Dielectrophoresis-Based SERS Sensors for the Detection of Cancer Cells in Microfluidic Chips
Abstract
1. Introduction
2. Materials and Methods
2.1. Cultivation and Preparation of Cancer Cells
2.2. Microfluidic System for Handling of Cancer Cells
2.3. Setup for DEP Deposition of CTCs
2.4. SERS Platforms and Measurements
2.5. Numerical Calculations
2.5.1. Theory of DEP
2.5.2. Crossover Frequencies (fc) and Distribution of Electric Field
- The pre-processing stage is where the fundamentals of the model are developed (e.g., geometrical structure) and parameters are determined (e.g., type of materials, domain and boundary conditions and meshing of the geometry). The geometrical dimensions for the calculations are taken from CAD/CAD designs or from SEM observations in the case of the steel needle (Figure 4).
- The solution stage uses mathematical equations (e.g., Partial Differential Equations) and information on the type of problem (stationary or time-dependent) to solve the problem.
- The post-processing stage is where the results are shown and the user can generate 1D, 2D or 3D plots.
2.6. Microscopic Characterization
3. Results
3.1. Numerical Calculation for the DEP Deposition of the CTCs
3.1.1. Crossover frequencies (fc) and pDEP Region
3.1.2. FEM Analysis of Electric Field Distribution
3.2. DEP-Based SERS System for Detection of CTCs
3.2.1. DEP Parameters and Its Influence on SERS Detection
3.2.2. The SERS-DEP Detection Sensitivity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Ethical Statements
References
- Descamps, L.; Le Roy, D.; Deman, A.L. Microfluidic-based technologies for CTC isolation: A review of 10 years of intense efforts towards liquid biopsy. Int. J. Mol. Sci. 2022, 23, 1981. [Google Scholar] [CrossRef]
- Cen, P.; Ni, X.; Yang, J.; Graham, D.Y.; Li, M. Circulating tumor cells in the diagnosis and management of pancreatic cancer. Biochim. Biophys. Acta Rev. Cancer 2012, 1826, 350–356. [Google Scholar] [CrossRef][Green Version]
- Qi, L.N.; De Xiang, B.; Wu, F.X.; Ye, J.Z.; Zhong, J.H.; Wang, Y.Y.; Chen, Y.Y.; Chen, Z.S.; Ma, L.; Chen, J.; et al. Circulating tumor cells undergoing emt provide a metric for diagnosis and prognosis of patients with hepatocellular carcinoma. Cancer Res. 2018, 78, 4731–4744. [Google Scholar] [CrossRef]
- Wang, S.; Liu, K.; Liu, J.; Yu, Z.T.F.; Xu, X.; Zhao, L.; Lee, T.; Lee, E.K.; Reiss, J.; Lee, Y.K.; et al. Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem. Int. Ed. 2011, 50, 3084–3088. [Google Scholar] [CrossRef]
- Edd, J.F.; Mishra, A.; Smith, K.C.; Kapur, R.; Maheswaran, S.; Haber, D.A.; Toner, M. Isolation of circulating tumor cells. iScience 2022, 25, 104696. [Google Scholar] [CrossRef]
- Bankó, P.; Lee, S.Y.; Nagygyörgy, V.; Zrínyi, M.; Chae, C.H.; Cho, D.H.; Telekes, A. Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48. [Google Scholar] [CrossRef]
- Coumans, F.A.W.; van Dalum, G.; Beck, M.; Terstappen, L.W.M.M. Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS ONE 2013, 8, e61770. [Google Scholar] [CrossRef]
- Andergassen, U.; Kölbl, A.C.; Mahner, S.; Jeschke, U. Real-time RT-PCR systems for CTC detection from blood samples of breast cancer and gynaecological tumour patients (Review). Oncol. Rep. 2016, 35, 1905–1915. [Google Scholar] [CrossRef]
- Hillig, T.; Nygaard, A.; Brems-Eskildsen, A.S.; Langkjer, S.T.T.; Sölétormos, G. circulating tumor cells analysed by scanning fluorescence microscopy in the xena trial. Ann. Oncol. 2014, 25, iv132. [Google Scholar] [CrossRef][Green Version]
- Bhagwat, N.; Dulmage, K.; Pletcher, C.H.; Wang, L.; DeMuth, W.; Sen, M.; Balli, D.; Yee, S.S.; Sa, S.; Tong, F.; et al. An integrated flow cytometry-based platform for isolation and molecular characterization of circulating tumor single cells and clusters. Sci. Rep. 2018, 8, 5035. [Google Scholar] [CrossRef]
- Lee, S.K.; Kim, G.S.; Wu, Y.; Kim, D.J.; Lu, Y.; Kwak, M.; Han, L.; Hyung, J.H.; Seol, J.K.; Sander, C.; et al. Nanowire substrate-based laser scanning cytometry for quantitation of circulating tumor cells. Nano Lett. 2012, 12, 2697–2704. [Google Scholar] [CrossRef][Green Version]
- Chen, W.; Zhang, J.; Huang, L.; Chen, L.; Zhou, Y.; Tang, D.; Xie, Y.; Wang, H.; Huang, C. Detection of HER2-positive circulating tumor cells using the liquid biopsy system in breast cancer. Clin. Breast Cancer 2019, 19, 239–285. [Google Scholar] [CrossRef]
- Baker, C.A.; Duong, C.T.; Grimley, A.; Roper, M.G. Recent advances in microfluidic detection systems. Bioanalysis 2009, 1, 967–975. [Google Scholar] [CrossRef]
- Low, W.S.; Wan Abas, W.A.B. Benchtop technologies for circulating tumor cells separation based on biophysical properties. Biomed. Res. Int. 2015, 2015, 239362. [Google Scholar] [CrossRef]
- Nasiri, R.; Shamloo, A.; Ahadian, S.; Amirifar, L.; Akbari, J.; Goudie, M.J.; Lee, K.J.; Ashammakhi, N.; Dokmeci, M.R.; Di Carlo, D.; et al. Microfluidic-based approaches in targeted cell/particle separation based on physical properties: Fundamentals and applications. Small 2020, 16, 2000171. [Google Scholar] [CrossRef]
- Ahn, J.C.; Teng, P.C.; Chen, P.J.; Posadas, E.; Tseng, H.R.; Lu, S.C.; Yang, J.D. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology 2021, 73, 422–436. [Google Scholar] [CrossRef]
- Russo, G.I.; Musso, N.; Romano, A.; Caruso, G.; Petralia, S.; Lanzanò, L.; Broggi, G.; Camarda, M. The role of dielectrophoresis for cancer diagnosis and prognosis. Cancers 2022, 14, 198. [Google Scholar] [CrossRef]
- Chiriac, E.; Avram, M.; Bălan, C. Manipulation of particles using dielectrophoresis in a microchannel. Rom. J. Inf. Sci. Technol. 2021, 24, 213–221. [Google Scholar]
- Shirmohammadli, V.; Manavizadeh, N. Application of differential electrodes in a dielectrophoresis-based device for cell separation. IEEE Trans. Electron. Devices 2019, 66, 4075–4080. [Google Scholar] [CrossRef]
- Aghaamoo, M.; Aghilinejad, A.; Chen, X.; Xu, J. On the design of deterministic dielectrophoresis for continuous separation of circulating tumor cells from peripheral blood cells. Electrophoresis 2019, 40, 1486–1493. [Google Scholar] [CrossRef]
- Farahinia, A.; Zhang, W.J.; Badea, I. Novel microfluidic approaches to circulating tumor cell separation and sorting of blood cells: A review. J. Sci. Adv. Mater. Devices 2021, 6, 303–320. [Google Scholar] [CrossRef]
- Chiu, T.K.; Chou, W.P.; Bin Huang, S.; Wang, H.M.; Lin, Y.C.; Hsieh, C.H.; Wu, M.H. Application of optically-induced-dielectrophoresis in microfluidic system for purification of circulating tumour cells for gene expression analysis-cancer cell line model. Sci. Rep. 2016, 6, 32851. [Google Scholar] [CrossRef]
- Moon, H.S.; Kwon, K.; Il Kim, S.; Han, H.; Sohn, J.; Lee, S.; Il Jung, H. continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125. [Google Scholar] [CrossRef]
- Hajba, L.; Guttman, A. Circulating tumor-cell detection and capture using microfluidic devices. TrAC Trends Anal. Chem. 2014, 59, 9–16. [Google Scholar] [CrossRef]
- Pødenphant, M.; Ashley, N.; Koprowska, K.; Mir, K.U.; Zalkovskij, M.; Bilenberg, B.; Bodmer, W.; Kristensen, A.; Marie, R. Separation of cancer cells from white blood cells by pinched flow fractionation. Lab Chip 2015, 15, 4598–4606. [Google Scholar] [CrossRef]
- Zhu, S.; Jiang, F.; Han, Y.; Xiang, N.; Ni, Z. Microfluidics for label-free sorting of rare circulating tumor cells. Analyst 2020, 145, 7103–7124. [Google Scholar] [CrossRef]
- Hu, D.; Liu, H.; Tian, Y.; Li, Z.; Cui, X. Sorting technology for circulating tumor cells based on microfluidics. ACS Comb. Sci. 2020, 22, 701–711. [Google Scholar] [CrossRef]
- Augustsson, P.; Magnusson, C.; Nordin, M.; Lilja, H.; Laurell, T. Microfluidic, label-free enrichment of prostate cancer cells in blood based on acoustophoresis. Anal. Chem. 2012, 84, 7954–7962. [Google Scholar] [CrossRef]
- Park, M.H.; Reátegui, E.; Li, W.; Tessier, S.N.; Wong, K.H.K.; Jensen, A.E.; Thapar, V.; Ting, D.; Toner, M.; Stott, S.L.; et al. Enhanced isolation and release of circulating tumor cells using nanoparticle binding and ligand exchange in a microfluidic chip. J. Am. Chem. Soc. 2017, 139, 2741–2749. [Google Scholar] [CrossRef]
- Pohl, H.A. The motion and precipitation of suspensoids in divergent electric fields. J. Appl. Phys. 1951, 22, 869–871. [Google Scholar] [CrossRef]
- Nerguizian, V.; Stiharu, I.; Al-Azzam, N.; Yassine-Diab, B.; Alazzam, A. The effect of dielectrophoresis on living cells: Crossover frequencies and deregulation in gene expression. Analyst 2019, 144, 3853–3860. [Google Scholar] [CrossRef]
- Yao, J.; Zhu, G.; Zhao, T.; Takei, M. Microfluidic device embedding electrodes for dielectrophoretic manipulation of cells-A. Electrophoresis 2019, 40, 1166–1177. [Google Scholar] [CrossRef]
- Jubery, T.Z.; Srivastava, S.K.; Dutta, P. Dielectrophoretic separation of bioparticles in microdevices: A review. Electrophoresis 2014, 35, 691–713. [Google Scholar] [CrossRef]
- Mathew, B.; Alazzam, A.; Abutayeh, M.; Stiharu, I. Model-based analysis of a dielectrophoretic microfluidic device for field-flow fractionation. J. Sep. Sci. 2016, 39, 3028–3036. [Google Scholar] [CrossRef]
- Adekanmbi, E.O.; Srivastava, S.K. Dielectrophoretic applications for disease diagnostics using lab-on-a-chip platforms. Lab Chip 2016, 16, 2148–2167. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S. Isolation of circulating tumor cells by dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef]
- Pethig, R. Review article—Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 022811. [Google Scholar] [CrossRef]
- Pohl, H.A.; Hawk, I. Separation of living and dead cells by dielectrophoresis. Science 1966, 152, 647–649. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Noshari, J.; Anderson, T.J.; Becker, F.F. Isolation of rare cells from cell mixtures by dielectrophoresis peter. Electrophoresis 2009, 30, 1388–1398. [Google Scholar] [CrossRef]
- Labeed, F.H.; Coley, H.M.; Thomas, H.; Hughes, M.P. Assessment of multidrug resistance reversal using dielectrophoresis and flow cytometry. Biophys. J. 2003, 85, 2028–2034. [Google Scholar] [CrossRef]
- Yang, F.; Yang, X.; Jiang, H.; Butler, W.M.; Wang, G. Dielectrophoretic separation of prostate cancer cells. Technol. Cancer Res. Treat. 2013, 12, 61–70. [Google Scholar] [CrossRef]
- Çağlayan Arslan, Z.; Demircan Yalçın, Y.; Külah, H. Label-free enrichment of MCF7 breast cancer cells from leukocytes using continuous flow dielectrophoresis. Electrophoresis 2022, 43, 1531–1544. [Google Scholar] [CrossRef]
- Cheng, I.F.; Huang, W.L.; Chen, T.Y.; Liu, C.W.; De Lin, Y.; Su, W.C. Antibody-free isolation of rare cancer cells from blood based on 3D lateral dielectrophoresis. Lab Chip 2015, 15, 2950–2959. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S.; Noshari, J.; Becker, F.F.; Stemke-Hale, K. Correlations between the dielectric properties and exterior morphology of cells revealed by dielectrophoretic field-flow fractionation. Wiley Online Libr. 2013, 34, 1042–1050. [Google Scholar] [CrossRef]
- Williams, A.; Balic, M.; Datar, R.; Cote, R. Size-based enrichment technologies for CTC detection and characterization. Recent Results Cancer Res. 2012, 195, 87–95. [Google Scholar] [CrossRef]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Di Size-selective collection of circulating tumor cells using Vortex technology. Lab Chip 2014, 14, 63. [Google Scholar] [CrossRef]
- De Giorgi, V.; Pinzani, P.; Salvianti, F.; Panelos, J.; Paglierani, M.; Janowska, A.; Grazzini, M.; Wechsler, J.; Orlando, C.; Santucci, M.; et al. Application of a Filtration- and Isolation-by-Size Technique for the Detection of Circulating Tumor Cells in Cutaneous Melanoma. J. Invest. Dermatol. 2010, 130, 2440–2447. [Google Scholar] [CrossRef]
- Mostert, B.; Sleijfer, S.; Foekens, J.A.; Gratama, J.W. Circulating tumor cells (CTCs): Detection methods and their clinical relevance in breast cancer. Cancer Treat. Rev. 2009, 35, 463–474. [Google Scholar] [CrossRef]
- Ma, Y.C.; Wang, L.; Yu, F.L. Recent advances and prospects in the isolation by size of epithelial tumor cells (ISET) methodology. Technol. Cancer Res. Treat. 2013, 12, 295–309. [Google Scholar] [CrossRef]
- Zhang, J.; Yuan, D.; Zhao, Q.; Yan, S.; Tang, S.Y.; Tan, S.H.; Guo, J.; Xia, H.; Nguyen, N.T.; Li, W. Tunable particle separation in a hybrid dielectrophoresis (DEP)- inertial microfluidic device. Sensors Actuators B Chem. 2018, 267, 14–25. [Google Scholar] [CrossRef]
- Murat Karabacak, N.; Spuhler, P.S.; Fachin, F.; Lim, E.J.; Pai, V.; Ozkumur, E.; Martel, J.M.; Kojic, N.; Smith, K.; Chen, P.; et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat. Protoc. 2013, 9, 694–710. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Amstislavskaya, T.G.; Chen, G.H.; Chang, H.H.; Chen, Y.H.; Jen, C.P. Selectively concentrating cervical carcinoma cells from red blood cells utilizing dielectrophoresis with circular ITO electrodes in stepping electric fields. J. Med. Biol. Eng. 2013, 33, 51–58. [Google Scholar] [CrossRef]
- Varmazyari, V.; Habibiyan, H.; Ghafoorifard, H.; Ebrahimi, M.; Ghafouri-Fard, S. A dielectrophoresis-based microfluidic system having double-sided optimized 3D electrodes for label-free cancer cell separation with preserving cell viability. Sci. Rep. 2022, 12, 12100. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.V.; Le Manh, T.; Nguyen, T.S.; Le, V.T.; Van Hieu, N. Applied electric field analysis and numerical investigations of the continuous cell separation in a dielectrophoresis-based microfluidic channel. J. Sci. Adv. Mater. Devices 2021, 6, 11–18. [Google Scholar] [CrossRef]
- Alshareef, M.; Metrakos, N.; Juarez Perez, E.; Azer, F.; Yang, F.; Yang, X.; Wang, G. Separation of tumor cells with dielec-trophoresis-based microfluidic chip. Biomicrofluidics 2013, 7, 011803. [Google Scholar] [CrossRef]
- DeMello, A.J. Control and detection of chemical reactions in microfluidic systems. Nature 2006, 442, 394–402. [Google Scholar] [CrossRef]
- Cho, S.K.; Moon, H.; Kim, C.J. Creating, transporting, cutting, and merging liquid droplets by electrowetting-based actuation for digital microfluidic circuits. J. Microelectromech. Syst. 2003, 12, 70–80. [Google Scholar] [CrossRef]
- Song, H.; Chen, D.L.; Ismagilov, R.F. Reactions in droplets in microfluidic channels. Angew. Chem. Int. Ed. 2006, 45, 7336–7356. [Google Scholar] [CrossRef]
- Teh, S.Y.; Lin, R.; Hung, L.H.; Lee, A.P. Droplet microfluidics. Lab Chip 2008, 8, 198–220. [Google Scholar] [CrossRef]
- Cha, H.; Fallahi, H.; Dai, Y.; Yuan, D.; An, H.; Nguyen, N.T.; Zhang, J. Multiphysics microfluidics for cell manipulation and separation: A review. Lab Chip 2022, 22, 423–444. [Google Scholar] [CrossRef]
- Kamińska, A.; Winkler, K.; Kowalska, A.; Witkowska, E.; Szymborski, T.; Janeczek, A.; Waluk, J. SERS-based immunoassay in a microfluidic system for the multiplexed recognition of interleukins from blood plasma: Towards picogram detection. Sci. Rep. 2017, 7, 10656. [Google Scholar] [CrossRef] [PubMed]
- Czaplicka, M.; Niciński, K.; Nowicka, A.; Szymborski, T.; Chmielewski, I.; Trzcińska-Danielewicz, J.; Girstun, A.; Kamińska, A. Effect of varying expression of EpCAM on the efficiency of CTCs detection by SERS-based immunomagnetic optofluidic device. Cancers 2020, 12, 3315. [Google Scholar] [CrossRef] [PubMed]
- Szymborski, T.; Stepanenko, Y.; Nicinski, K.; Piecyk, P.; Berus, S.M.; Adamczyk-Poplawska, M.; Kaminska, A. Ultrasensitive SERS platform made via femtosecond laser micromachining for biomedical applications. J. Mater. Res. Technol. 2021, 12, 1496–1507. [Google Scholar] [CrossRef]
- Pethig, R. Dielectrophoresis, 1st ed.; Wiley: Hoboken, NJ, USA, 2017; ISBN 9781118671450. [Google Scholar]
- Cottet, J.; Fabregue, O.; Berger, C.; Buret, F.; Renaud, P.; Frénéa-Robin, M. MyDEP: A new computational tool for dielectric modeling of particles and cells. Biophys. J. 2019, 116, 12–18. [Google Scholar] [CrossRef]
- Luna-Pineda, T.; Soto-Feliciano, K.; De La Cruz-Montoya, E.; Pacheco Londoño, L.C.; Ríos-Velázquez, C.; Hernández-Rivera, S.P. Spectroscopic characterization of biological agents using FTIR, normal Raman and surface-enhanced Raman spectroscopies. Chem. Biol. Sens. VIII 2007, 6554, 65540K. [Google Scholar] [CrossRef]
- Demirel, M.C.; Kao, P.; Malvadkar, N.; Wang, H.; Gong, X.; Poss, M.; Allara, D.L. Bio-organism sensing via surface enhanced raman spectroscopy on controlled metal/polymer nanostructured substrates. Biointerphases 2009, 4, 35–41. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Uzunbajakava, N.; Lenferink, A.; Kraan, Y.; Willekens, B.; Vrensen, G.; Greve, J.; Otto, C. Nonresonant Raman imaging of protein distribution in single human cells. Biopolym. Biospectroscopy Sect. 2003, 72, 1–9. [Google Scholar] [CrossRef]
- Notingher, I.; Verrier, S.; Romanska, H.; Bishop, A.E.; Polak, J.M.; Hench, L.L. In situ characterisation of living cells by Raman spectroscopy. Spectroscopy 2002, 16, 43–51. [Google Scholar] [CrossRef]
- Taleb, A.; Diamond, J.; McGarvey, J.J.; Beattie, J.R.; Toland, C.; Hamilton, P.W. Raman microscopy for the chemometric analysis of tumor cells. J. Phys. Chem. B 2006, 110, 19625–19631. [Google Scholar] [CrossRef]
- El-Said, W.A.; Kim, T.H.; Kim, H.; Choi, J.W. Analysis of intracellular state based on controlled 3D nanostructures mediated surface enhanced Raman scattering. PLoS ONE 2011, 6, e15836. [Google Scholar] [CrossRef] [PubMed]
- Henslee, E.A.; Sano, M.B.; Rojas, A.D.; Schmelz, E.M.; Davalos, R.V. Selective concentration of human cancer cells using contactless dielectrophoresis. Electrophoresis 2011, 32, 2523–2529. [Google Scholar] [CrossRef]
- Wang, X.B.; Huang, Y.; Wang, X.; Becker, F.F.; Gascoyne, P.R.C. Dielectrophoretic manipulation of cells with spiral electrodes. Biophys. J. 1997, 72, 1887–1899. [Google Scholar] [CrossRef]

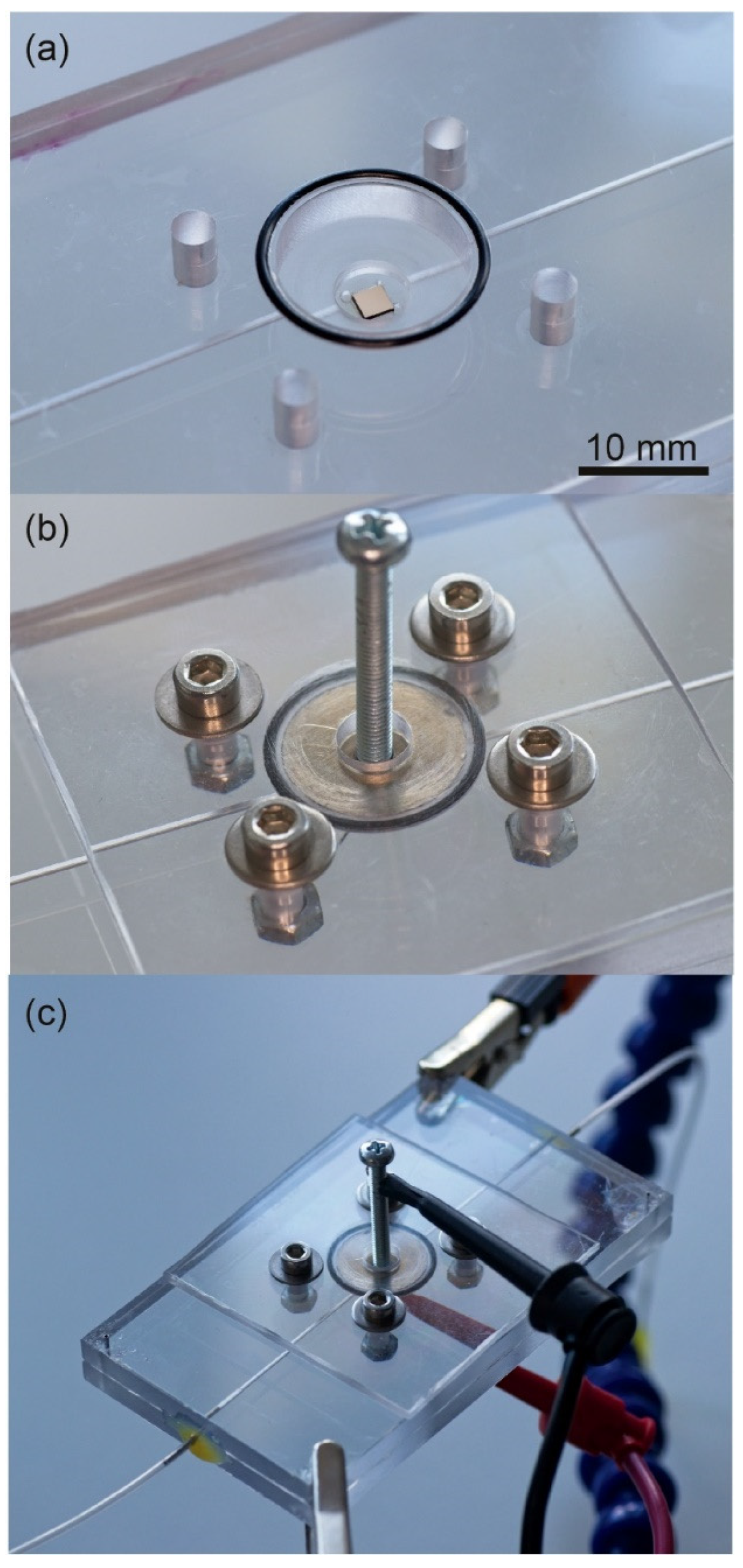
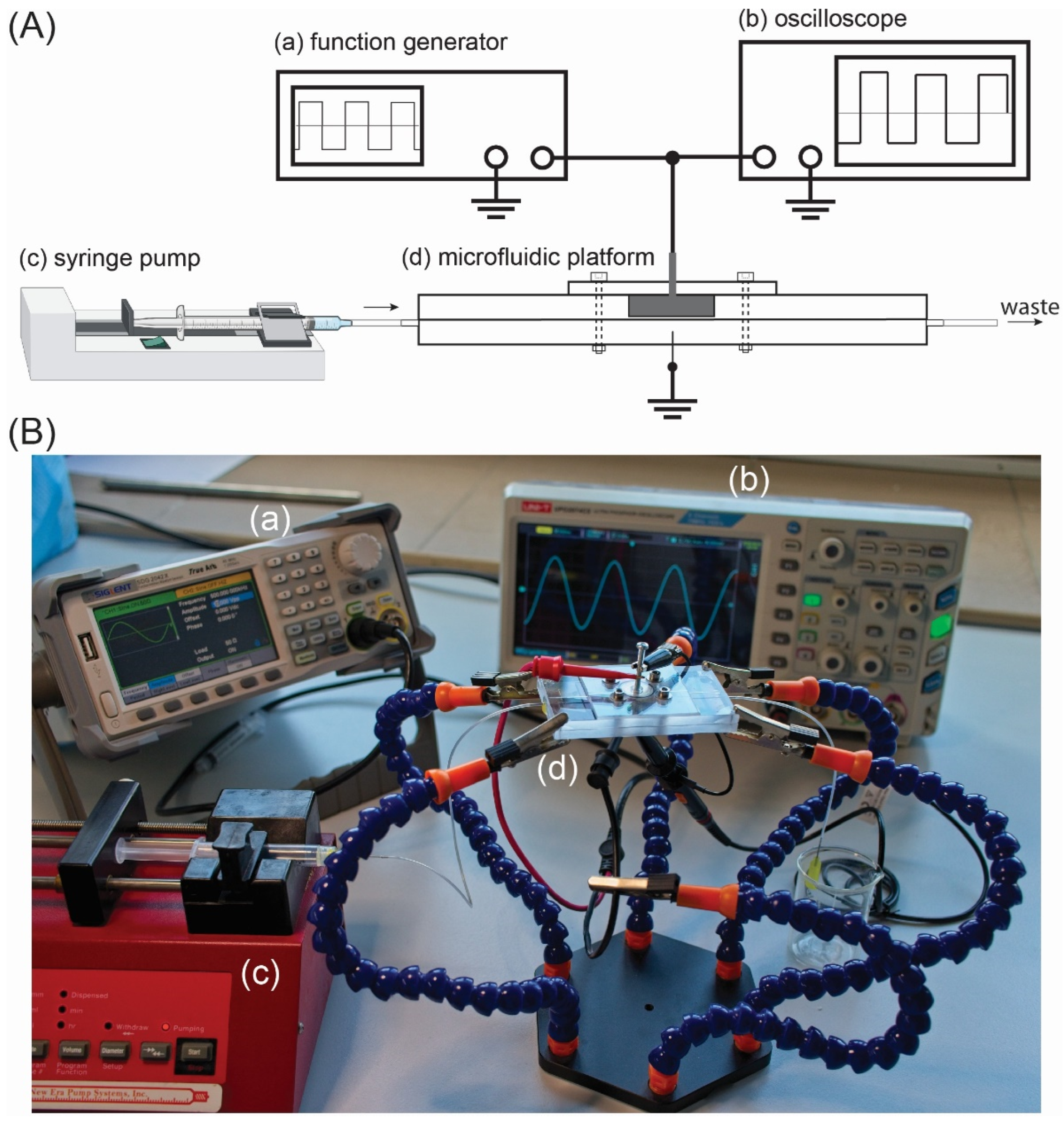
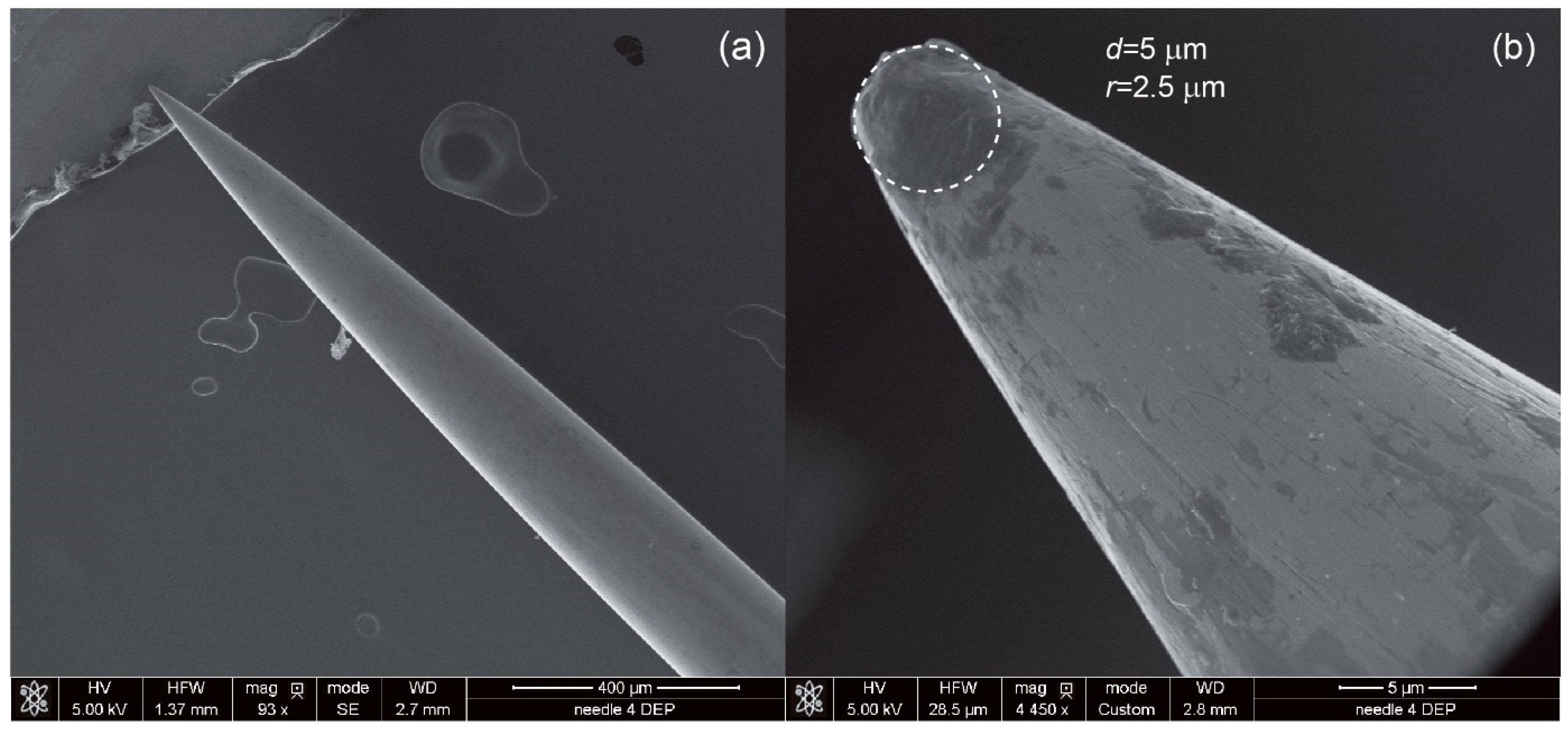
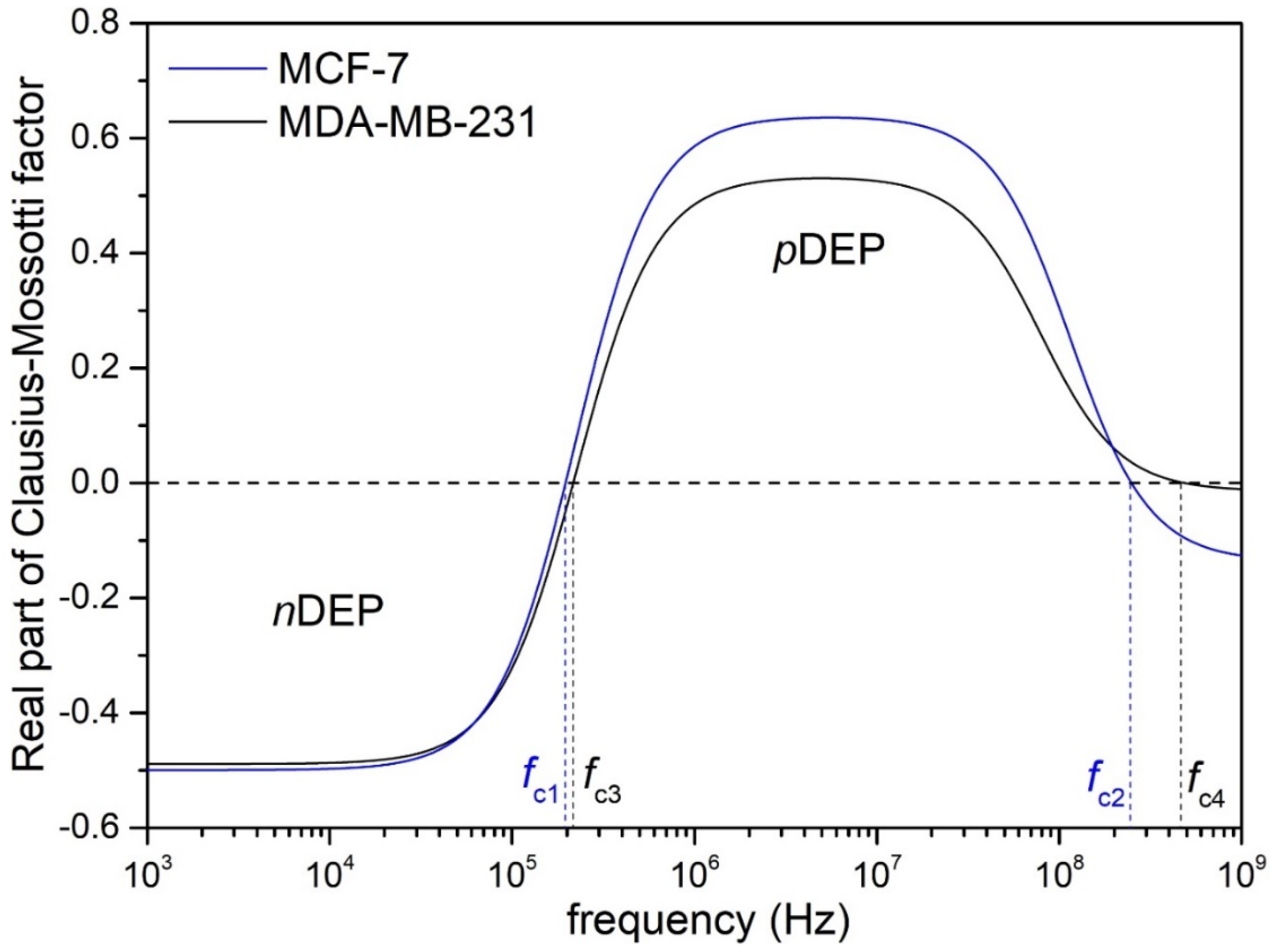
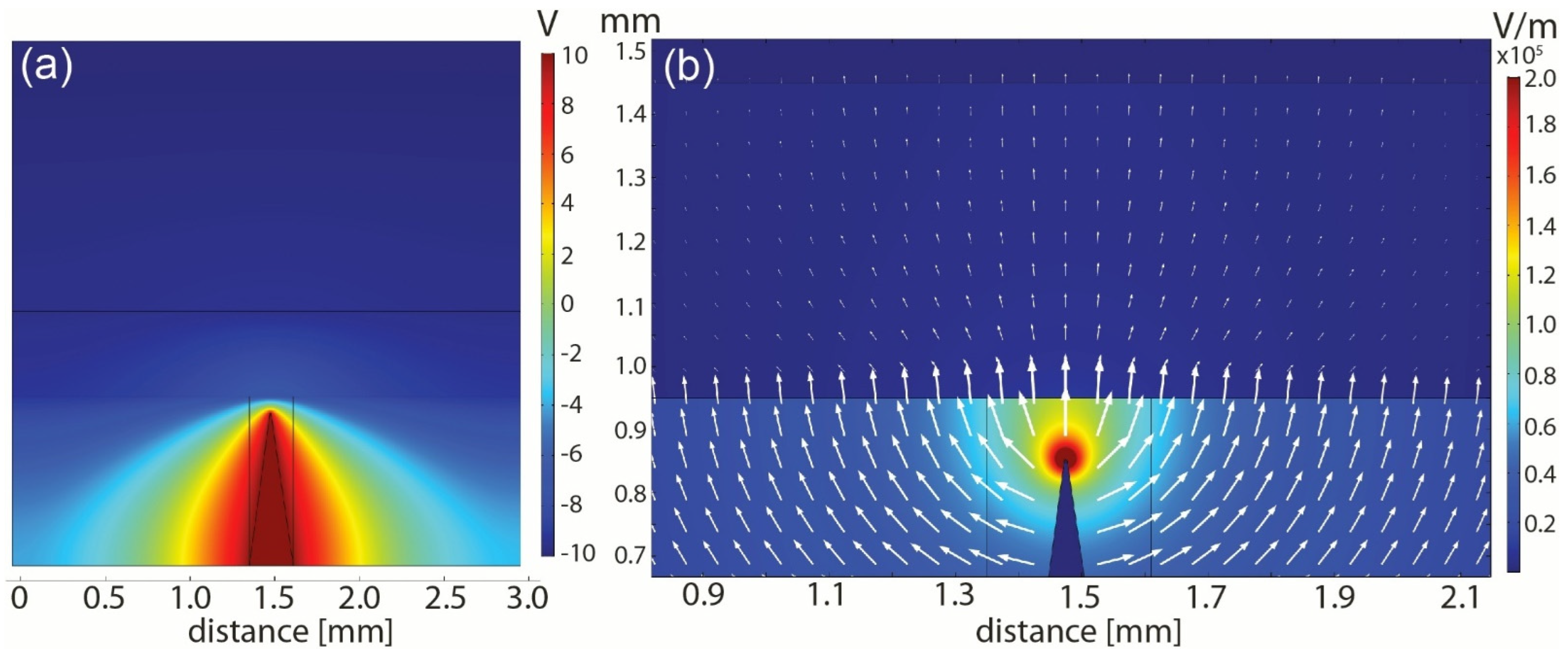


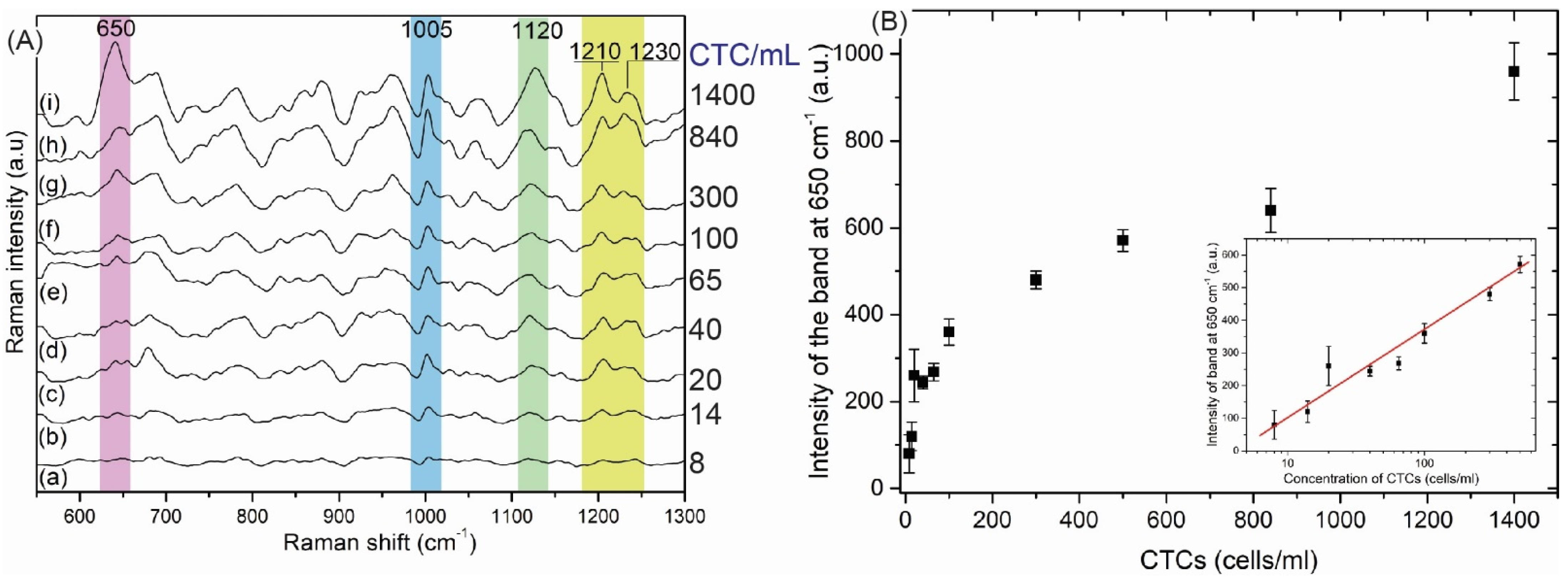
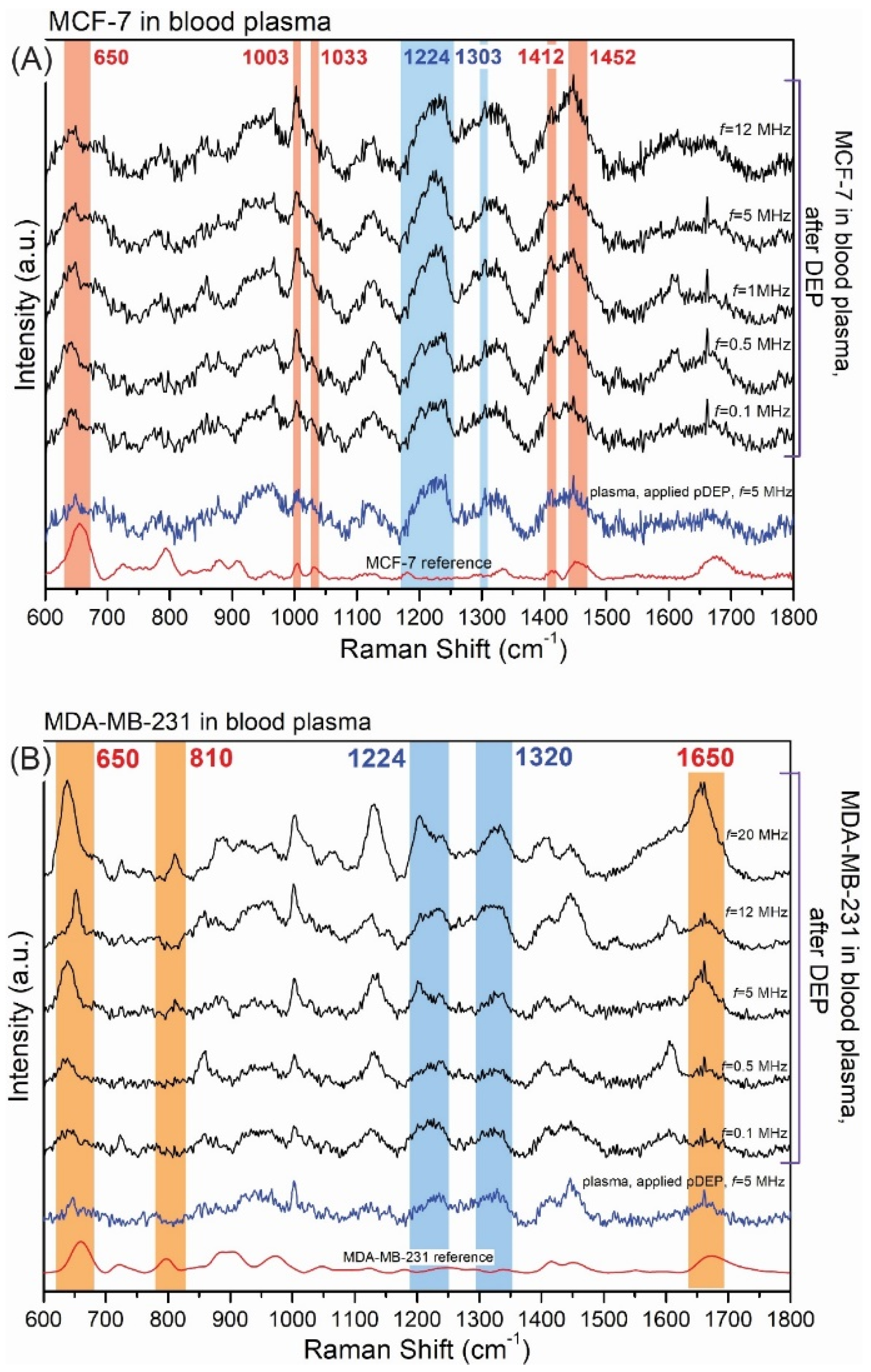
| Type of CTCs | fc1 (kHz) | fc2 (MHz) |
|---|---|---|
| MCF-7 | 196.83 | 248.64 |
| MDA-MB-231 | 216.82 | 482.33 |
| Observed SERS Band (cm−1) | Protein | Lipids | Nucleic Acid | MCF-7 | MDA-MB-231 |
|---|---|---|---|---|---|
| 650 | Tyr (C-C twist) | +++ | +++ | ||
| 720–730 | Trp | C-N head group choline (H3C)3N+ | A | +++ | +++ |
| 790 | PO2 symm | +++ | +++ | ||
| 827 | Structural protein modes of tumors | + | ++ | ||
| 850–880 | Tyr, Pro | +++ | ++ | ||
| 910 | C-C str alpha-helix, Pro, Val | +++ | + | ||
| 960 | CH3 def | CH3 def | + | ++ | |
| 1005 | Phe | ++ | +++ | ||
| 1030 | Phe | CH2CH3 bending modes of lipids | ++ | ++ | |
| 1090 | C-N stretch | CC str chain, C-O str | PO2 symm | ++ | + |
| 1120 | C-N str bk | porphyrin | ++ | ++ | |
| 1210 | C-C6H5 str in phenylalanine tyrosine and Amide III (beta sheet) | + | ++ | ||
| 1230–1240 | Amide III | ++ | ++ | ||
| 1265–1270 | Amide III (random coil) | =CH def | |||
| 1300–1308 | CH3 def, collagen | CH3CH2 twist | G | ++ | ++ |
| 1335 | A, G | ++ | ++ | ||
| 1414–1418 | C=C stretching in quinoid ring | ++ | ++ | ||
| 1445–1450 | structural protein modes of tumors | +++ | +++ | ||
| 1552 | A, G | + | - | ||
| 1585–1600 | Phe, Tyr | + | + | ||
| 1650–1665 | Amide I | C=C str | +++ | ++ |
| Cancer Cell | Intensity of Band at 650 cm−1 (cps) | Aspect Ratio (IpDEP/InoDEP) | |
|---|---|---|---|
| pDEP | no DEP | ||
| MCF-7 | 3800 | 290 | 13.1 |
| MDA-MB-231 | 1600 | 610 | 2.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szymborski, T.R.; Czaplicka, M.; Nowicka, A.B.; Trzcińska-Danielewicz, J.; Girstun, A.; Kamińska, A. Dielectrophoresis-Based SERS Sensors for the Detection of Cancer Cells in Microfluidic Chips. Biosensors 2022, 12, 681. https://doi.org/10.3390/bios12090681
Szymborski TR, Czaplicka M, Nowicka AB, Trzcińska-Danielewicz J, Girstun A, Kamińska A. Dielectrophoresis-Based SERS Sensors for the Detection of Cancer Cells in Microfluidic Chips. Biosensors. 2022; 12(9):681. https://doi.org/10.3390/bios12090681
Chicago/Turabian StyleSzymborski, Tomasz R., Marta Czaplicka, Ariadna B. Nowicka, Joanna Trzcińska-Danielewicz, Agnieszka Girstun, and Agnieszka Kamińska. 2022. "Dielectrophoresis-Based SERS Sensors for the Detection of Cancer Cells in Microfluidic Chips" Biosensors 12, no. 9: 681. https://doi.org/10.3390/bios12090681
APA StyleSzymborski, T. R., Czaplicka, M., Nowicka, A. B., Trzcińska-Danielewicz, J., Girstun, A., & Kamińska, A. (2022). Dielectrophoresis-Based SERS Sensors for the Detection of Cancer Cells in Microfluidic Chips. Biosensors, 12(9), 681. https://doi.org/10.3390/bios12090681







