RT-LAMP-Based Molecular Diagnostic Set-Up for Rapid Hepatitis C Virus Testing
Abstract
1. Introduction
2. Materials and Methods
2.1. HCV RT-LAMP Primer Design and Testing
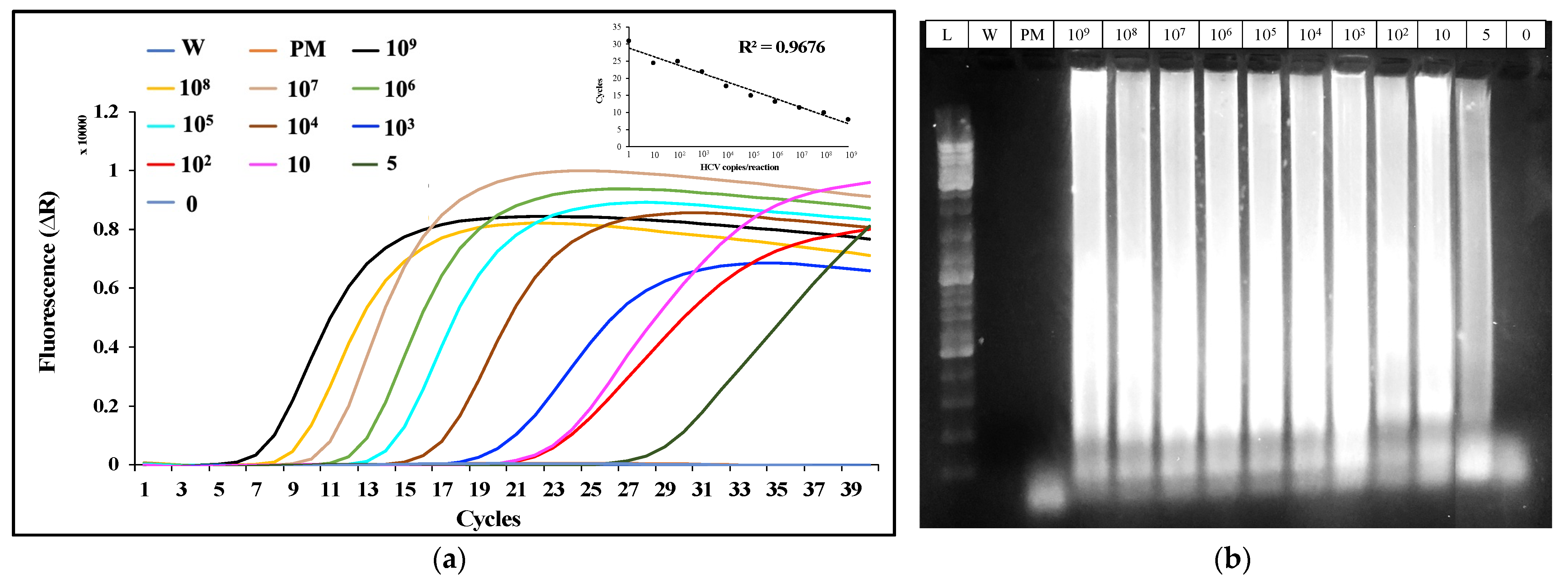
2.2. Analysis of the HCV-Spiked Samples by RT-LAMP
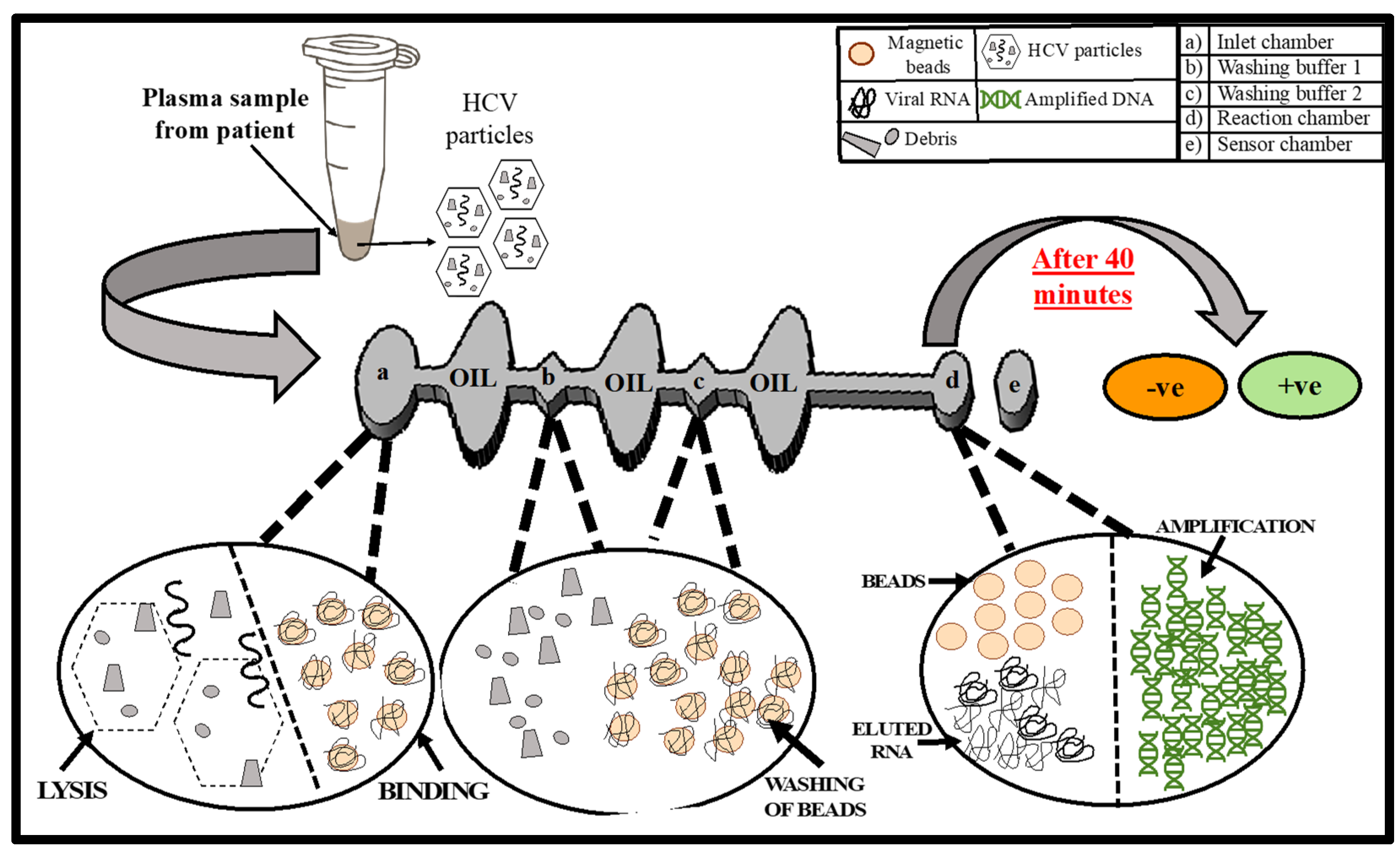
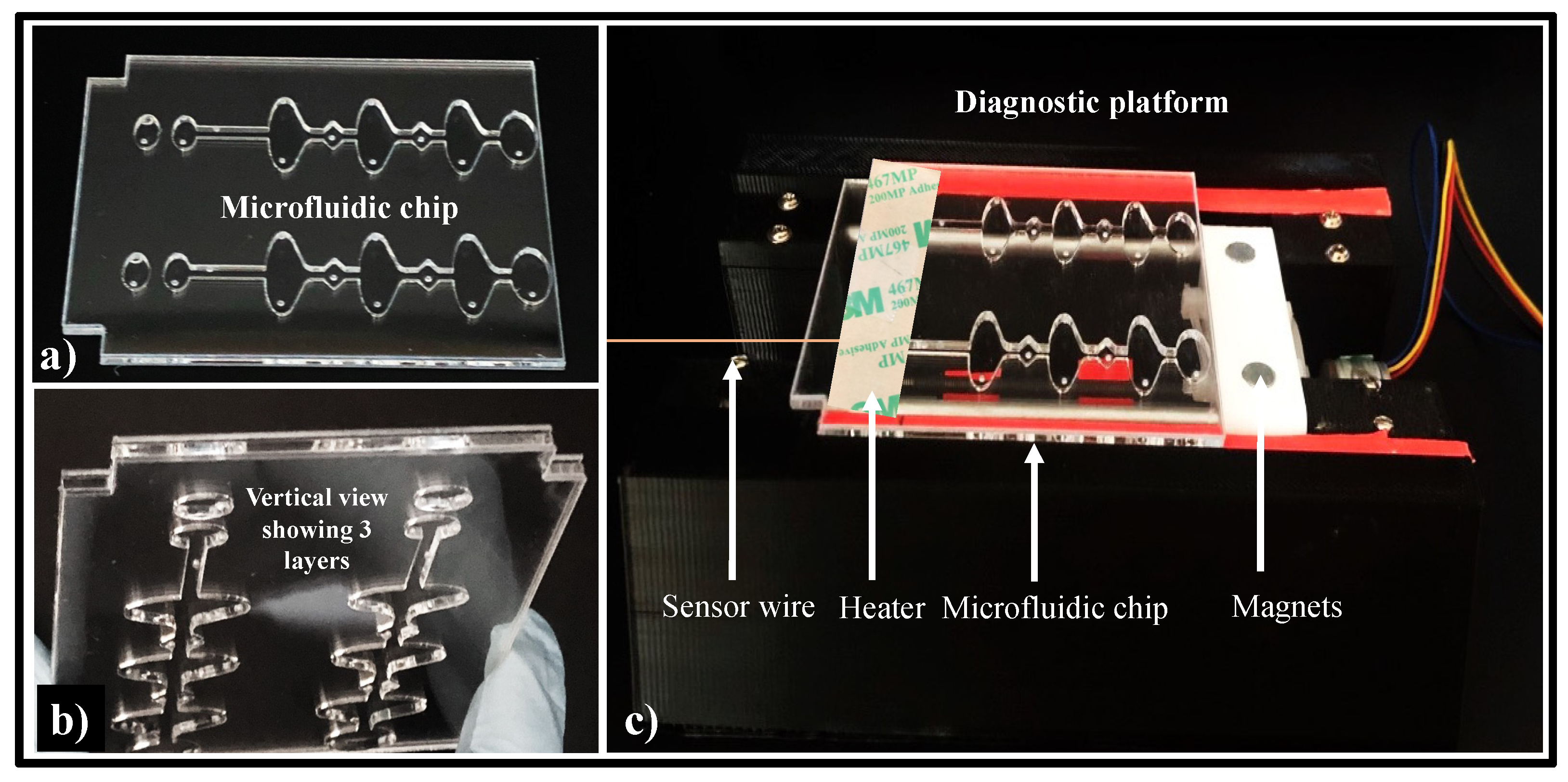
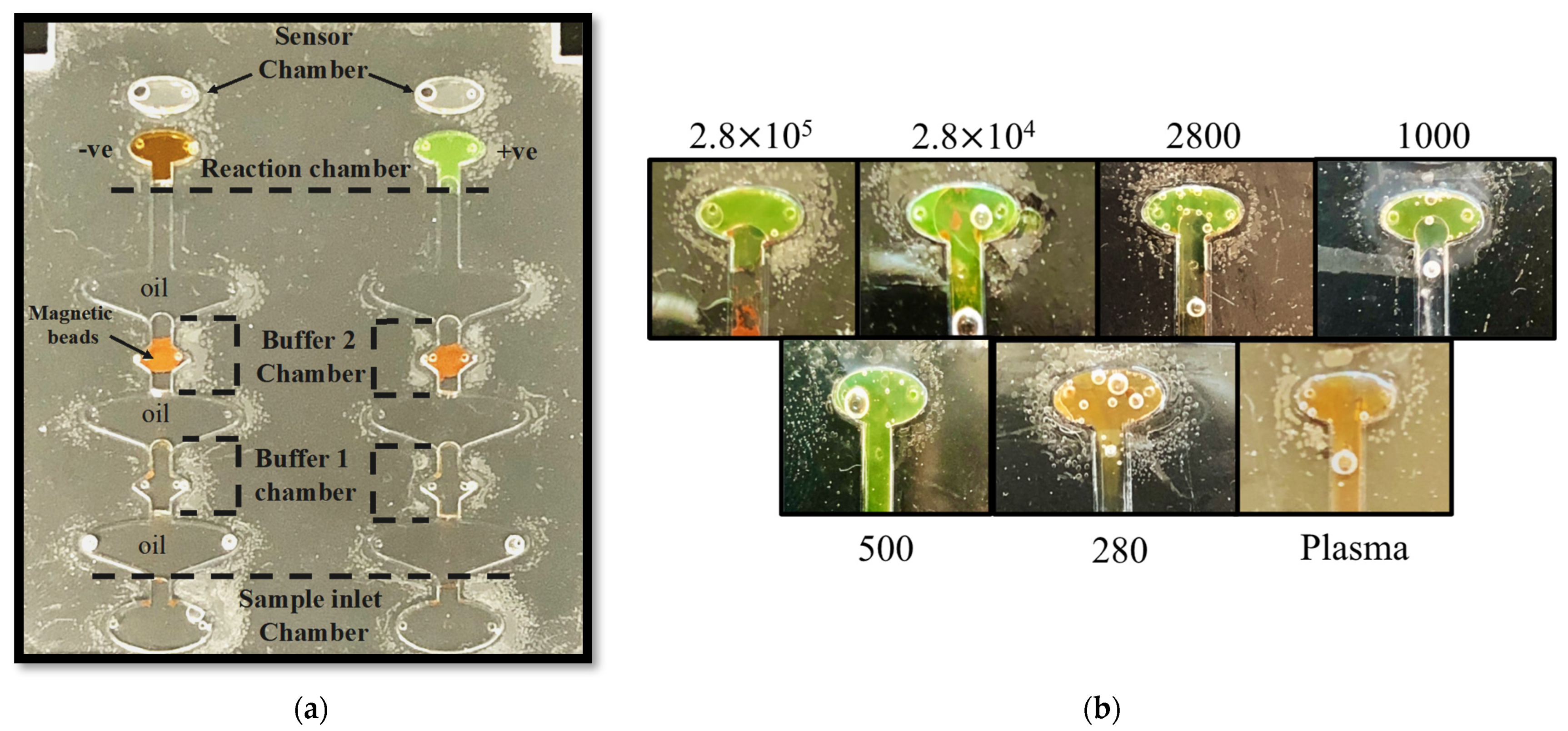
2.3. Microfluidic Chip Design
2.4. Diagnostic Platform Set-Up
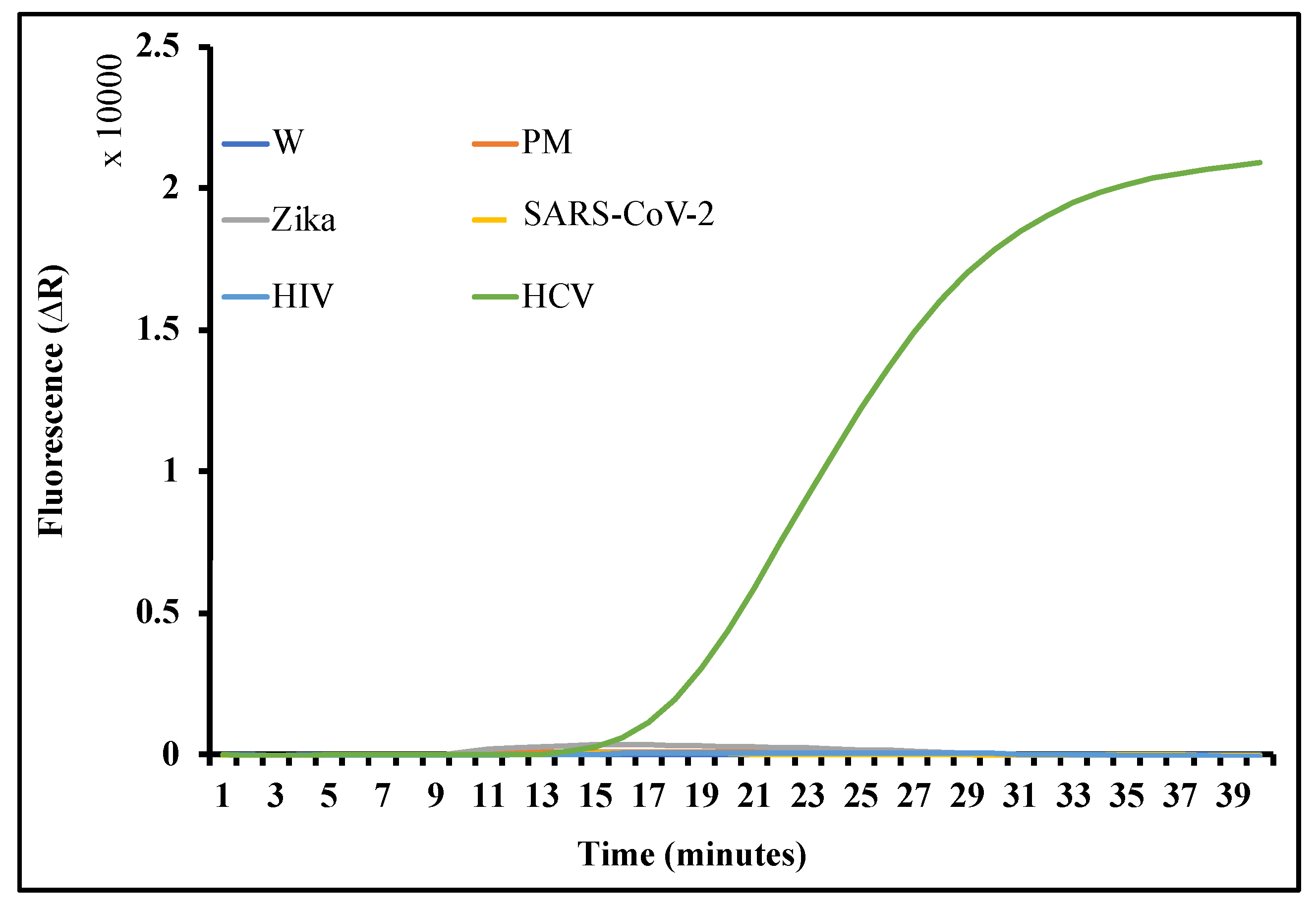
2.5. On-Chip Detection of HCV from Human Plasma
2.6. Results and Discussion
3. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hepatitis, C. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 15 November 2021).
- Hepatitis Can’t Wait—WHO Commemorates World Hepatitis Day. 2021. Available online: https://www.who.int/news/item/28-07-2021-hepatitis-can-t-wait---who-commemorates-world-hepatitis-day-2021 (accessed on 15 November 2021).
- 2019 Viral Hepatitis Surveillance Report/CDC. Available online: https://www.cdc.gov/hepatitis/statistics/2019surveillance/index.htm (accessed on 15 November 2021).
- Gupta, E.; Bajpai, M.; Choudhary, A. Hepatitis C virus: Screening, diagnosis, and interpretation of laboratory assays. Asian J. Trans. Sci. 2014, 8, 19–25. [Google Scholar]
- Rau, M.; Baur, K.; Geier, A. Host Genetic Variants in the Pathogenesis of Hepatitis C. Viruses 2012, 4, 3281–3302. [Google Scholar] [CrossRef] [PubMed]
- Colombo, M.; Rumi, M.G.; Donato, M.F.; Tommasini, M.A.; del Ninno, E.; Ronchi, G.; Kuo, G.; Houghton, M. Hepatitis C Antibody in Patients with Chronic Liver Disease and Hepatocellular Carcinoma. Dig. Dis. Sci. 1991, 36, 1130–1133. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M. Era of Direct Acting Anti-Viral Agents for the Treatment of Hepatitis C. World J. Hepatol. 2018, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Catlett, B.; Hajarizadeh, B.; Cunningham, E.; Stofko, B.; Wheeler, A.; Hussain, B.; Feld, J.; Martro, E.; Chevaliez, S.; Pawlotskey, J.; et al. Diagnostic accuracy of assay using point-of-care testing or dried blood spot samples for the determination of HCV RNA: A systematic review. J. Inf. Dis. 2022. [Google Scholar] [CrossRef]
- Vermehren, J.; Sarrazin, C. The Role of Resistance in HCV Treatment. Best Pract. Res. Clin. Gastroenterol. 2012, 26, 487–503. [Google Scholar] [CrossRef]
- Muljono, D.H. Effective Drugs on the Road to HCV Elimination and a Therapeutic Gap to Close. Lancet Gastroenterol. Hepatol. 2019, 4, 86–88. [Google Scholar] [CrossRef]
- Fiore, V.; de Matteis, G.; Ranieri, R.; Saderi, L.; Pontali, E.; Muredda, A.; Ialungo, A.M.; Caruso, R.; Madeddu, G.; Sotgiu, G.; et al. HCV Testing and Treatment Initiation in an Italian Prison Setting: A Step-by-Step Model to Micro-Eliminate Hepatitis C. Int. J. Drug Policy 2021, 90, 103055. [Google Scholar] [CrossRef]
- Hellard, M.E.; Chou, R.; Easterbrook, P. WHO guidelines on testing for hepatitis B and C—Meeting targets for testing. BMC Infect. Dis. 2017, 17, 703. [Google Scholar] [CrossRef]
- Global Surveillance and Control of Hepatitis C. Report of a WHO Consultation Organized in Collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. Available online: https://pubmed.ncbi.nlm.nih.gov/10847128/ (accessed on 15 November 2021).
- Alter, M.J. Epidemiology of Hepatitis C Virus Infection. World J. Gastroenterol. 2007, 13, 2436. [Google Scholar] [CrossRef]
- Geane, L.F.; Jurema, C.M.; Larissa, T.S.A.; Renata, S.L.; Francisco, I.B.; Livia, M.V. Performance of HCV Antigen Testing for the Diagnosis and Monitoring of Antiviral Treatment: A Systematic Review and Meta-Analysis. Biomed. Res. Int. 2022, 1–17. [Google Scholar]
- Thedja, M.D.; Wibowo, D.P.; El-Khobar, K.E.; Ie, S.I.; Turyadi; Setiawan, L.; Murti, I.S.; Muljono, D.H. Improving Linkage to Care of Hepatitis C: Clinical Validation of GeneXpert® HCV Viral Load Point-of-Care Assay in Indonesia. Am. J. Trop. Med. Hyg. 2021, 105, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Schnuriger, A.; Dominguez, S.; Valantin, M.A.; Tubiana, R.; Duvivier, C.; Ghosn, J.; Simon, A.; Katlama, C.; Thibault, V. Early Detection of Hepatitis C Virus Infection by Use of a New Combined Antigen-Antibody Detection Assay: Potential Use for High-Risk Individuals. J. Clin. Microbiol. 2006, 44, 1561. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, J.T.; Diepolder, H.M.; Zachoval, R.; Gruener, N.H.; Jung, M.C.; Ulsenheimer, A.; Schraut, W.W.; Schirren, C.A.; Waechtler, M.; Backmund, M.; et al. High Rate of Both Spontaneous and Treatment-Induced Viral Clearance. Gastroenterology 2003, 125, 80–88. [Google Scholar] [CrossRef]
- Testing for HCV Infection: An Update of Guidance for Clinicians and Laboratorians. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm6218a5.htm (accessed on 15 November 2021).
- Kabir, M.A.; Soto-Acosta, R.; Sharma, S.; Bradrick, S.S.; Garcia-Blanco, M.A.; Caputi, M.; Asghar, W. An Antibody Panel for Highly Specific Detection and Differentiation of Zika Virus. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Mustafa, M.A.; Al-Samarraie, M.Q.; Ahmed, M.T. Molecular Techniques of Viral Diagnosis. Sci. Arch. 2020, 1, 98–101. [Google Scholar]
- Sharma, S.; Kabir, M.A.; Asghar, W. Lab-on-a-Chip Zika Detection With Reverse Transcription Loop-Mediated Isothermal Amplification-Based Assay for Point-of-Care Settings. Arch. Pathol. Lab. Med. 2020, 144, 1335–1343. [Google Scholar] [CrossRef]
- Vermeersch, P.; van Ranst, M.; Lagrou, K. Validation of a Strategy for HCV Antibody Testing with Two Enzyme Immunoassays in a Routine Clinical Laboratory. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2008, 42, 394–398. [Google Scholar] [CrossRef]
- Saludes, V.; González, V.; Planas, R.; Matas, L.; Ausina, V.; Martró, E. Tools for the Diagnosis of Hepatitis C Virus Infection and Hepatic Fibrosis Staging. World J. Gastroenterol. 2014, 20, 3431. [Google Scholar] [CrossRef]
- Cadieux, G.; Campbell, J.; Dendukuri, N. Systematic Review of the Accuracy of Antibody Tests Used to Screen Asymptomatic Adults for Hepatitis C Infection. CMAJ Open 2016, 4, E737. [Google Scholar] [CrossRef]
- Sharma, S.; Zhuang, R.; Long, M.; Pavlovic, M.; Kang, Y.; Ilyas, A.; Asghar, W. Circulating Tumor Cell Isolation, Culture, and Downstream Molecular Analysis. Biotechnol. Adv. 2018, 36, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Strader, D.B.; Wright, T.; Thomas, D.L.; Seeff, L.B. Diagnosis, Management, and Treatment of Hepatitis C. Hepatology 2004, 39, 1147–1171. [Google Scholar] [CrossRef] [PubMed]
- Narayanamurthy, V.; Jeroish, Z.E.; Bhuvaneshwari, K.S.; Samsuri, F. Hepatitis C Virus (HCV) Diagnosis via Microfluidics. Anal. Methods Adv. Methods Appl. 2021, 13, 740–763. [Google Scholar] [CrossRef] [PubMed]
- Smookler, D.; Vanderhoff, A.; Biondi, M.J.; Valencia, J.; Ryan, P.; Karkada, J.; Hong, R.; Sattar, I.; Mandel, E.; Gjevori, M.; et al. Reducing Read Time of Point-of-Care Test Does Not Affect Detection of Hepatitis C Virus and Reduces Need for Reflex RNA. Clin. Gastroenterol. Hepatol. Off. Clin. Pract. J. Am. Gastroenterol. Assoc. 2021, 19, 1451–1458.e4. [Google Scholar] [CrossRef]
- Prabdial-Sing, N.; Gaelejwe, L.; Makhathini, L.; Thaver, J.; Manamela, M.J.; Malfeld, S.; Spearman, C.W.; Sonderup, M.; Scheibe, A.; Young, K.; et al. The performance of hepatitis C virus (HCV) antibody point-of-care tests on oral fluid or whole blood and dried blood spot testing for HCV serology and viral load among individuals at higher risk for HCV in South Africa. Health Sci. Rep. 2021, 4, e229. [Google Scholar] [CrossRef]
- Roger, S.; Ducancelle, A.; Le Guillou-Guillemette, H.; Gaudy, C.; Lunel, F. HCV virology and diagnosis. Clin. Res. Hepatol. Gastroenterol. 2021, 45, 101626. [Google Scholar] [CrossRef]
- Yoo, S.J.; Wang, L.L.; Ning, H.C.; Tao, C.M.; Hirankarn, N.; Kuakarn, S.; Yang, R.; Han, T.H.; Chan, R.C.; Hussain, B.M.; et al. Evaluation of the Elecsys® Anti-HCV II Assay for Routine Hepatitis C Virus Screening of Different Asian Pacific Populations and Detection of Early Infection. J. Clin. Virol. 2015, 64, 20–27. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, M.; Hang, L.; Kong, F.; Yan, H.; Zhang, Y.; Feng, X.; Gao, Y.; Wang, C.; Ma, H.; et al. Evaluation of a New Point-of-Care Oral Anti-HCV Test for Screening of Hepatitis C Virus Infection. Virol. J. 2020, 17, 1–11. [Google Scholar] [CrossRef]
- Catlett, B.; Carrera, A.; Starr, M.; Applegate, T.L.; Lowe, P.; Cunningham, H.P.; Grebely, J.J. Performance Evaluation of the Hologic Aptima HCV Quant Dx Assay for Detection of HCV RNA from Dried Blood Spots. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2019, 112, 40–44. [Google Scholar] [CrossRef]
- Lee, S.R.; Kardos, K.W.; Schiff, E.; Berne, C.A.; Mounzer, K.; Banks, A.T.; Tatum, H.A.; Friel, T.J.; DeMicco, M.P.; Lee, W.M.; et al. Evaluation of a New, Rapid Test for Detecting HCV Infection, Suitable for Use with Blood or Oral Fluid. J. Virol. Methods 2011, 172, 27–31. [Google Scholar] [CrossRef]
- Lamoury, F.; Njouom, R.; Amougou-Atsama, M.; Mfopou, E.Y.; Berishvili, N.; Sologashvili, M.; Fajardo, E.; Malobela, A.; Macé, A.; Chirehwa, M.; et al. Diagnostic Performance and Usability of the Genedrive® HCV ID Kit in Two Decentralized Settings in Cameroon and Georgia. Diagnostics 2021, 11, 746. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-Mediated Isothermal Amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-Chip: Revising Microfluidic Platforms for Loop-Mediated DNA Amplification. TrAC Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Gao, J.; Chen, T.; Dong, C.; Li, H.; Wen, Y.Z.; Lun, Z.R.; Jia, Y.; Mak, P.I.; Martins, R.P. A Handheld Digital Microfluidic Device for Loop-Mediated Isothermal Amplification (LAMP). Biomed. Microdevices 2019, 21, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Zhao, N.; Jing, W.; Liu, Q.; Lu, H.; Zhao, W.; Zhao, W.; Yuan, Z.; Xia, H.; Sui, G. A Self-Powered Rapid Loading Microfluidic Chip for Vector-Borne Viruses Detection Using RT-LAMP. Sens. Actuators B Chem. 2021, 333, 129521. [Google Scholar] [CrossRef]
- Sharma, S.; Kabir, A.; Asghar, W. Selection of healthy sperm based on positive rheotaxis using a microfluidic device. Analyst 2022, 147, 1589–1597. [Google Scholar] [CrossRef]
- Zhou, L.; Chen, Y.; Fang, X.; Liu, Y.; Du, M.; Lu, X.; Li, Q.; Sun, Y.; Ma, J.; Lan, T. Microfluidic-RT-LAMP Chip for the Point-of-Care Detection of Emerging and Re-Emerging Enteric Coronaviruses in Swine. Anal. Chim. Acta 2020, 1125, 57–65. [Google Scholar] [CrossRef]
- Jogezai, N.; Shabbir, M.I. Hand-Held Device for Rapid Single Tube Detection of Hepatitis-C Virus. Anal. Methods 2018, 10, 4233–4241. [Google Scholar] [CrossRef]
- Hongjaisee, S.; Doungjinda, N.; Khamduang, W.; Carraway, T.S.; Wipasa, J.; Debes, J.D.; Supparatpinyo, K. Rapid Visual Detection of Hepatitis C Virus Using a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. Int. J. Infect. Dis. 2021, 102, 440–445. [Google Scholar] [CrossRef]
- Detection of HCV Gene by Reverse Transcription Loop—Mediated Isothermal Amplification Method—Abstract—Europe PMC. Available online: https://europepmc.org/article/cba/623387 (accessed on 7 March 2022).
- Zhao, N.; Liu, J.; Sun, D. Detection of HCV Genotypes 1b and 2a by a Reverse Transcription Loop-Mediated Isothermal Amplification Assay. J. Med. Virol. 2017, 89, 1048–1054. [Google Scholar] [CrossRef]
- Choi, G.; Song, D.; Shrestha, S.; Miao, J.; Cui, L.; Guan, W. A Field-Deployable Mobile Molecular Diagnostic System for Malaria at the Point of Need. Lab A Chip 2016, 16, 4341–4349. [Google Scholar] [CrossRef] [PubMed]
- Mosley, O.; Melling, L.; Tarn, M.D.; Kemp, C.; Esfahani, M.M.N.; Pamme, N.; Shaw, K.J. Sample Introduction Interface for On-Chip Nucleic Acid-Based Analysis of Helicobacter Pylori from Stool Samples. Lab A Chip 2016, 16, 2108–2115. [Google Scholar] [CrossRef] [PubMed]
- Ganguli, A.; Ornob, A.; Yu, H.; Damhorst, G.; Chen, W.; Sun, F.; Bhuiya, A.; Cunningham, B.T.; Bashir, R. Hands-free smartphone-based diagnostics for simultaneous detection of Zika, Chikungunya, and Dengue at point-of-care. Biomed. Microdevices 2017, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Mauk, M.G.; Hackett, B.A.; Cherry, S.; Bau, H.H.; Liu, C. Instrument-Free Point-of-Care Molecular Detection of Zika Virus. Anal. Chem. 2016, 88, 7289–7294. [Google Scholar] [CrossRef]
- Velders, A.H.; Schoen, C.; Saggiomo, V. Loop-Mediated Isothermal Amplification (LAMP) Shield for Arduino DNA Detection. BMC Res. Notes 2018, 11, 93. [Google Scholar] [CrossRef]
- Wakita, T.; Pietschmann, T.; Kato, T.; Date, T.; Miyamoto, M.; Zhao, Z.; Murthy, K.; Habermann, A.; Kräusslich, H.G.; Mizokami, M.; et al. Production of Infectious Hepatitis C Virus in Tissue Culture from a Cloned Viral Genome. Nat. Med. 2005, 11, 791–796. [Google Scholar] [CrossRef]
- Kabir, M.A.; Zilouchian, H.; Sher, M.; Asghar, W. Development of a Flow-Free Automated Colorimetric Detection Assay Integrated with Smartphone for Zika NS1. Diagnostics 2020, 10, 42. [Google Scholar] [CrossRef]
- Sher, M.; Asghar, W. Development of a Multiplex Fully Automated Assay for Rapid Quantification of CD4+ T Cells from Whole Blood. Biosens. Bioelectron. 2019, 142, 111490. [Google Scholar] [CrossRef]
- Coarsey, C.; Coleman, B.; Kabir, M.A.; Sher, M.; Asghar, W. Development of a Flow-Free Magnetic Actuation Platform for an Automated Microfluidic ELISA. RSC Adv. 2019, 9, 8159–8168. [Google Scholar] [CrossRef]
- Kamal, S.M.; Nasser, I.A. Hepatitis C Genotype 4: What We Know and What We Don’t yet Know. Hepatology 2008, 47, 1371–1383. [Google Scholar] [CrossRef]
- Kato, N.; Hijikata, M.; Ootsuyama, Y.; Nakagawa, M.; Ohkoshi, S.; Sugimura, T.; Shimotohno, K. Molecular Cloning of the Human Hepatitis C Virus Genome from Japanese Patients with Non-A, Non-B Hepatitis. Proc. Natl. Acad. Sci. USA 1990, 87, 9524–9528. [Google Scholar] [CrossRef] [PubMed]
- Rehermann, B.; Nascimbeni, M. Molecular Cloning of the Human Hepatitis C Virus Genome from Japanese Patients with Non-A, Non-B Hepatitis. Nat. Rev. Immunol. 2005, 5, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Zein, N.N. Clinical Significance of Hepatitis C Virus Genotypes. Clin. Microbiol. Rev. 2000, 13, 223–235. [Google Scholar] [CrossRef] [PubMed]
- Mauger, D.M.; Golden, M.; Yamane, D.; Williford, S.; Lemon, S.M.; Martin, D.P.; Weeks, K.M. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. USA 2015, 112, 3692–3697. [Google Scholar] [CrossRef] [PubMed]
- Moretti, F.; Bolcic, F.; Mammana, L.; Bouzas, M.B.; Laufer, N.; Quarleri, J. The Hepatitis C Virus 5′UTR Genomic Region Remains Highly Conserved Under HAART: A 4- to 8-Year Longitudinal Study from HCV/HIV Co-Infected Patients. AIDS Res. Hum. Retrovir. 2010, 26, 527–532. [Google Scholar] [CrossRef]
- BEI Reagent Search. Available online: https://www.beiresources.org/Catalog/BEINucleicAcids/NR-52347.aspx (accessed on 15 November 2021).
- Item Details. Available online: https://www.hivreagentprogram.org/Catalog/Clones/ARP-114.aspx (accessed on 15 November 2021).
- Dragan, A.I.; Pavlovic, R.; McGivney, J.B.; Casas-Finet, J.R.; Bishop, E.S.; Strouse, R.J.; Schenerman, M.A.; Geddes, C.D. SYBR Green I: Fluorescence Properties and Interaction with DNA. J. Fluoresc. 2012, 22, 1189–1199. [Google Scholar] [CrossRef]
- Chevaliez, S.; Wlassow, M.; Volant, J.; Roudot-Thoraval, F.; Bachelard, A.; Poiteau, L.; Trabut, J.-B.; Hézode, C.; Bourdel, A.; Dominguez, S. Assessing Molecular Point-of-Care Testing and Dried Blood Spot for Hepatitis C Virus Screening in People Who Inject Drugs. Open Forum Infect. Dis. 2020, 7, 196. [Google Scholar] [CrossRef]
- Uusküla, A.; Talu, A.; Rannap, J.; Barnes, D.M.; Jarlais, D.D. Rapid Point-of-Care (POC) Testing for Hepatitis C Antibodies in a Very High Prevalence Setting: Persons Injecting Drugs in Tallinn, Estonia. Harm Reduct. J. 2021, 18, 1–6. [Google Scholar] [CrossRef]
- Fennell, R.D.; Sher, M.; Asghar, W. Development of a Microfluidic Device for CD4 T Cell Isolation and Automated Enumeration from Whole Blood. Biosensors 2022, 12, 12. [Google Scholar] [CrossRef]
- Neelakanta, P.; Sharma, S. Estimating Maximum Plausible Conserved Synteny between Orthologous Genomes of a Species Pair Compared in an Exhaustive Search-Space. Whites Sci. J. 2016, 3, 1–9. [Google Scholar]
- Kabir, M.A.; Zilouchian, H.; Younas, M.A.; Asghar, W. Dengue Detection: Advances in Diagnostic Tools from Conventional Technology to Point of Care. Biosensors 2021, 11, 206. [Google Scholar] [CrossRef] [PubMed]
- Qualls, R.G.; Johnson, D. Bioassay and Dose Measurement in UV Disinfection. Appl. Environ. Mic. 1983, 45, 872–877. [Google Scholar] [CrossRef]
- Grist, K.L.; Taylor, T.; Augenstein, L. The Inactivation of Enzymes by Ultraviolet Light. V. The Disruption of Specific Cystines in Ribonuclease. Radiat. Res. 1965, 26, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Kodzius, R.; Xiao, K.; Wu, J.; Yi, X.; Gong, X.; Foulds, I.G.; Wen, W. Inhibitory Effect of Common Microfluidic Materials on PCR Outcome. Sens. Actuators B Chem. 2012, 161, 349–358. [Google Scholar] [CrossRef]
- Roux, K.H. Optimization and Troubleshooting in PCR. Cold Spring Harb. J. Protoc. 2009, 2009, 66. [Google Scholar] [CrossRef]
- Kuffel, A.; Gray, A.; Daeid, N.N. Impact of Metal Ions on PCR Inhibition and RT-PCR Efficiency. Int. J. Leg. Med. 2021, 135, 63–72. [Google Scholar] [CrossRef]
- Dietrich, E.A.; Replogle, A.J.; Sheldon, S.W.; Petersen, J.M. Simultaneous Detection and Differentiation of Clinically Relevant Relapsing Fever Borrelia with Semimultiplex Real-Time PCR. J. Clin. Microbiol. 2021, 59, e0298120. [Google Scholar] [CrossRef]
- Vázquez-Morón, S.; Ryan, P.; Ardizone-Jiménez, B.; Martín, D.; Troya, J.; Cuevas, G.; Valencia, J.; Jimenez-Sousa, M.A.; Avellón, A.; Resino, S. Evaluation of Dried Blood Spot Samples for Screening of Hepatitis C and Human Immunodeficiency Virus in a Real-World Setting. Sci. Rep. 2018, 8, 1–6. [Google Scholar]
- Sarrazin, C.; Teuber, G.; Kokka, R.; Rabenau, H.; Zeuzem, S. Detection of Residual Hepatitis C Virus RNA by Transcription-Mediated Amplification in Patients with Complete Virologic Response According to Polymerase Chain Reaction–Based Assays. Hepatology 2000, 32, 818–823. [Google Scholar] [CrossRef]
- Ji, M.H.; Hu, G.F.; Zheng, Y.; Gu, D.Y.; Long, J.; Lu, W.P.; He, J.A.; Tan, S.Q.; Shi, L.; Liu, C.X.; et al. Surface Plasmon Resonance Technology Combined with Rolling Circle Amplification for Detection of Hepatitis C Virus. J. Shanghai Jiaotong Univ. (Med. Sci. ) 2012, 32, 693. [Google Scholar]
- Firdaus, R.; Saha, K.; Biswas, A.; Sadhukhan, P.C. Current Molecular Methods for the Detection of Hepatitis C Virus in High Risk Group Population: A Systematic Review. World J. Virol. 2015, 4, 25. [Google Scholar] [CrossRef]
- Pauly, M.D.; Kamili, S.; Hayden, T.M. Impact of Nucleic Acid Extraction Platforms on Hepatitis Virus Genome Detection. J. Virol. Methods 2019, 273, 113715. [Google Scholar] [CrossRef] [PubMed]
- Warkad, S.D.; Song, K.S.; Pal, D.; Nimse, S.B. Developments in the HCV Screening Technologies Based on the Detection of Antigens and Antibodies. Sensors 2019, 19, 4257. [Google Scholar] [CrossRef] [PubMed]
- Sam, S.S.; Steinmetz, H.B.; Tsongalis, G.J.; Tafe, L.J.; Lefferts, J.A. Validation of a Solid-Phase Electrochemical Array for Genotyping Hepatitis C Virus. Exp. Mol. Pathol. 2013, 95, 18–22. [Google Scholar] [CrossRef]
- Lu, M.; Xu, L.; Zhang, X.; Xiao, R.; Wang, Y. Ag(I)-Coordinated Hairpin DNA for Homogenous Electronic Monitoring of Hepatitis C Virus Accompanying Isothermal Cycling Signal Amplification Strategy. Biosens. Bioelectron. 2015, 73, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Yin, J.; Lv, S.; Wang, B.; Mu, Y. Advanced “Lab-on-a-Chip” to Detect Viruses—Current Challenges and Future Perspectives. Biosens. Bioelectron. 2020, 163, 112291. [Google Scholar] [CrossRef] [PubMed]
- Llibre, A.; Shimakawa, Y.; Mottez, E.; Ainsworth, S.; Buivan, T.P.; Firth, R.; Harrison, E.; Rosenberg, A.R.; Meritet, J.F.; Fontanet, A.; et al. Development and Clinical Validation of the Genedrive Point-of-Care Test for Qualitative Detection of Hepatitis C Virus. Gut 2018, 67, 2017–2024. [Google Scholar] [CrossRef] [PubMed]
- Monteail, S.; Casson, A.J.; Jones, S.T. Electronic and electrochemical viral detection for point-of-care use: A systematic review. PLoS ONE 2021, 16, e0258002. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Talbot, E.A.; Loring, C.H.; Power, J.J.; Dionne-Odom, J.; Alroy-Preis, S.; Jackson, P.; Bean, C.L. Performance of the OraQuick HCV Rapid Antibody Test for Screening Exposed Patients in a Hepatitis C Outbreak Investigation. J. Clin. Microbiol. 2014, 52, 2650. [Google Scholar] [CrossRef]
- Xu, R.; Gan, X.; Fang, Y.; Zheng, S.; Dong, Q. A Simple, Rapid, and Sensitive Integrated Protein Microarray for Simultaneous Detection of Multiple Antigens and Antibodies of Five Human Hepatitis Viruses (HBV, HCV, HDV, HEV, and HGV). Anal. Biochem. 2007, 362, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.D.; Teshale, E.; Jewett, A.; Weinbaum, C.M.; Neaigus, A.; Hagan, H.; Jenness, S.M.; Melville, S.K.; Burt, R.; Thiede, H.; et al. Performance of Premarket Rapid Hepatitis C Virus Antibody Assays in 4 National Human Immunodeficiency Virus Behavioral Surveillance System Sites. Clin. Infect. Dis. 2011, 53, 780–786. [Google Scholar] [CrossRef]
- Yu, Y.C.; Wang, Y.; He, C.L.; Wang, M.R.; Wang, Y.M. Management of Hepatitis C Virus Infection in Hemodialysis Patients. World J. Hepatol. 2014, 6, 419. [Google Scholar] [CrossRef][Green Version]
- Leary, T.P.; Gutierrez, R.A.; Muerhoff, A.S.; Birkenmeyer, L.G.; Desai, S.M.; Dawson, G.J. A Chemiluminescent, Magnetic Particle-Based Immunoassay for the Detection of Hepatitis C Virus Core Antigen in Human Serum or Plasma. J. Med. Virol. 2006, 78, 1436–1440. [Google Scholar] [CrossRef] [PubMed]
- Timurdogan, E.; Alaca, B.E.; Kavakli, I.H.; Urey, H. MEMS Biosensor for Detection of Hepatitis A and C Viruses in Serum. Biosens. Bioelectron. 2011, 28, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Fourati, S.; Feld, J.J.; Chevaliez, S.; Luhmann, N. Approaches for Simplified HCV Diagnostic Algorithms. J. Int. AIDS Soc. 2018, 21, e25058. [Google Scholar] [CrossRef] [PubMed]

| Sequence Name | Sequence 5′ to 3′ | Concentration | Tm | 5′d ΔG | 3′d ΔG |
|---|---|---|---|---|---|
| F3 | AAACCCACTCTATGCCCG | 4 | 58.3 | −5.11 | −7.53 |
| B3 | TACTCCGCCAACGATCTG | 4 | 57.9 | −4.43 | −4.25 |
| FIP | GCCCTATCAGGCAGTACCACAAGCAAGACTGCTAGCCGAG | 32 | |||
| BIP | GCACCATGAGCACAAATCCTGGGAACTTAACGTCTTCTGG | 32 | |||
| LF | GCCTTTCGCAACCCAACGCTA | 16 | 65.6 | −5.35 | −5.65 |
| LB | AAACCAAAAGAAACACCAACCGTCG | 16 | 64.6 | −4.67 | −6.96 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, S.; Thomas, E.; Caputi, M.; Asghar, W. RT-LAMP-Based Molecular Diagnostic Set-Up for Rapid Hepatitis C Virus Testing. Biosensors 2022, 12, 298. https://doi.org/10.3390/bios12050298
Sharma S, Thomas E, Caputi M, Asghar W. RT-LAMP-Based Molecular Diagnostic Set-Up for Rapid Hepatitis C Virus Testing. Biosensors. 2022; 12(5):298. https://doi.org/10.3390/bios12050298
Chicago/Turabian StyleSharma, Sandhya, Emmanuel Thomas, Massimo Caputi, and Waseem Asghar. 2022. "RT-LAMP-Based Molecular Diagnostic Set-Up for Rapid Hepatitis C Virus Testing" Biosensors 12, no. 5: 298. https://doi.org/10.3390/bios12050298
APA StyleSharma, S., Thomas, E., Caputi, M., & Asghar, W. (2022). RT-LAMP-Based Molecular Diagnostic Set-Up for Rapid Hepatitis C Virus Testing. Biosensors, 12(5), 298. https://doi.org/10.3390/bios12050298






