolution detection of non-polar small molecular species. Lab Chip 2011, 11, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Stewart, K.D.; Tan, W.; Park, J.Y. Aptamer selection for detecting molecular target using Cell-SELEX (Systematic evolution of ligands by exponential enrichment) technology. Methods Mol. Biol. 2019, 2054, 223–241. [Google Scholar] [CrossRef]
- Yan, J.; Xiong, H.; Cai, S.; Wen, N.; He, Q.; Liu, Y.; Peng, D.; Liu, Z. Advances in aptamer screening technologies. Talanta 2019, 200, 124–144. [Google Scholar] [CrossRef]
- Kuwahara, M.; Li, Y.; Rozners, E.; Murakami, H. Artificially created nucleic acids and peptides/proteins in chemical biology. J. Nucleic Acids 2013, 2013, 219263. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ulrich, H.; Martins, A.H.B.; Pesquero, J.B. RNA and DNA aptamers in cytomics analysis. Curr. Protoc. Cytom. 2005, 33, 7–28. [Google Scholar] [CrossRef]
- Liu, R.; Zhang, F.; Sang, Y.; Katouzian, I.; Jafari, S.M.; Wang, X.; Li, W.; Wang, J.; Mohammadi, Z. Screening, identification, and application of nucleic acid aptamers applied in food safety biosensing. Trends Food Sci. Technol. 2022, 123, 355–375. [Google Scholar] [CrossRef]
- McKeague, M.; Velu, R.; Hill, K.; Bardóczy, V.; Mészáros, T.; De Rosa, M.C. Selection and characterization of a novel DNA aptamer for label-free fluorescence biosensing of ochratoxin A. Toxins 2014, 6, 2435–2452. [Google Scholar] [CrossRef]
- Zhang, F.; Liu, M.; Liu, R.; Li, J.; Sang, Y.; Tang, Y.; Wang, X.; Wang, S. A broad-spectrum sensing strategy for the tetracycline family of antibiotics based on an ovalbumin-stabilized gold nanocluster and its application in a pump-free microfluidic sensing platform. Biosens. Bioelectron. 2021, 171, 9. [Google Scholar] [CrossRef]
- Purschke, W.G.; Radtke, F.; Kleinjung, F.; Klussmann, S. A DNA Spiegelmer to staphylococcal enterotoxin B. Nucleic Acids Res. 2003, 31, 3027–3032. [Google Scholar] [CrossRef]
- Li, W.; Luo, Y.; Gao, T.; Yang, L.; Wang, J.; Pei, R. In vitro selection of DNA aptamers for a small-molecule porphyrin by gold nanoparticle-based SELEX. J. Mol. Evol. 2019, 87, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Kou, Q.; Wu, P.; Sun, Q.; Wu, J.; Le, T. Selection and application of DNA aptamers against sulfaquinoxaline assisted by graphene oxide-based SELEX. Food Anal. Methods 2021, 14, 250–259. [Google Scholar] [CrossRef]
- Bao, Y.; Zhu, D.; Zhao, Y.; Li, X.; Gu, C.; Yu, H. Selection and identification of high-affinity aptamer of Kunitz trypsin inhibitor and their application in rapid and specific detection. Food Sci. Nutr. 2022, 10, 953–963. [Google Scholar] [CrossRef] [PubMed]
- Hamedani, N.S.; Müller, J. Capillary electrophoresis for the selection of DNA aptamers recognizing activated protein C. Methods Mol. Biol. 2016, 1380, 61–75. [Google Scholar] [CrossRef]
- Ștefan, G.; Hosu, O.; De Wael, K.; Lobo-Castañón, M.J.; Cristea, C. Aptamers in biomedicine: Selection strategies and recent advances. Electrochim. Acta 2021, 376, 25. [Google Scholar] [CrossRef]
- Wei, H.; Cai, R.; Yue, H.; Tian, Y.; Zhou, N. Screening and application of a truncated aptamer for high-sensitive fluorescent detection of metronidazole. Anal. Chim. Acta 2020, 1128, 203–210. [Google Scholar] [CrossRef]
- Yue, H.; Chen, J.; Chen, X.; Wang, X.; Zhang, Y.; Zhou, N. Systematic screening and optimization of single-stranded DNA aptamer specific for N-acetylneuraminic acid: A comparative study. Sens. Actuators B-Chem. 2021, 344, 10. [Google Scholar] [CrossRef]
- Darmostuk, M.; Rimpelova, S.; Gbelcova, H.; Ruml, T. Current approaches in SELEX: An update to aptamer selection technology. Biotechnol. Adv. 2015, 33, 1141–1161. [Google Scholar] [CrossRef]
- Sadeghi, A.S.; Mohsenzadeh, M.; Abnous, K.; Taghdisi, S.M.; Ramezani, M. Development and characterization of DNA aptamers against florfenicol: Fabrication of a sensitive fluorescent aptasensor for specific detection of florfenicol in milk. Talanta 2018, 182, 193–201. [Google Scholar] [CrossRef]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Avci-Adali, M.; Paul, A.; Wilhelm, N.; Ziemer, G.; Wendel, H.P. Upgrading SELEX technology by using lambda exonuclease digestion for single-stranded DNA generation. Molecules 2010, 15, 1–11. [Google Scholar] [CrossRef]
- Gao, J.; Liu, N.; Zhang, X.; Yang, E.; Song, Y.; Zhang, J.; Han, Q. Utilizing the DNA aptamer to determine lethal alpha-amanitin in mushroom samples and urine by Magnetic Bead-ELISA (MELISA). Molecules 2022, 27, 538. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.-H.; Wang, J.-Y.; Li, J. Screening and structure analysis of florfenicol based aptamers based on graphene oxide SELEX technologytitis. Chin. J. Prev. Vet. Med. 2021, 43, 468–476. (In Chinese) [Google Scholar] [CrossRef]
- Li, R.; Lin, Z.-J.; Yang, J.-Y.; Xu, Z.-L.; Wang, H.; Lei, H.-T.; Sun, Y.-M.; Shen, Y.-D. An indirect competitive enzyme-linked immunosorbent assay for simultaneous determination of florfenicol and thiamphenicol in animal meat and urine. Chin. J. Anal. Chem. 2018, 46, 1321–1327. [Google Scholar] [CrossRef]
- Han, J.; Hu, L.; Yi, Y.; Liu, M.; Xia, J.; Xu, G.; Luo, K.; Wang, Q.; Lai, W. Development of colloidal gold lateral flow immunoassay for quantitative detection of florfenicol. Chin. J. Anal. Chem. 2017, 45, 1188–1194. [Google Scholar] [CrossRef]
- Chae, W.-S.; Yoo, C.-Y.; Tutkun, L.; Kim, S.; Lee, H.-J. Determination of florfenicol residues in swine tissues using high-performance liquid chromatography with ultraviolet photometric detector. J. Prev. Vet. Med. 2018, 42, 171–176. [Google Scholar] [CrossRef]
- Cheng, G.; Li, S.; Wu, K.; Deng, A.; Li, J. Highly sensitive competitive electrochemiluminescence immunosensor based on ABEI-H2O2 system with cobalt hydroxide nanosheets and bimetal PdAg as co-enhancer for detection of florfenicol. Microchim. Acta 2022, 189, 9. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Zheng, Y.; Peng, T.; Yao, K.; Xie, S.; Zhang, X.; Xia, X.; Li, J.; Jiang, H. Development of a quantitative fluorescence-based lateral flow immunoassay for determination of chloramphenicol, thiamphenicol and florfenicol in milk. Food Agric. Immunol. 2018, 29, 56–66. [Google Scholar] [CrossRef]
- Xu, M.; Qian, M.; Zhang, H.; Ma, J.; Wang, J.; Wu, H. Simultaneous determination of florfenicol with its metabolite based on modified quick, easy, cheap, effective, rugged, and safe sample pretreatment and evaluation of their degradation behavior in agricultural soils. J. Sep. Sci. 2015, 38, 211–217. [Google Scholar] [CrossRef]

| Methods | Detection Range (ng/mL) | LOD (ng/mL) | Reference |
|---|---|---|---|
| icELISA | 0.31–5.61 | 0.12 | [34] |
| Colloidal Gold Lateral Flow Immunoassay | 0.1–1.5 | 0.08 | [35] |
| UPLC | 12.5–1000 | 12 | [36] |
| Electrochemiluminescence immunoassay | 0.0001–100 | 3.1 × 10−5 | [37] |
| FMs lateral flow assay | 1.25–80 | 1.9 | [38] |
| HPLC-MS-MS | 0.1–500 | 2.9 × 10−3 | [39] |
| Nanogold Colorimetry | 0.00128–500 | 1.28 × 10−3 | This work |
| Sample | Spiked (ng/mL) | Found (ng/mL) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 0.1 | 0.11 | 112.2 | 5.6 | |
| Milk | 1 | 1.23 | 123.1 | 2.3 |
| 5 | 4.45 | 88.9 | 2.7 | |
| 0.1 | 0.09 | 92.4 | 5.1 | |
| Egg | 1 | 0.84 | 84.0 | 1.6 |
| 5 | 5.61 | 112.2 | 4.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, M.; Liu, R.; Zhang, F.; Chitrakar, B.; Wang, X. Screening of Single-Stranded DNA Aptamer Specific for Florfenicol and Application in Detection of Food Safety. Biosensors 2022, 12, 701. https://doi.org/10.3390/bios12090701
Shi M, Liu R, Zhang F, Chitrakar B, Wang X. Screening of Single-Stranded DNA Aptamer Specific for Florfenicol and Application in Detection of Food Safety. Biosensors. 2022; 12(9):701. https://doi.org/10.3390/bios12090701
Chicago/Turabian StyleShi, Minghui, Ruobing Liu, Fuyuan Zhang, Bimal Chitrakar, and Xianghong Wang. 2022. "Screening of Single-Stranded DNA Aptamer Specific for Florfenicol and Application in Detection of Food Safety" Biosensors 12, no. 9: 701. https://doi.org/10.3390/bios12090701
APA StyleShi, M., Liu, R., Zhang, F., Chitrakar, B., & Wang, X. (2022). Screening of Single-Stranded DNA Aptamer Specific for Florfenicol and Application in Detection of Food Safety. Biosensors, 12(9), 701. https://doi.org/10.3390/bios12090701





