Portable Electrochemical Biosensors Based on Microcontrollers for Detection of Viruses: A Review
Abstract
1. Introduction
| LAMP | CRISPR | ELISA | qPCR | Electrochemical | |
|---|---|---|---|---|---|
| Temperature | 65 °C | 37 °C | Room temperature | 3 temperatures: 95 °C, 60 °C and 70 °C | Room temperature |
| Material to enhance electrochemical signal | Fluorescent dye, active nanoparticles, magnetic beads or redox materials | Viral protein | Viral nucleic acid | Viral particle/nucleic acid | |
| Nucleic acid primer bases | Specific forward and backward internal primers with at least 100 bases in length to match with the virus-targeted gene sequence | CRISPR RNA designed to match a specific sequence of the desired target gene or region in the virus | Antibody and ligand binding | 3–4 h Forward and reverse labelled primers | Less than 1 h Hybridization of complementary DNA probe |
| Advantages | Low cost Specificity Highly effective detection Rapid monitoring strategies | Highly effective detection and rapid monitoring strategies | Specificity through antibody affinity | Specific and highly sensitive signal, shows real-time infection | Low cost, i.e., it can be reprinted and reused. Specificity through full-match hybridization, simplicity of miniaturization |
| Disadvantages | A heat block is needed for on-site detection, which might be solved by embedding a fluidic channel or mini reactor coupled with a heat regulator | Expensive RNA unstable in nature, setting for enzymatic reaction, Well-trained personnel | Not at the time of infection; time is required for antibody production | A qPCR terminal cycler is needed for on-site detection, setting for enzymatic reaction, Well-trained personnel | Requires a portable detector for on-site application, orientation of the immobilized ssDNA influence the result |
| References | [8,10,20,21,22] | [14,15,16,17,18] | [12,13] | [3] | [23,24,25] |
2. Detection of Viruses Using Electrochemical Biosensors
2.1. Theory of Electrochemical Biosensors
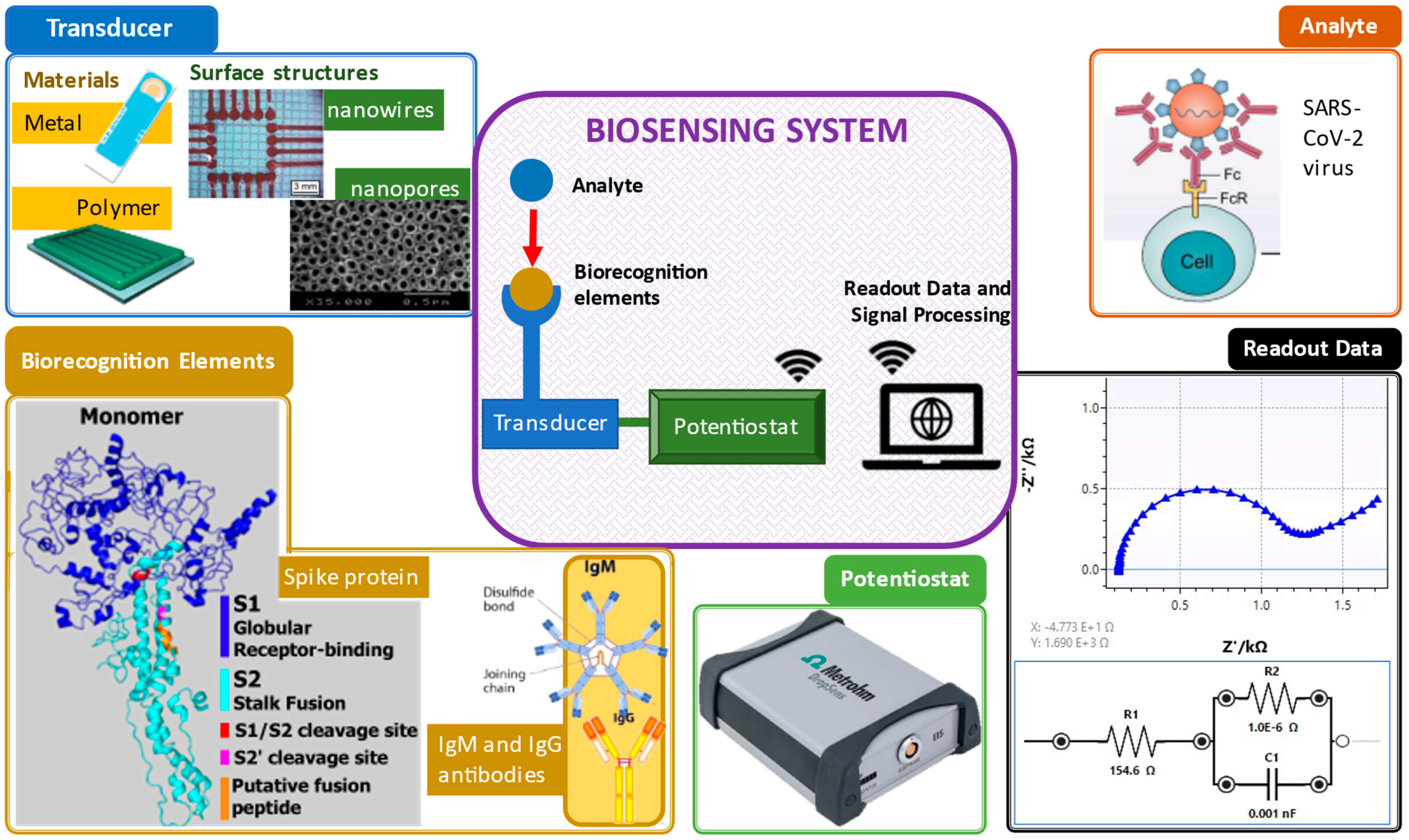
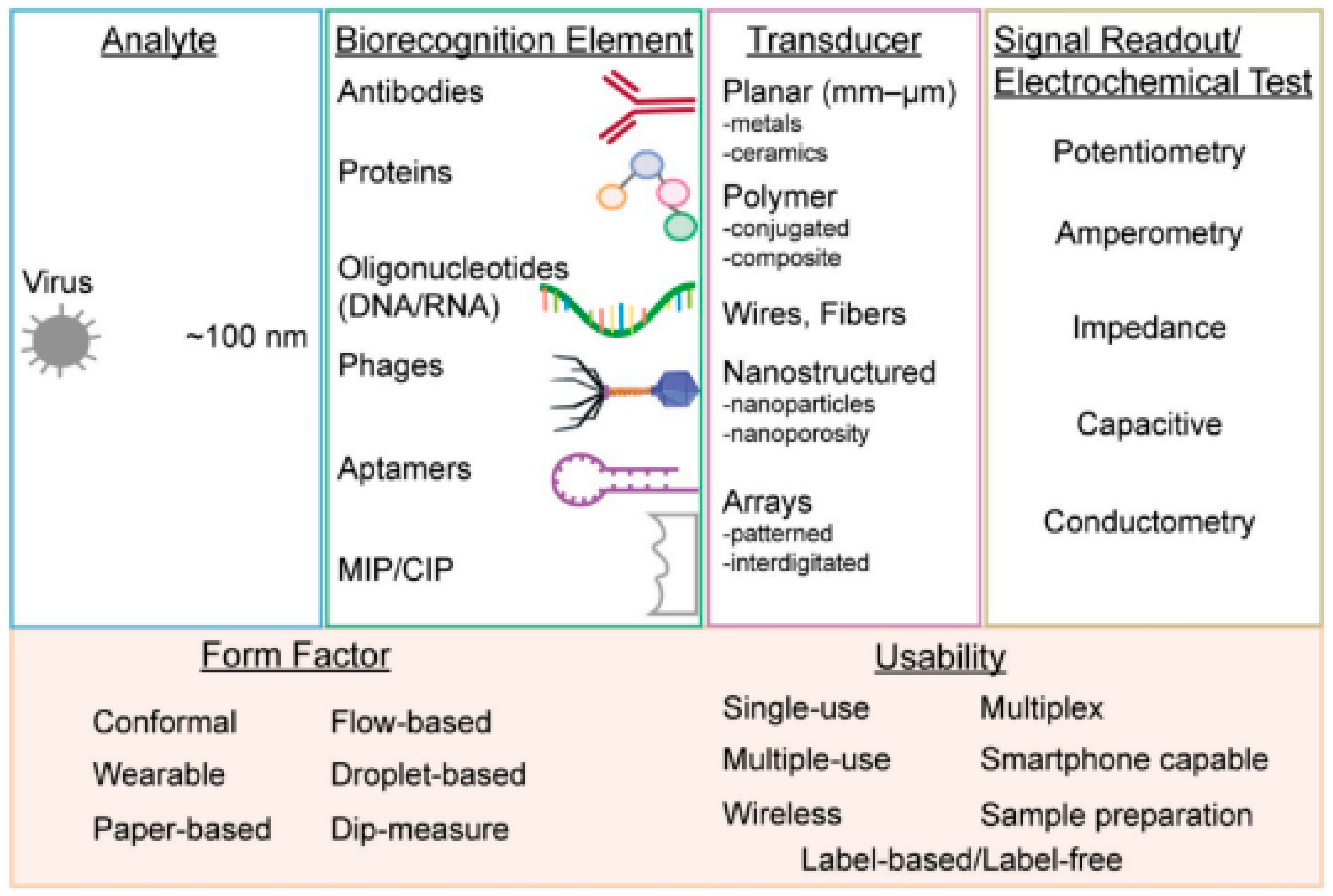
2.2. Important Design Considerations for Detection of Viruses
2.2.1. Transducing Elements
2.2.2. Biorecognition Elements
3. Point-of-Care Testing for Viruses Using Portable Potentiostats
3.1. Measurement Techniques
3.2. Wireless versus Wired Potentiostats
3.3. Microcontroller
4. Microfluidic Systems for Electrochemical Measurement Using a Portable Potentiostat
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bedenham, G.; Kirk, A.; Luhano, U.; Shields, A. The importance of biodiversity risks: Link to zoonotic diseases. Br. Actuar. J. 2022, 27, 1–9. [Google Scholar] [CrossRef]
- Roy, B.; Dhillon, J.; Habib, N.; Pugazhandhi, B. Global variants of COVID-19: Current understanding. J. Biomed. Sci. 2021, 8, 8–11. [Google Scholar] [CrossRef]
- Long, Q.-X.; Liu, B.-Z.; Deng, H.-J.; Wu, G.-C.; Deng, K.; Chen, Y.-K.; Liao, P.; Qiu, J.-F.; Lin, Y.; Cai, X.-F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef]
- Zhao, Z.; Huang, C.; Huang, Z.; Lin, F.; He, Q.; Tao, D.; Jaffrezic-Renault, N.; Guo, Z. Advancements in electrochemical biosensing for respiratory virus detection: A review. TrAC Trends Anal. Chem. 2021, 139, 116253. [Google Scholar] [CrossRef] [PubMed]
- Park, G.-S.; Ku, K.; Baek, S.-H.; Kim, S.-J.; Kim, S.I.; Kim, B.-T.; Maeng, J.-S. Development of Reverse Transcription Loop-Mediated Isothermal Amplification Assays Targeting Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 2020, 22, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Sun, X.; Dai, Z.; Gao, Y.; Gong, X.; Zhou, B.; Wu, J.; Wen, W. Point-of-care testing detection methods for COVID-19. Lab Chip 2021, 21, 1634–1660. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef]
- Parida, M.M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef]
- Nzelu, C.O.; Gomez, E.A.; Cáceres, A.G.; Sakurai, T.; Martini-Robles, L.; Uezato, H.; Mimori, T.; Katakura, K.; Hashiguchi, Y.; Kato, H. Development of a loop-mediated isothermal amplification method for rapid mass-screening of sand flies for Leishmania infection. Acta Trop. 2014, 132, 1–6. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Ibarra-Meneses, A.V.; Cruz, I.; Chicharro, C.; Sánchez, C.; Biéler, S.; Broger, T.; Moreno, J.; Carrillo, E. Evaluation of fluorimetry and direct visualization to interpret results of a loop-mediated isothermal amplification kit to detect Leishmania DNA. Parasites Vectors 2018, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Cui, F.; Zhou, H.S. Diagnostic methods and potential portable biosensors for coronavirus disease 2019. Biosens. Bioelectron. 2020, 165, 112349. [Google Scholar] [CrossRef]
- Huang, L.; Ding, L.; Zhou, J.; Chen, S.; Chen, F.; Zhao, C.; Xu, J.; Hu, W.; Ji, J.; Xu, H.; et al. One-step rapid quantification of SARS-CoV-2 virus particles via low-cost nanoplasmonic sensors in generic microplate reader and point-of-care device. Biosens. Bioelectron. 2021, 171, 112685. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Kant, R.; Behera, S.P.; Dwivedi, G.R.; Singh, R. Next-Generation Diagnostic with CRISPR/Cas: Beyond Nucleic Acid Detection. Int. J. Mol. Sci. 2022, 23, 6052. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-Y.; Cheng, Q.-X.; Wang, J.-M.; Li, X.-Y.; Zhang, Z.-L.; Gao, S.; Cao, R.-B.; Zhao, G.-P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef]
- Lee, R.A.; Herigon, J.C.; Benedetti, A.; Pollock, N.R.; Denkinger, C.M. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. J. Clin. Microbiol. 2021, 59, e02881-20. [Google Scholar] [CrossRef]
- Shinoda, H.; Taguchi, Y.; Nakagawa, R.; Makino, A.; Okazaki, S.; Nakano, M.; Muramoto, Y.; Takahashi, C.; Takahashi, I.; Ando, J.; et al. Amplification-free RNA detection with CRISPR–Cas13. Commun. Biol. 2021, 4, 476. [Google Scholar] [CrossRef]
- Heo, W.; Lee, K.; Park, S.; Hyun, K.-A.; Jung, H.-I. Electrochemical biosensor for nucleic acid amplification-free and sensitive detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA via CRISPR/Cas13a trans-cleavage reaction. Biosens. Bioelectron. 2022, 201, 113960. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.; Lei, Y. Mini review: Recent progress in RT-LAMP enabled COVID-19 detection. Sens. Actuators Rep. 2020, 2, 100017. [Google Scholar] [CrossRef] [PubMed]
- Kaci, K.; del Caño, R.; Luna, M.; Milán-Rois, P.; Castellanos, M.; Abreu, M.; Cantón, R.; Galán, J.C.; Somoza, Á.; Miranda, R.; et al. Paving the way to point of care (POC) devices for SARS-CoV-2 detection. Talanta 2022, 247, 123542. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, V.; Boni, A.; Bassoli, M.; Giannetto, M.; Fortunati, S.; Careri, M.; De Munari, I. IoT and Biosensors: A Smart Portable Potentiostat With Advanced Cloud-Enabled Features. IEEE Access 2021, 9, 141544. [Google Scholar] [CrossRef]
- Amine Smaini, M.; Maallah, R.; Smaini, I.; Eddine El Qouatli, S.; Najih, R.; Chtaini, A. Electrochemical Biosensor for the Detection of Viruses and Possibly Covid-19. J. Immunol. Tech. Infect Dis. 2021, 3, 2. [Google Scholar]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Hiesgen, R.; Haiber, J. MEASUREMENT METHODS | Structural Properties: Atomic Force Microscopy. In Encyclopedia of Electrochemical Power Sources; Elsevier: Amsterdam, The Netherlands, 2009; pp. 696–717. [Google Scholar] [CrossRef]
- Jenkins, D.M.; Lee, B.E.; Jun, S.; Reyes-De-Corcuera, J.; McLamore, E.S. ABE-Stat, a Fully Open-Source and Versatile Wireless Potentiostat Project Including Electrochemical Impedance Spectroscopy. J. Electrochem. Soc. 2019, 166, B3056–B3065. [Google Scholar] [CrossRef]
- Cook, J. SIMstat: Hardware Design Considerations for Implementing a Low-Cost, Portable Potentiostat. Bachelor’s Thesis, Honors College, Oregon State University, Corvallis, OR, USA, 2020. [Google Scholar]
- Hoilett, O.S.; Walker, J.F.; Balash, B.M.; Jaras, N.J.; Boppana, S.; Linnes, J.C. KickStat: A Coin-Sized Potentiostat for High-Resolution Electrochemical Analysis. Sensors 2020, 20, 2407. [Google Scholar] [CrossRef]
- Valizadeh, A.; Sohrabi, N.; Badrzadeh, F. Electrochemical detection of HIV-1 by nanomaterials. Artif. Cells Nanomed. Biotechnol. 2017, 45, 1467–1477. [Google Scholar] [CrossRef]
- Yang, L.; Liu, S.; Liu, J.; Zhang, Z.; Wan, X.; Huang, B.; Chen, Y.; Zhang, Y. COVID-19: Immunopathogenesis and Immunotherapeutics. Signal Transduct. Target. Ther. 2020, 5, 128. [Google Scholar] [CrossRef]
- Stat-i 400 Bipotentiostat/Galvanostat/Impedance Analyzer (EIS)|Metrohm. Available online: https://www.metrohm.com/en_my/products/s/tat-/stat-i-400.html (accessed on 8 July 2022).
- Belouzard, S.; Millet, J.K.; Licitra, B.N.; Whittaker, G.R. Mechanisms of Coronavirus Cell Entry Mediated by the Viral Spike Protein. Viruses 2012, 4, 1011–1033. [Google Scholar] [CrossRef] [PubMed]
- Antibody Types: IgM, IgA, IgD, IgG, IgE and Camelid Antibodies. Available online: https://www.news-medical.net/life-sciences/Types-of-Antibodies.aspx (accessed on 17 June 2022).
- Screen-Printed Gold Electrode (Aux.:Au; Ref.:Ag)/Ink AT/Work in Solution. Available online: https://www.metrohm.com/id_id/products/c/220a/c220at.html (accessed on 17 June 2022).
- Sekine, S.; Ido, Y.; Miyake, T.; Nagamine, K.; Nishizawa, M. Conducting Polymer Electrodes Printed on Hydrogel. J. Am. Chem. Soc. 2010, 132, 13174–13175. [Google Scholar] [CrossRef] [PubMed]
- Chapman, C.A.R.; Cuttaz, E.A.; Tahirbegi, B.; Novikov, A.; Petkos, K.; Koutsoftidis, S.; Drakakis, E.M.; Goding, J.A.; Green, R.A. Flexible Networks of Patterned Conducting Polymer Nanowires for Fully Polymeric Bioelectronics. Adv. NanoBiomed. Res. 2022, 2, 2100102. [Google Scholar] [CrossRef]
- Strnad, G.; German-Sallo, Z.; Jakab-Farkas, L.; Petrovan, C.; Portan, D. Influence of electrical parameters on morphology of nanostructured TiO2layers developed by electrochemical anodization. MATEC Web Conf. 2017, 112, 4021. [Google Scholar] [CrossRef]
- Zhang, H.; Miller, B.L. Immunosensor-based label-free and multiplex detection of influenza viruses: State of the art. Biosens. Bioelectron. 2019, 141, 111476. [Google Scholar] [CrossRef]
- Xiao, Y.; Piorek, B.D.; Plaxco, K.W.; Heeger, A.J. A Reagentless Signal-On Architecture for Electronic, Aptamer-Based Sensors via Target-Induced Strand Displacement. J. Am. Chem. Soc. 2005, 127, 17990–17991. [Google Scholar] [CrossRef]
- Elgrishi, N.; Rountree, K.J.; McCarthy, B.D.; Rountree, E.S.; Eisenhart, T.T.; Dempsey, J.L. A Practical Beginner’s Guide to Cyclic Voltammetry. J. Chem. Educ. 2018, 95, 197–206. [Google Scholar] [CrossRef]
- Ronkainen, N.J.; Halsall, H.B.; Heineman, W.R. Electrochemical biosensors. Chem. Soc. Rev. 2010, 39, 1747–1763. [Google Scholar] [CrossRef]
- Batchelor-Mcauley, C.; Kätelhçn, E.; Barnes, E.O.; Compton, R.G.; Aborda, E.; Molina, A. Recent Advances in Voltammetry. ChemistryOpen 2015, 4, 224–260. [Google Scholar] [CrossRef]
- Nossol, E.; Muñoz, R.A.A.; Richter, E.M.; de Souza Borges, P.H.; Silva, S.C.; Rocha, D.P. Sensing Materials: Graphene. Ref. Modul. Biomed. Sci. 2021. [Google Scholar] [CrossRef]
- Harvey, D. Effect of Scan Rate in Cyclic Voltammetry. Available online: https://community.asdlib.org/imageandvideoexchangeforum/2013/07/31/effect-of-scan-rate-in-cyclic-voltammetry/ (accessed on 7 January 2022).
- Orazem, M.E.; Tribollet, B. Electrochemical Impedance Spectroscopy. New Jersey 2008, 383–389. [Google Scholar] [CrossRef]
- Schroll, C.A.; Cohen, S.M. Electrochemical Impedance Spectroscopy. In Experiment-EIS-Teacher-Edition; Gamry Instruments Inc.: Warminster, PA, USA, 2016; ISBN 0-9983378-1-1. [Google Scholar]
- Sebastian, B.; Cg, T. Impedance, Electrical and Dielectric behaviour of Tin Oxide Nanoparticle doped with Graphite, Graphene Oxide and Reduced Graphene Oxide. Int. J. Electrochem. Sci. 2021, 16, 210810. [Google Scholar] [CrossRef]
- Suni, I.I. Impedance methods for electrochemical sensors using nanomaterials. TrAC Trends Anal. Chem. 2008, 27, 604–611. [Google Scholar] [CrossRef]
- Kaiser, G. 10.2: Size and Shapes of Viruses—Biology LibreTexts. Available online: https://bio.libretexts.org/Bookshelves/Microbiology/Book%3A_Microbiology_(Kaiser)/Unit_4%3A_Eukaryotic_Microorganisms_and_Viruses/10%3A_Viruses/10.02%3A_Size_and_Shapes_of_Viruses (accessed on 9 June 2022).
- Parker, N.; Schneegurt, M.; Tu, A.-H.T.; Forster, B.M.; Lister, P.; Allen, S.; Auman, A.; Brelles-Mariño, G.; Feldman, M.A.; Flowers, P.; et al. Microbiology; OpenStax: Houston, TX, USA, 2016; ISBN 1-947172-23-9. [Google Scholar]
- Navakul, K.; Warakulwit, C.; Yenchitsomanus, P.-T.; Panya, A.; Lieberzeit, P.A.; Sangma, C. A novel method for dengue virus detection and antibody screening using a graphene-polymer based electrochemical biosensor. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Arshak, K.; Velusamy, V.; Korostynska, O.; Oliwa-Stasiak, K.; Adley, C. Conducting Polymers and Their Applications to Biosensors: Emphasizing on Foodborne Pathogen Detection. IEEE Sens. J. 2009, 9, 1942–1951. [Google Scholar] [CrossRef]
- Guimard, N.K.; Gomez, N.; Schmidt, C.E. Conducting polymers in biomedical engineering. Prog. Polym. Sci. 2007, 32, 876–921. [Google Scholar] [CrossRef]
- Wei, D.; Bailey, M.J.A.; Andrew, P.; Ryhänen, T. Electrochemical biosensors at the nanoscale. Lab Chip 2009, 9, 2123–2131. [Google Scholar] [CrossRef]
- Singh, K.V.; Whited, A.M.; Ragineni, Y.; Barrett, T.W.; King, J.; Solanki, R. 3D nanogap interdigitated electrode array biosensors. Anal. Bioanal. Chem. 2010, 397, 1493–1502. [Google Scholar] [CrossRef]
- Patolsky, F.; Lieber, C.M. Nanowire nanosensors. Mater. Today 2005, 8, 20–28. [Google Scholar] [CrossRef]
- El Sharif, H.F.; Dennison, S.R.; Tully, M.; Crossley, S.; Mwangi, W.; Bailey, D.; Graham, S.P.; Reddy, S.M. Evaluation of electropolymerized molecularly imprinted polymers (E-MIPs) on disposable electrodes for detection of SARS-CoV-2 in saliva. Anal. Chim. Acta 2022, 1206, 339777. [Google Scholar] [CrossRef]
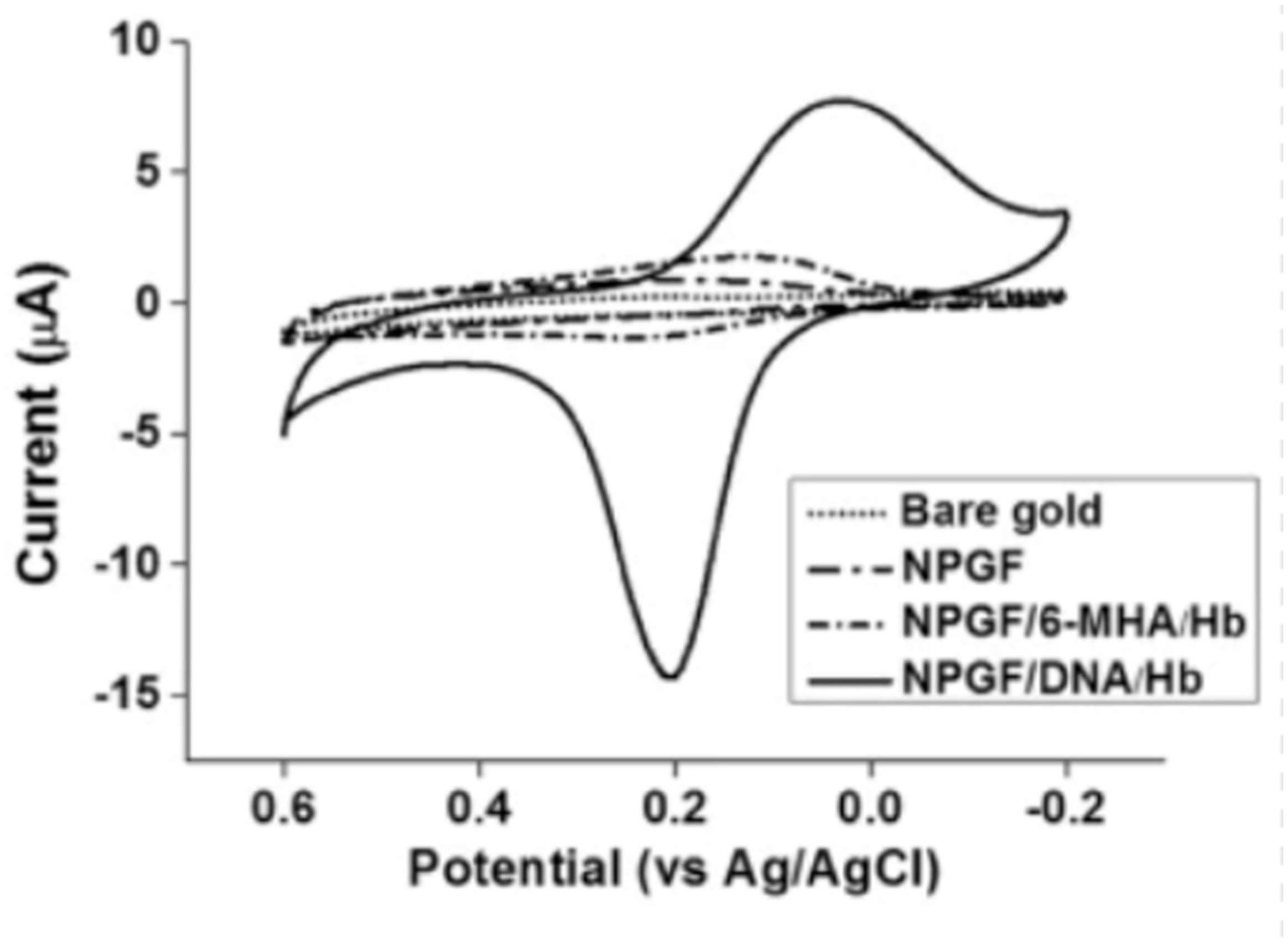
- Jo, J.; Yoon, J.; Lee, T.; Cho, H.-Y.; Lee, J.-Y.; Choi, J.-W. H2O2 biosensor consisted of hemoglobin-DNA conjugate on nanoporous gold thin film electrode with electrochemical signal enhancement. Nano Converg. 2019, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Dutta, N.; Lillehoj, P.B.; Estrela, P.; Dutta, G. Electrochemical Biosensors for Cytokine Profiling: Recent Advancements and Possibilities in the Near Future. Biosensors 2021, 11, 94. [Google Scholar] [CrossRef]
- Songa, E.A.; Somerset, V.S.; Waryo, T.; Baker, P.G.L.; Iwuoha, E.I. Amperometric nanobiosensor for quantitative determination of glyphosate and glufosinate residues in corn samples. Pure Appl. Chem. 2009, 81, 123–139. [Google Scholar] [CrossRef]
- Hong, P.; Li, W.; Li, J. Applications of Aptasensors in Clinical Diagnostics. Sensors 2012, 12, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Patris, S.; Vandeput, M.; Kauffmann, J.-M. Antibodies as target for affinity biosensors. TrAC Trends Anal. Chem. 2016, 79, 239–246. [Google Scholar] [CrossRef]
- Cooper, M.A. Label-Free Biosensors: Techniques and Applications; Cambridge University Press: Cambridge, MA, USA, 2009; pp. 1–28. [Google Scholar] [CrossRef]
- Sang, S.; Wang, Y.; Feng, Q.; Wei, Y.; Ji, J.; Zhang, W. Progress of new label-free techniques for biosensors: A review. Crit. Rev. Biotechnol. 2015, 36, 465–481. [Google Scholar] [CrossRef]
- Kumar, N.; Shetti, N.P.; Jagannath, S.; Aminabhavi, T.M. Electrochemical sensors for the detection of SARS-CoV-2 virus. Chem. Eng. J. 2022, 430, 132966. [Google Scholar] [CrossRef]
- Alafeef, M.; Dighe, K.; Moitra, P.; Pan, D. Rapid, Ultrasensitive, and Quantitative Detection of SARS-CoV-2 Using Antisense Oligonucleotides Directed Electrochemical Biosensor Chip. ACS Nano 2020, 14, 17028–17045. [Google Scholar] [CrossRef]
- Vadlamani, B.S.; Uppal, T.; Verma, S.C.; Misra, M. Functionalized TiO2 Nanotube-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2. Sensors 2020, 20, 5871. [Google Scholar] [CrossRef]
- Chaibun, T.; Puenpa, J.; Ngamdee, T.; Boonapatcharoen, N.; Athamanolap, P.; O’Mullane, A.P.; Vongpunsawad, S.; Poovorawan, Y.; Lee, S.Y.; Lertanantawong, B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021, 12, 802. [Google Scholar] [CrossRef]
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-based electrochemical biosensor for diagnosing COVID-19: Detection of SARS-CoV-2 antibodies and antigen. Biosens. Bioelectron. 2021, 176, 112912. [Google Scholar] [CrossRef]
- Lakhin, A.V.; Tarantul, V.Z.; Gening, L.V. Aptamers: Problems, Solutions and Prospects. Acta Nat. 2013, 5, 34–43. [Google Scholar] [CrossRef]
- Stoltenburg, R.; Reinemann, C.; Strehlitz, B. SELEX—A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [Google Scholar] [CrossRef]
- Parashar, N.C.; Poddar, J.; Chakrabarti, S.; Parashar, G. Repurposing of SARS-CoV nucleocapsid protein specific nuclease resistant RNA aptamer for therapeutics against SARS-CoV-2. Infect. Genet. Evol. 2020, 85, 104497. [Google Scholar] [CrossRef]
- Dalirirad, S.; Steckl, A.J. Lateral flow assay using aptamer-based sensing for on-site detection of dopamine in urine. Anal. Biochem. 2020, 596, 113637. [Google Scholar] [CrossRef]
- Lum, J.; Wang, R.; Hargis, B.; Tung, S.; Bottje, W.; Lu, H.; Li, Y. An Impedance Aptasensor with Microfluidic Chips for Specific Detection of H5N1 Avian Influenza Virus. Sensors 2015, 15, 18565–18578. [Google Scholar] [CrossRef]
- Ghanbari, K.; Roushani, M.; Azadbakht, A. Ultra-sensitive aptasensor based on a GQD nanocomposite for detection of hepatitis C virus core antigen. Anal. Biochem. 2017, 534, 64–69. [Google Scholar] [CrossRef]
- Piletska, E.V.; Guerreiro, A.R.; Whitcombe, M.J.; Piletsky, S.A. Influence of the Polymerization Conditions on the Performance of Molecularly Imprinted Polymers. Macromolecules 2009, 42, 4921–4928. [Google Scholar] [CrossRef]
- Scorrano, S.; Mergola, L.; Del Sole, R.; Vasapollo, G. Synthesis of Molecularly Imprinted Polymers for Amino Acid Derivates by Using Different Functional Monomers. Int. J. Mol. Sci. 2011, 12, 1735–1743. [Google Scholar] [CrossRef]
- Chatterjee, T.N.; Bandyopadhyay, R. A Molecularly Imprinted Polymer-Based Technology for Rapid Testing of COVID-19. Trans. Indian Natl. Acad. Eng. 2020, 5, 225–228. [Google Scholar] [CrossRef]
- Belbruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 119, 94–119. [Google Scholar] [CrossRef]
- Arshad, R.; Rhouati, A.; Hayat, A.; Nawaz, M.H.; Yameen, M.A.; Mujahid, A.; Latif, U. MIP-Based Impedimetric Sensor for Detecting Dengue Fever Biomarker. Appl. Biochem. Biotechnol. 2020, 191, 1384–1394. [Google Scholar] [CrossRef] [PubMed]
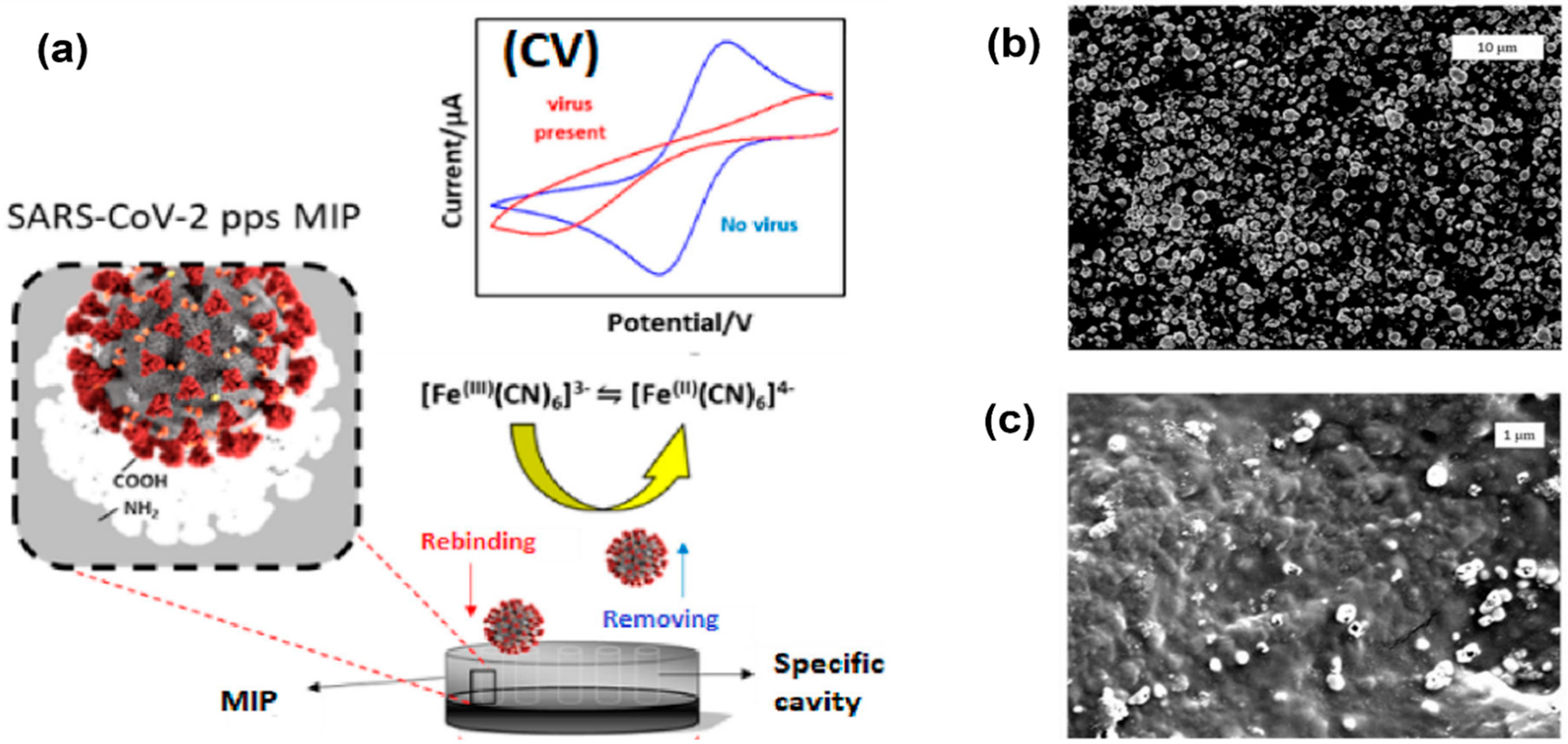
- Ayankojo, A.G.; Boroznjak, R.; Reut, J.; Öpik, A.; Syritski, V. Molecularly imprinted polymer based electrochemical sensor for quantitative detection of SARS-CoV-2 spike protein. Sens. Actuators B Chem. 2022, 353, 131160. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Yao, B.; Ding, Y.; Zhao, J.; Xie, M.; Zhang, K. Entropy-driven amplified electrochemiluminescence biosensor for RdRp gene of SARS-CoV-2 detection with self-assembled DNA tetrahedron scaffolds. Biosens. Bioelectron. 2021, 178, 113015. [Google Scholar] [CrossRef]
- Perdomo, S.A.; Ortega, V.; Jaramillo-Botero, A.; Mancilla, N.; Mosquera-DeLaCruz, J.H.; Valencia, D.P.; Quimbaya, M.; Contreras, J.D.; Velez, G.E.; Loaiza, O.A.; et al. SenSARS: A Low-Cost Portable Electrochemical System for Ultra-Sensitive, Near Real-Time, Diagnostics of SARS-CoV-2 Infections. IEEE Trans. Instrum. Meas. 2021, 70, 1–10. [Google Scholar] [CrossRef]
- Teengam, P.; Siangproh, W.; Tontisirin, S.; Jiraseree-Amornkun, A.; Chuaypen, N.; Tangkijvanich, P.; Henry, C.S.; Ngamrojanavanich, N.; Chailapakul, O. NFC-enabling smartphone-based portable amperometric immunosensor for hepatitis B virus detection. Sens. Actuators B Chem. 2021, 326, 128825. [Google Scholar] [CrossRef]
- Vásquez, V.; Navas, M.-C.; Jaimes, J.A.; Orozco, J. SARS-CoV-2 electrochemical immunosensor based on the spike-ACE2 complex. Anal. Chim. Acta 2022, 1205, 339718. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.D.T.; de Araujo, W.R.; de Lima, L.F.; Ferreira, A.L.; de la Fuente-Nunez, C. Low-cost biosensor for rapid detection of SARS-CoV-2 at the point of care. Matter 2021, 4, 2403–2416. [Google Scholar] [CrossRef]
- Salahandish, R.; Jalali, P.; Tabrizi, H.O.; Hyun, J.E.; Haghayegh, F.; Khalghollah, M.; Zare, A.; Berenger, B.M.; Niu, Y.D.; Ghafar-Zadeh, E.; et al. A compact, low-cost, and binary sensing (BiSense) platform for noise-free and self-validated impedimetric detection of COVID-19 infected patients. Biosens. Bioelectron. 2022, 213, 114459. [Google Scholar] [CrossRef]
- Campbell, J.M.; Balhoff, J.B.; Landwehr, G.M.; Rahman, S.M.; Vaithiyanathan, M.; Melvin, A.T. Microfluidic and Paper-Based Devices for Disease Detection and Diagnostic Research. Int. J. Mol. Sci. 2018, 19, 2731. [Google Scholar] [CrossRef]
- Nguyen, N.-T.; Wereley, S.T.; Shaegh, S.A.M. Fabrication Techniques for Microfluidics; Artech House: Norwood, MA, USA, 2019; ISBN 9781630813659. [Google Scholar]
- Cui, Z.; Chang, H.; Wang, H.; Lim, B.; Hsu, C.-C.; Yu, Y.; Jia, H.; Wang, Y.; Zeng, Y.; Ji, M.; et al. Development of a rapid test kit for SARS-CoV-2: An example of product design. Bio-Des. Manuf. 2020, 3, 83–86. [Google Scholar] [CrossRef]
- Roy, S.; Mohd-Naim, N.F.; Safavieh, M.; Ahmed, M.U. Colorimetric Nucleic Acid Detection on Paper Microchip Using Loop Mediated Isothermal Amplification and Crystal Violet Dye. ACS Sens. 2017, 2, 1713–1720. [Google Scholar] [CrossRef]
- Song, J.; Mauk, M.G.; Hackett, B.A.; Cherry, S.; Bau, H.H.; Liu, C. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 2016, 88, 7289–7294. [Google Scholar] [CrossRef]
- Ganguli, A.; Ornob, A.; Yu, H.; Damhorst, G.L.; Chen, W.; Sun, F.; Bhuiya, A.; Cunningham, B.T.; Bashir, R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices 2017, 19, 73. [Google Scholar] [CrossRef]
- Safavieh, M.; Kaul, V.; Khetani, S.; Singh, A.; Dhingra, K.; Kanakasabapathy, M.K.; Draz, M.S.; Memic, A.; Kuritzkes, D.R.; Shafiee, H. Paper microchip with a graphene-modified silver nano-composite electrode for electrical sensing of microbial pathogens. Nanoscale 2017, 9, 1852–1861. [Google Scholar] [CrossRef]
- Kaarj, K.; Akarapipad, P.; Yoon, J.-Y. Simpler, Faster, and Sensitive Zika Virus Assay Using Smartphone Detection of Loop-mediated Isothermal Amplification on Paper Microfluidic Chips. Sci. Rep. 2018, 8, 12438. [Google Scholar] [CrossRef]
- Lin, X.; Huang, X.; Urmann, K.; Xie, X.; Hoffmann, M.R. Digital Loop-Mediated Isothermal Amplification on a Commercial Membrane. ACS Sens. 2019, 4, 242–249. [Google Scholar] [CrossRef]
- Huang, X.; Tang, G.; Ismail, N.; Wang, X. Developing RT-LAMP Assays for Rapid Diagnosis of SARS-CoV-2 in Saliva. EBioMedicine 2022, 75, 103736. [Google Scholar] [CrossRef]
- Erickson, D.; Serey, X.; Chen, Y.-F.; Mandal, S. Nanomanipulation using near field photonics. Lab Chip 2011, 11, 995–1009. [Google Scholar] [CrossRef]
- Xu, Z.; Song, W.; Crozier, K.B. Optical Trapping of Nanoparticles Using All-Silicon Nanoantennas. ACS Photonics 2018, 5, 4993–5001. [Google Scholar] [CrossRef]
- Conteduca, D.; Brunetti, G.; Pitruzzello, G.; Tragni, F.; Dholakia, K.; Krauss, T.F.; Ciminelli, C. Exploring the Limit of Multiplexed Near-Field Optical Trapping. ACS Photonics 2021, 8, 2060–2066. [Google Scholar] [CrossRef]
- Hua, P.; Luff, B.J.; Quigley, G.R.; Wilkinson, J.S.; Kawaguchi, K. Integrated optical dual Mach–Zehnder interferometer sensor. Sens. Actuators B Chem. 2002, 87, 250–257. [Google Scholar] [CrossRef][Green Version]
- Xie, Y.; Zhang, M.; Dai, D. Design Rule of Mach-Zehnder Interferometer Sensors for Ultra-High Sensitivity. Sensors 2020, 20, 2640. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X. Optical ring resonators for biochemical and chemical sensing. Anal. Bioanal. Chem. 2010, 399, 205–211. [Google Scholar] [CrossRef]
- Ciminelli, C.; Innone, F.; Brunetti, G.; Conteduca, D.; Dell’Olio, F.; Tatoli, T.; Armenise, M.N. Rigorous model for the design of ultra-high Q-factor resonant cavities. In Proceedings of the 18th International Conference on Transparent Optical Networks 2016, Trento, Italy, 10–14 July 2016. [Google Scholar] [CrossRef]



| Author | Instrument | Detection Method | Analysis Time | Limit of Detection | Remarks |
|---|---|---|---|---|---|
| Alafeef et al. [69] | Graphene-based electrochemical biosensor (integration of thiol-modified antisense oligonucleotide probes with AuNP caps) | Protein detection: nucleocapsid (N gene) of COVID-19 | Less than 5 min | 6.9 copies/μL | At present, the device is expected to be integrated with a portable mobile platform that can be used for instantaneous diagnosis of positive COVID-19 cases. |
| Vadlamani et al. [70] | Cobalt-functionalized TiO2 nanotube (Co-TNT)-based electrochemical sensor | Protein detection: spike protein receptor-binding domain (RBD) | ~30 s | ~0.7 nM levels | This assay has a tendency to be applied for the diagnosis of other recognized respiratory viruses or in conjunction with a suitable metallic element to ensure TNT function. |
| Chaibun et al. [71] | Multiplex rolling circle amplification (RCA) | Protein detection: nucleocapsid and spike glycoprotein | No data available | 1 copy/μL for both N and S genes | The RNA extraction procedure for this assay is currently being optimized, in addition to integration of a smartphone-based biosensor device. Researchers are also working on minimizing the turnaround time for the assay and simplifying the test method. |
| Yakoh et al. [72] | Label-free, paper-based electrochemical platform | Antibody detection of IgM and IgG | 30 min | 1 ng/mL | Researchers are working on the direct detection of COVID-19 spike protein of with this assay. |
| Fan et al. [85] | Entropy-driven amplified electrochemiluminescence (ECL) biosensor | Nucleic acid detection includes the RdRp gene of COVID-19 | 30 min | As low as 2.67 fM | Not available |
| Ayankojo et al. [78] | MIP-based electrochemical sensor | S1 component of the S protein (nCovS1) | Not available | 4.8 pg/mL | This sensor demonstrates high selectivity to SARS-CoV-2 spike protein compared to other proteins. The sensor is compatible with portable potentiostats and suitable for point-of-care diagnosis. |
| Reference | Potentiostat/Microcontroller | Brief Description | Detection Method | Size | Voltage Supply | Virus | Limit of Detection (LOD) |
|---|---|---|---|---|---|---|---|
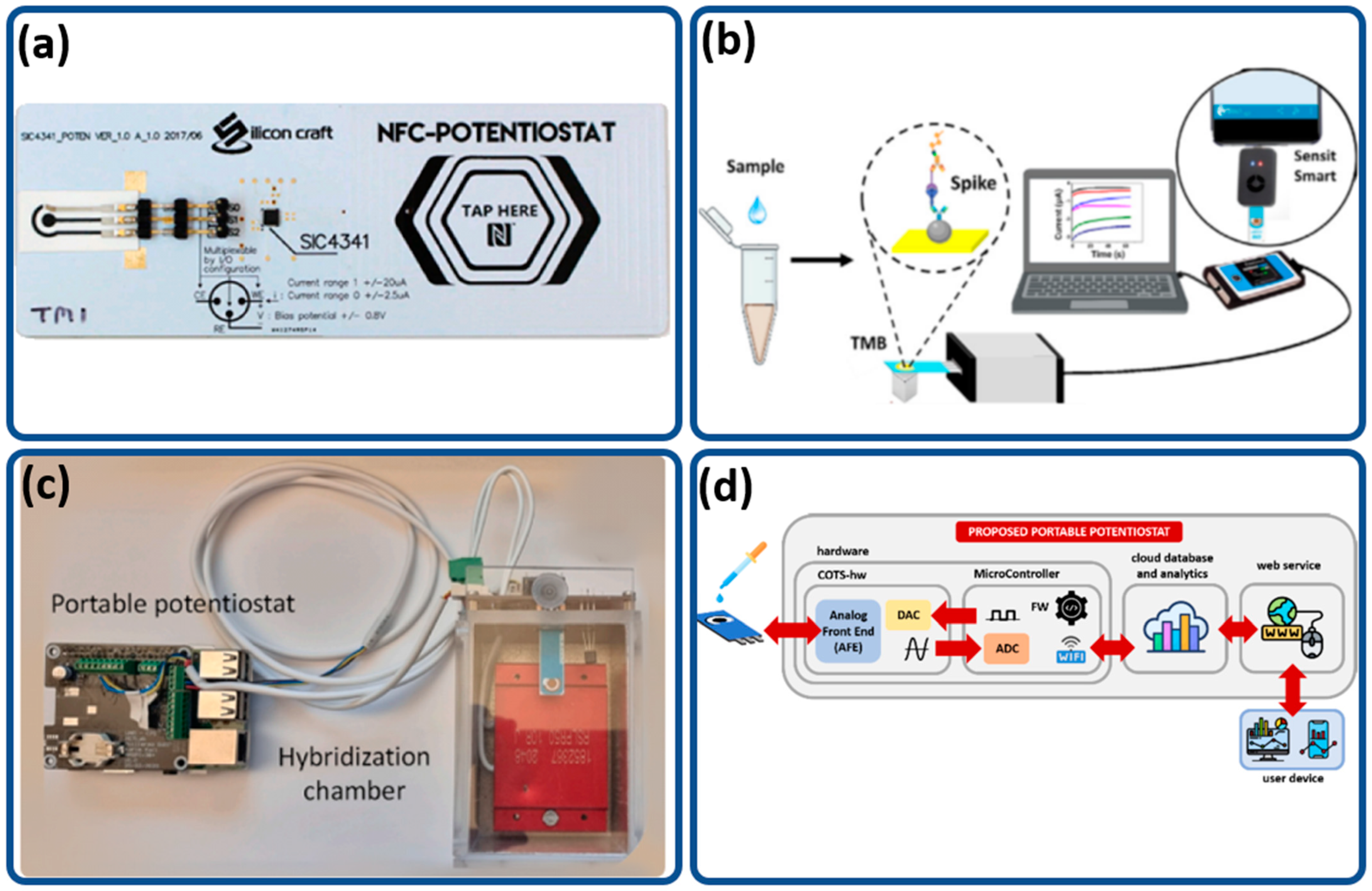
| [87] | SIC4341 | Smartphone-controlled sensor that can operate through an NFC tag sensor. It is unparalleled in terms of portability, with the smallest form factor. | Cyclic voltammetry, amperometry | 52 × 18 × 1 mm | 1.8V to 3.3V | Hepatitis B Virus | 0.17 μg/mL |
| [88] | PalmSens4 | The PalmSens4 potentiostat is used with a sensor linked to anti-ACE2 protein to detect SARS-CoV-2 virus | Amperometry | 15.7 × 9.7 × 3.5 cm | 5 V | SARS-CoV-2 | 22.5 ng/mL |
| [23] | Raspberry Pi 3B LMP91000 | Portable point of care (POC) with low cost and high portability designed with a simple user interface for ease of use. | Differential pulse voltammetry | 10 × 8 × 4 cm (main module) 12 × 7 × 7 cm (hybridization chamber) | 5 V | SARS-CoV-2 | 22.1 fM |
| [24] | LMP91000 | Portable potentiostat with machine learning, achieving an accuracy of 98.23%. | Differential pulse voltammetry | Not available | 5V | Hepatitis C virus | Not specified |
| [86] | Raspberry Pi 4B | SenSARS: accurate, low cost and portable potentiostat with EIS as the main technique used for viral detection. | Electrochemical impedance spectroscopy | Not specified | 5V | SARS-CoV-2 | 1.065 fg/mL |
| [89] | Palmsens Sensit | RAPID 1.0 uses Palmsens Sensit as the reader for the sensor with EIS. Other than viral detection, it can also be used to detect bacterial and fungal infection. | Electrochemical impedance spectroscopy | 43 × 25 × 11 mm | 5 V | SARS-CoV-2 | 2.8 fg/mL |
| [90] | Bisense | Bisense is a potentiostat developed to perform EIS on custom-designed dual working electrodes with rapid readings within 1.5 min. | Electrochemical impedance spectroscopy | 18 × 15 × 9 cm | Not specified | SARS-CoV-2 | 56 fg/mL |
| Authors | Detection Methods | Detected Virus | Remarks |
|---|---|---|---|
| Song et al. [95] | RT-LAMP and RT-qPCR | Zika virus | High sensitivity at a low cost. Within 40 min, the electricity-free point-of-care diagnostic system detects ZIKV in saliva with a sensitivity of less than 5 PFU of ZIKV per sample. |
| Ganguli et al. [96] | RT-LAMP and optical sensor | Zika, chikungunya and dengue viruses | Clinically relevant sensitivity. Zika virus detection as low as 1.56 × 105 PFU/mL from whole blood; low reagent consumption. Readout on smartphone. |
| Safavieh et al. [97] | RT-LAMP and impedance spectroscopy | HIV | Disposable, flexible, low-cost, light, high sensitivity and specificity, rapid amplification, increased stability and low complexity. |
| Kaarj et al. [98] | RT-LAMP and optical sensor | Zika virus | Limit of detection: 1 copy/μL; simple, quick (15 min) and easily quantifiable with a smartphone. |
| Lin et al. [99] | RT-LAMP and optical sensor | MS2 virus | Simple to use, low cost, fluorescence intensities 100 times greater than other methods for distinguishing between positive and negative pores. |
| Huang et al. [100] | RT-LAMP and colorimetric sensor | SARS-CoV-2 | Detects SARS-CoV-2 particles in saliva at levels as low as 1.5 copies/µL without the need for RNA isolation. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdul Ghani, M.A.; Nordin, A.N.; Zulhairee, M.; Che Mohamad Nor, A.; Shihabuddin Ahmad Noorden, M.; Muhamad Atan, M.K.F.; Ab Rahim, R.; Mohd Zain, Z. Portable Electrochemical Biosensors Based on Microcontrollers for Detection of Viruses: A Review. Biosensors 2022, 12, 666. https://doi.org/10.3390/bios12080666
Abdul Ghani MA, Nordin AN, Zulhairee M, Che Mohamad Nor A, Shihabuddin Ahmad Noorden M, Muhamad Atan MKF, Ab Rahim R, Mohd Zain Z. Portable Electrochemical Biosensors Based on Microcontrollers for Detection of Viruses: A Review. Biosensors. 2022; 12(8):666. https://doi.org/10.3390/bios12080666
Chicago/Turabian StyleAbdul Ghani, Muhammad Afiq, Anis Nurashikin Nordin, Munirah Zulhairee, Adibah Che Mohamad Nor, Mohd Shihabuddin Ahmad Noorden, Muhammad Khairul Faisal Muhamad Atan, Rosminazuin Ab Rahim, and Zainiharyati Mohd Zain. 2022. "Portable Electrochemical Biosensors Based on Microcontrollers for Detection of Viruses: A Review" Biosensors 12, no. 8: 666. https://doi.org/10.3390/bios12080666
APA StyleAbdul Ghani, M. A., Nordin, A. N., Zulhairee, M., Che Mohamad Nor, A., Shihabuddin Ahmad Noorden, M., Muhamad Atan, M. K. F., Ab Rahim, R., & Mohd Zain, Z. (2022). Portable Electrochemical Biosensors Based on Microcontrollers for Detection of Viruses: A Review. Biosensors, 12(8), 666. https://doi.org/10.3390/bios12080666







