Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis
Abstract
1. Introduction
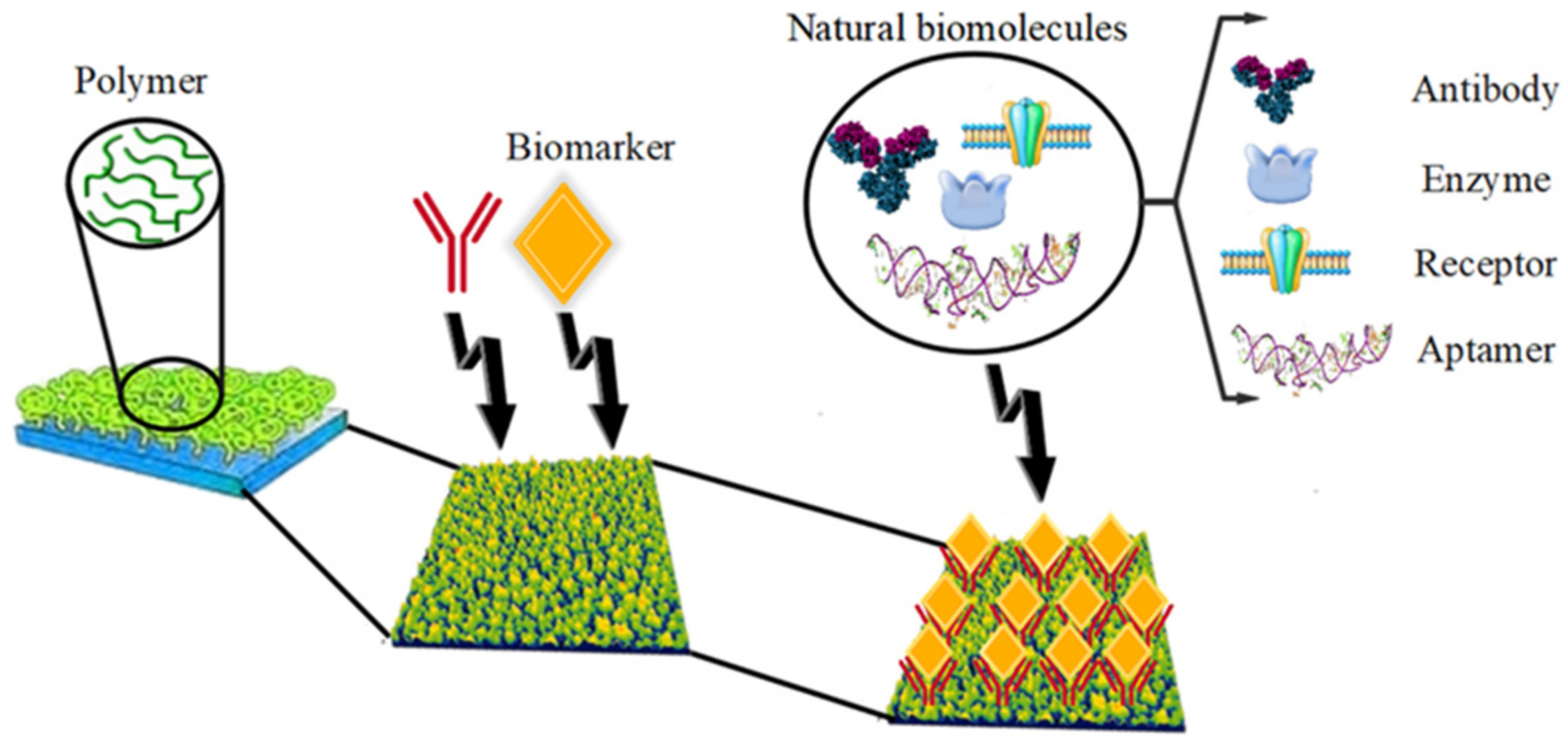
2. Polymer Biomarkers
3. Plasma-Engineered Polymer
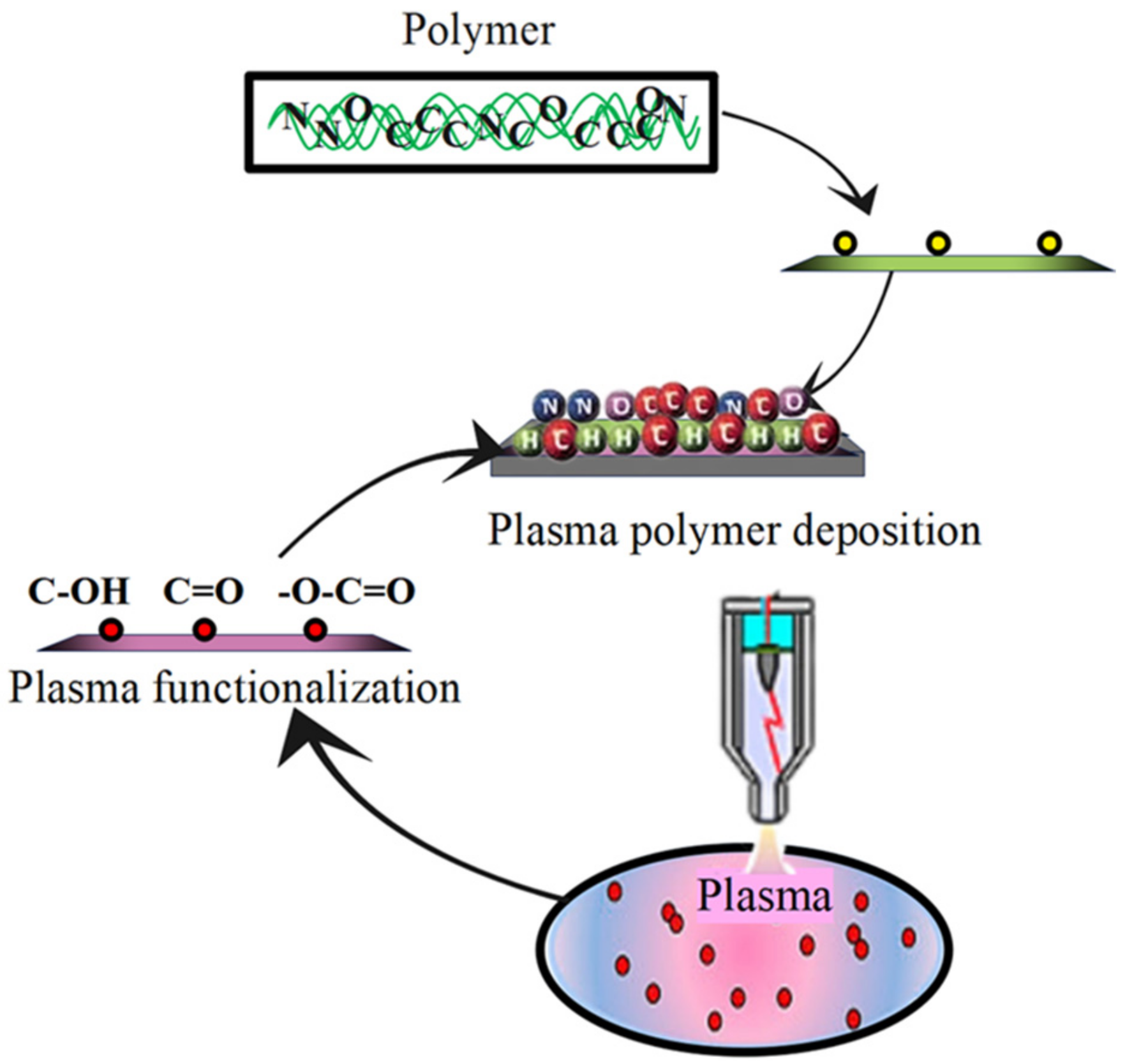
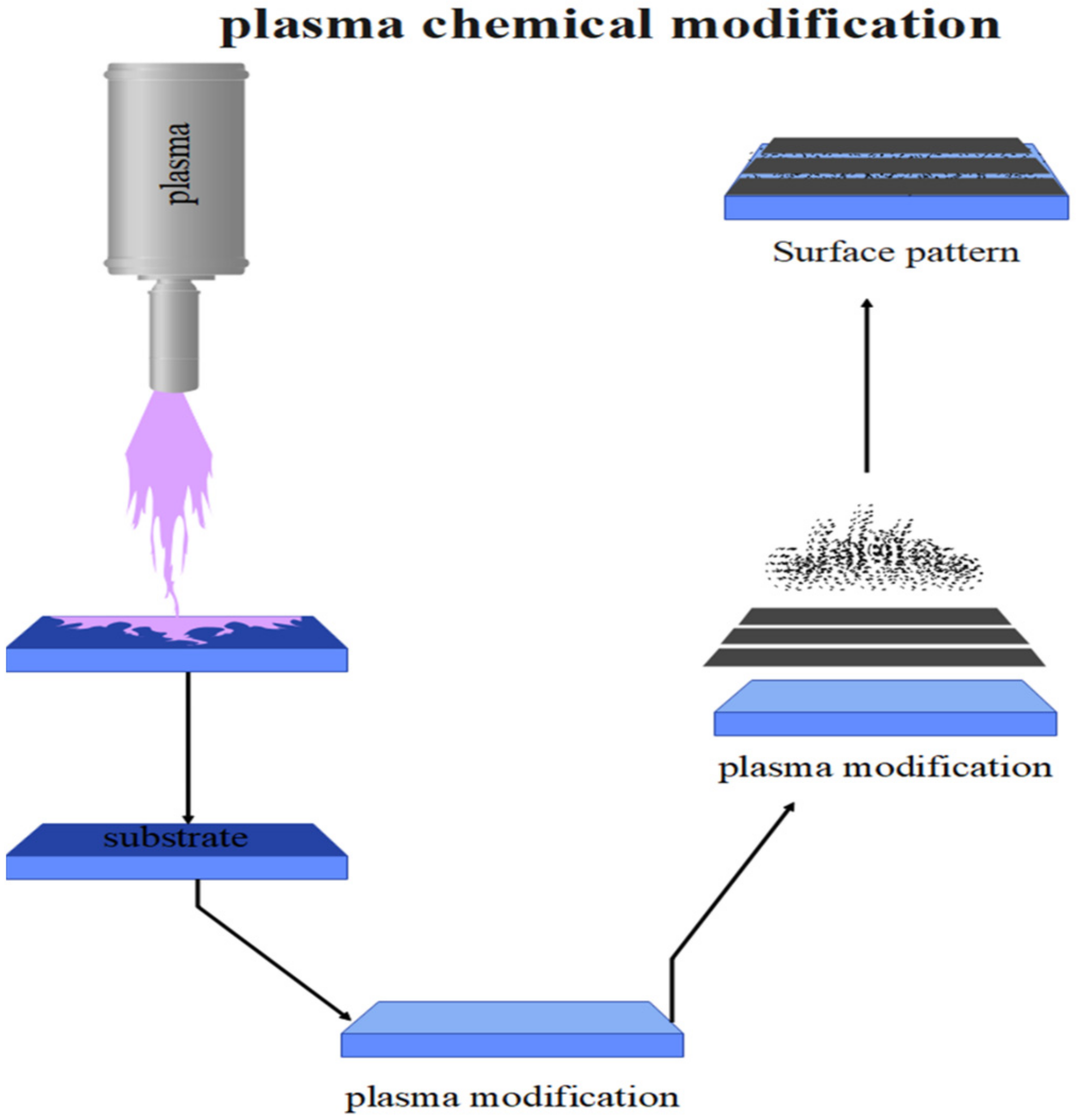
3.1. Plasma Processing of Surface Morphology for Polymers
3.2. Plasma Processing of Surface Chemistry for Polymers
4. Highly Multiplexed Analysis for Viral Infections
5. Latest Applications of Plasma-Engineered Polymer-Based Biomarker for Viral Detection
5.1. HIV & HCV
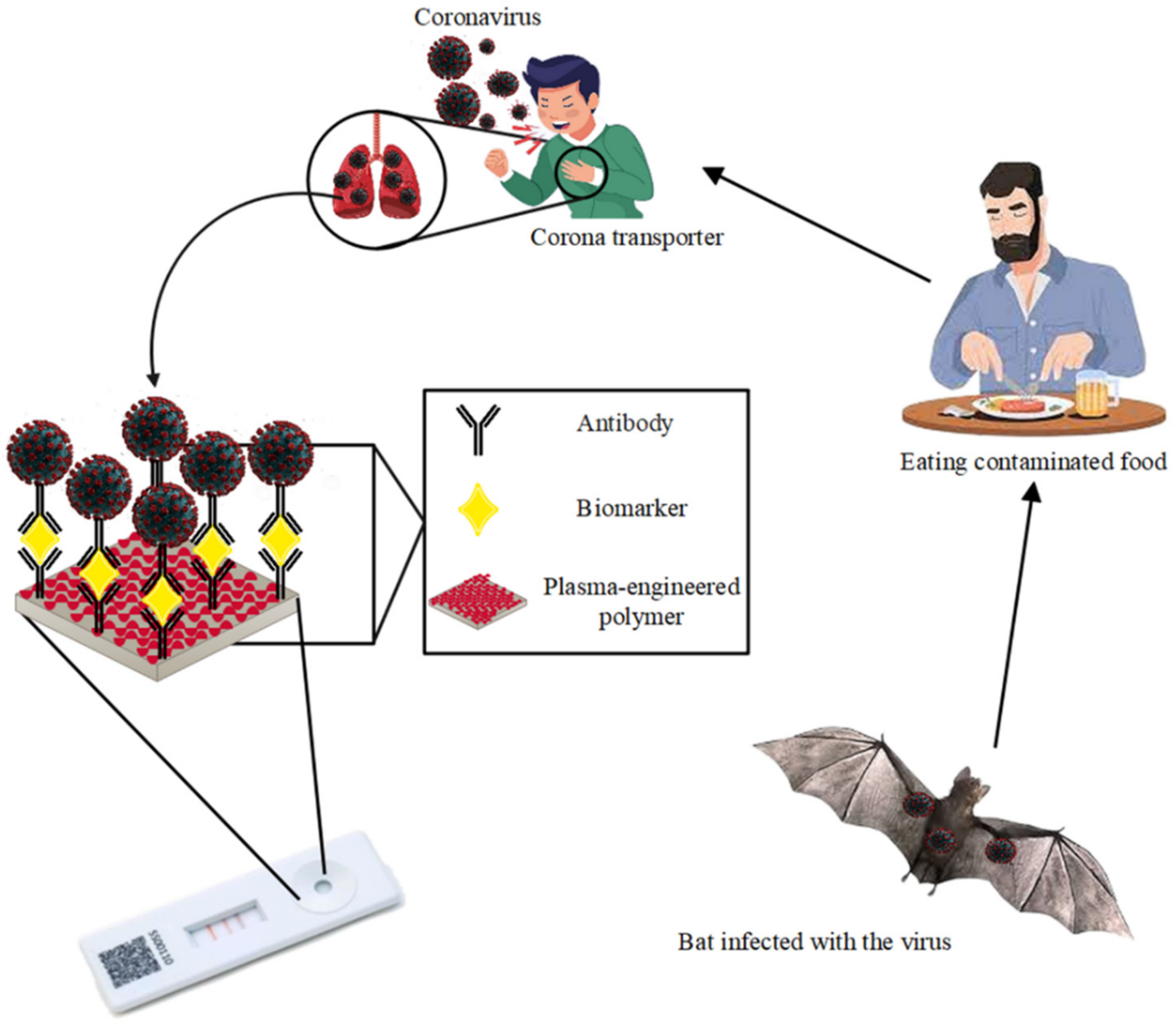
5.2. COVID-19
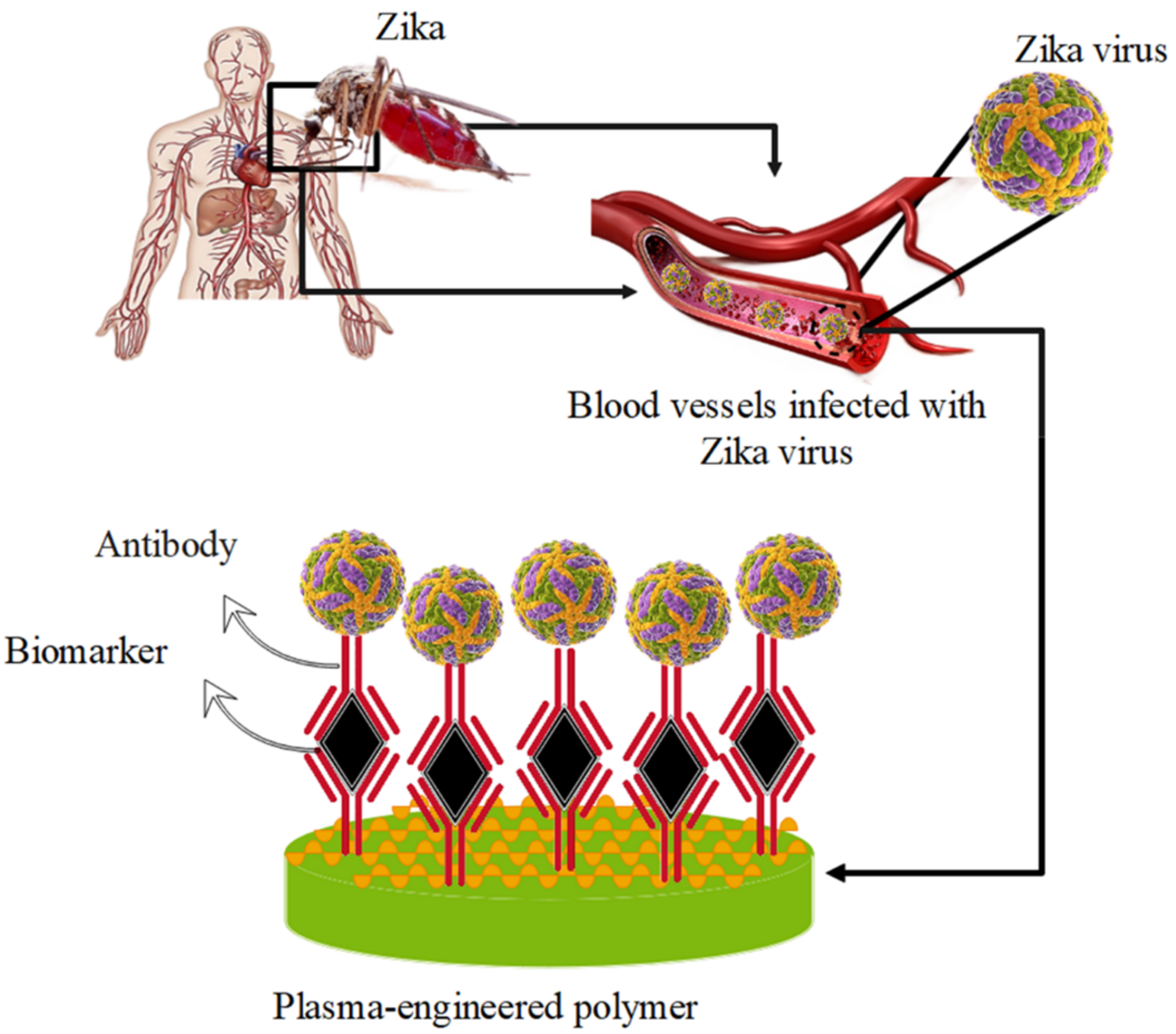
5.3. Zika
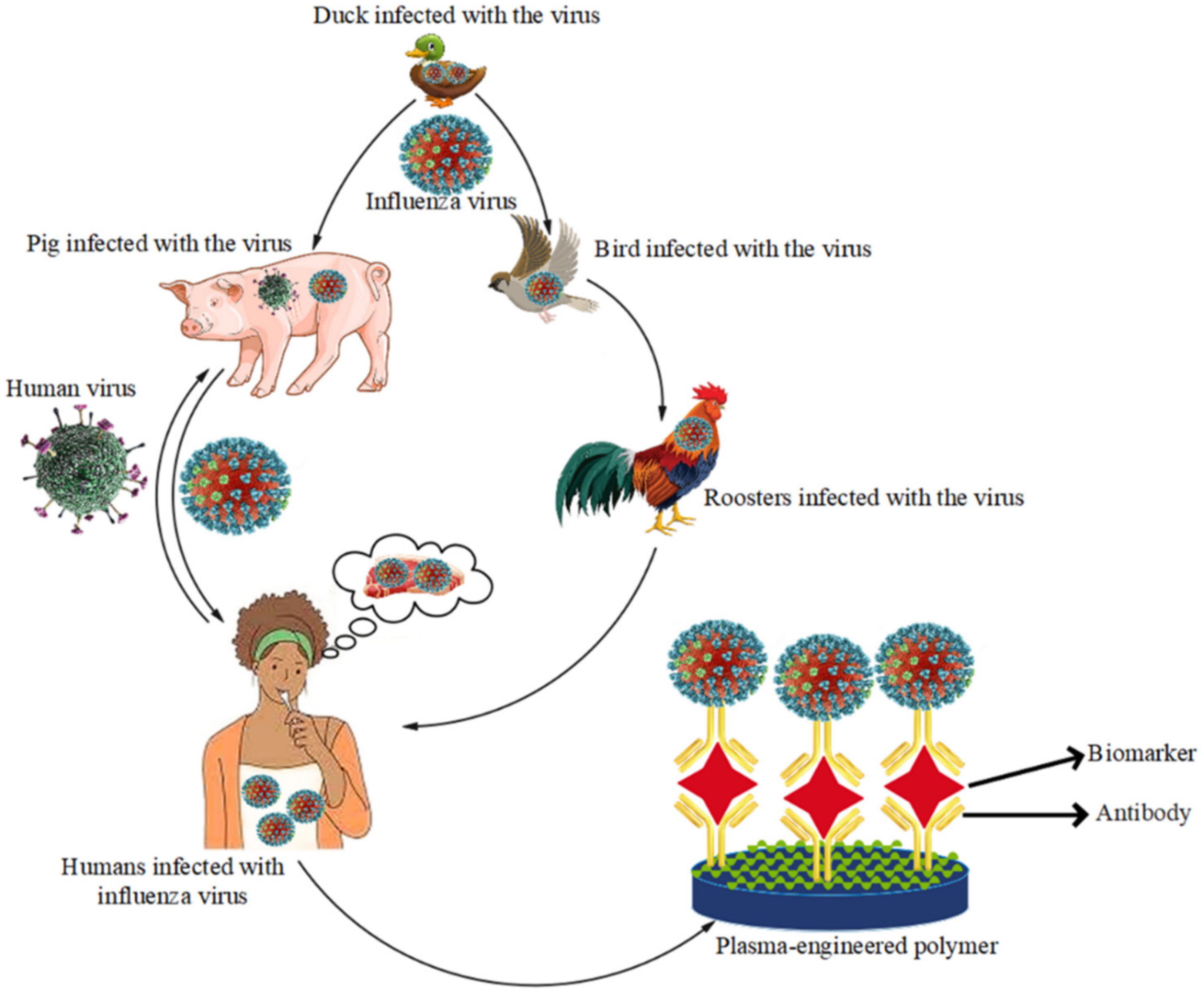
5.4. Influenza
6. Future and Perspectives
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scott, J.C. History of conductive polymers. In Nanostructured Conductive Polymers; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 1–17. [Google Scholar]
- Malik, A.A.; Nantasenamat, C.; Piacham, T. Molecularly imprinted polymer for human viral pathogen detection. Mater. Sci. Eng. C 2017, 77, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Hotez, P.J. Blue Marble Health and “the Big Three Diseases”: HIV/AIDS, Tuberculosis, and Malaria. Microbes Infect. 2015, 17, 539–541. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.K.; Gack, M.U. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016, 14, 360–373. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.; Mujahid, A.; Schirhagl, R.; Bajwa, S.Z.; Latif, U.; Feroz, S. Gravimetric viral diagnostics: QCM based biosensors for early detection of viruses. Chemosensors 2017, 5, 7. [Google Scholar] [CrossRef]
- Berretta, R.; Moscato, P. Cancer biomarker discovery: The entropic hallmark. PLoS ONE 2010, 5, e12262. [Google Scholar] [CrossRef]
- Prensner, J.R.; Chinnaiyan, A.M.; Srivastava, S. Systematic, evidence-based discovery of biomarkers at the NCI. Clin. Exp. Metastasis 2012, 29, 645–652. [Google Scholar] [CrossRef]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463. [Google Scholar] [CrossRef]
- Wagner, P.D.; Srivastava, S. New paradigms in translational science research in cancer biomarkers. Transl. Res. 2012, 159, 343–353. [Google Scholar] [CrossRef]
- Koehler, J.W.; Douglas, C.E.; Minogue, T.D. A highly multiplexed broad pathogen detection assay for infectious disease diagnostics. PLoS Negl. Trop. Dis. 2018, 12, e0006889. [Google Scholar] [CrossRef]
- Gerdes, M.J.; Sevinsky, C.J.; Sood, A.; Adak, S.; Bello, M.O.; Bordwell, A.; Can, A.; Corwin, A.; Dinn, S.; Filkins, R.J. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. USA 2013, 110, 11982–11987. [Google Scholar] [CrossRef]
- Credle, J.J.; Robinson, M.L.; Gunn, J.; Monaco, D.; Sie, B.; Tchir, A.; Hardick, J.; Zheng, X.; Shaw-Saliba, K.; Rothman, R.E. Highly multiplexed oligonucleotide probe-ligation testing enables efficient extraction-free SARS-CoV-2 detection and viral genotyping. Mod. Pathol. 2021, 34, 1093–1103. [Google Scholar] [CrossRef] [PubMed]
- Shaffer, S.M.; Joshi, R.P.; Chambers, B.S.; Sterken, D.; Biaesch, A.G.; Gabrieli, D.J.; Li, Y.; Feemster, K.A.; Hensley, S.E.; Issadore, D. Multiplexed detection of viral infections using rapid in situ RNA analysis on a chip. Lab Chip 2015, 15, 3170–3182. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, S.A.; Mousavi, S.M.; Bahrani, S.; Ramakrishna, S.; Babapoor, A.; Chiang, W.-H. Coupled graphene oxide with hybrid metallic nanoparticles as potential electrochemical biosensors for precise detection of ascorbic acid within blood. Anal. Chim. Acta 2020, 1107, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Gast, A.P.; Adamson, A.W. Physical Chemistry of Surfaces; Wiley New York: New York, NY, USA, 1997. [Google Scholar]
- Levchenko, I.; Xu, S.; Baranov, O.; Bazaka, O.; Ivanova, E.P.; Bazaka, K. Plasma and polymers: Recent progress and trends. Molecules 2021, 26, 4091. [Google Scholar] [CrossRef] [PubMed]
- Mei, X.; Ye, D.; Zhang, F.; Di, C.a. Implantable application of polymer-based biosensors. J. Polym. Sci. 2022, 60, 328–347. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Zarei, M.; Hashemi, S.A.; Babapoor, A.; Amani, A.M. A conceptual review of rhodanine: Current applications of antiviral drugs, anticancer and antimicrobial activities. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1132–1148. [Google Scholar] [CrossRef]
- Egitto, F.D. Plasma etching and modification of organic polymers. Pure Appl. Chem. 1990, 62, 1699–1708. [Google Scholar] [CrossRef]
- Chapman, B. Glow Discharge Processes: Sputtering and Plasma Etching; The University of Michigan: Ann Arbor, MI, USA, 1980. [Google Scholar]
- Chan, C.-M.; Ko, T.-M.; Hiraoka, H. Polymer surface modification by plasmas and photons. Surf. Sci. Rep. 1996, 24, 1–54. [Google Scholar] [CrossRef]
- Gomathi, N.; Sureshkumar, A.; Neogi, S. RF plasma-treated polymers for biomedical applications. Curr. Sci. 2008, 94, 1478–1486. [Google Scholar]
- Gancarz, I.; Bryjak, J.; Poźniak, G.; Tylus, W. Plasma modified polymers as a support for enzyme immobilization II. Amines plasma. Eur. Polym. J. 2003, 39, 2217–2224. [Google Scholar] [CrossRef]
- Bahrani, S.; Hashemi, S.A.; Mousavi, S.M.; Azhdari, R. Zinc-based metal–organic frameworks as nontoxic and biodegradable platforms for biomedical applications: Review study. Drug Metab. Rev. 2019, 51, 356–377. [Google Scholar] [CrossRef] [PubMed]
- Witjes, C.D.; van Aalten, S.M.; Steyerberg, E.W.; Borsboom, G.J.; de Man, R.A.; Verhoef, C.; IJzermans, J.N. Recently introduced biomarkers for screening of hepatocellular carcinoma: A systematic review and meta-analysis. Hepatol. Int. 2013, 7, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Waidely, E.; Al-Yuobi, A.-R.O.; Bashammakh, A.; El-Shahawi, M.S.; Leblanc, R.M. Serum protein biomarkers relevant to hepatocellular carcinoma and their detection. Analyst 2016, 141, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Rachner, T.D.; Thiele, S.; Göbel, A.; Browne, A.; Fuessel, S.; Erdmann, K.; Wirth, M.P.; Fröhner, M.; Todenhöfer, T.; Muders, M.H. High serum levels of Dickkopf-1 are associated with a poor prognosis in prostate cancer patients. BMC Cancer 2014, 14, 649. [Google Scholar] [CrossRef] [PubMed]
- Bazié, W.W.; Boucher, J.; Traoré, I.T.; Kania, D.; Somé, D.Y.; Alary, M.; Gilbert, C. Vesicular MicroRNA as Potential Biomarkers of Viral Rebound. Cells 2022, 11, 859. [Google Scholar] [CrossRef] [PubMed]
- Price, R.W.; Peterson, J.; Fuchs, D.; Angel, T.E.; Zetterberg, H.; Hagberg, L.; Spudich, S.; Smith, R.D.; Jacobs, J.M.; Brown, J.N. Approach to cerebrospinal fluid (CSF) biomarker discovery and evaluation in HIV infection. J. Neuroimmune Pharmacol. 2013, 8, 1147–1158. [Google Scholar] [CrossRef]
- Boslaugh, S. Encyclopedia of Epidemiology; Sage Publications: Thousand Oaks, CA, USA, 2007. [Google Scholar]
- Zhang, L.; Guo, H. Biomarkers of COVID-19 and technologies to combat SARS-CoV-2. Adv. Biomark. Sci. Technol. 2020, 2, 1–23. [Google Scholar] [CrossRef]
- Lauc, G.; Sinclair, D. Biomarkers of biological age as predictors of COVID-19 disease severity. Aging 2020, 12, 6490. [Google Scholar] [CrossRef]
- Song, G.; Rho, H.-S.; Pan, J.; Ramos, P.; Yoon, K.-J.; Medina, F.A.; Lee, E.M.; Eichinger, D.; Ming, G.-l.; Muñoz-Jordan, J.L. Multiplexed biomarker panels discriminate Zika and Dengue virus infection in humans. Mol. Cell. Proteom. 2018, 17, 349–356. [Google Scholar] [CrossRef]
- Peng, F.; Loo, J.F.C.; Kong, S.K.; Li, B.; Gu, D. Identification of serum MicroRNAs as diagnostic biomarkers for influenza H7N9 infection. Virol. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Zarei Ghobadi, M.; Mozhgani, S.-H.; Farzanehpour, M.; Behzadian, F. Identifying novel biomarkers of the pediatric influenza infection by weighted co-expression network analysis. Virol. J. 2019, 16, 124. [Google Scholar] [CrossRef] [PubMed]
- Bhakta, S.; Mishra, P. Molecularly imprinted polymer-based sensors for cancer biomarker detection. Sens. Actuators Rep. 2021, 3, 100061. [Google Scholar] [CrossRef]
- O’Connell, P.J.; Guilbault, G.G. Future trends in biosensor research. Anal. Lett. 2001, 34, 1063–1078. [Google Scholar] [CrossRef]
- Boyer, P.D.; Krebs, E.G. The Enzymes; Academic Press: Cambridge, MA, USA, 1986. [Google Scholar]
- Kapingidza, A.B.; Kowal, K.; Chruszcz, M. Antigen–antibody complexes. In Vertebrate and Invertebrate Respiratory Proteins, Lipoproteins and other Body Fluid Proteins; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 465–497. [Google Scholar]
- Huber, R. Structural basis for antigen-antibody recognition. Science 1986, 233, 702–703. [Google Scholar] [CrossRef] [PubMed]
- Abdelghani, R.; Hassan, H.S.; Morsi, I.; Kashyout, A. Nano-architecture of highly sensitive SnO2–based gas sensors for acetone and ammonia using molecular imprinting technique. Sens. Actuators B Chem. 2019, 297, 126668. [Google Scholar] [CrossRef]
- Altintas, Z.; Gittens, M.; Guerreiro, A.; Thompson, K.-A.; Walker, J.; Piletsky, S.; Tothill, I.E. Detection of waterborne viruses using high affinity molecularly imprinted polymers. Anal. Chem. 2015, 87, 6801–6807. [Google Scholar] [CrossRef] [PubMed]
- Wulff, G. Molecular imprinting in cross-linked materials with the aid of molecular templates—A way towards artificial antibodies. Angew. Chem. Int. Ed. Engl. 1995, 34, 1812–1832. [Google Scholar] [CrossRef]
- Kriz, D.; Ramström, O.; Mosbach, K. Peer reviewed: Molecular imprinting: New possibilities for sensor technology. Anal. Chem. 1997, 69, 345A–349A. [Google Scholar] [CrossRef]
- BelBruno, J.J. Molecularly imprinted polymers. Chem. Rev. 2018, 119, 94–119. [Google Scholar] [CrossRef]
- Park, R.; Jeon, S.; Jeong, J.; Park, S.-Y.; Han, D.-W.; Hong, S.W. Recent Advances of Point-of-Care Devices Integrated with Molecularly Imprinted Polymers-Based Biosensors: From Biomolecule Sensing Design to Intraoral Fluid Testing. Biosensors 2022, 12, 136. [Google Scholar] [CrossRef]
- Akagi, T.; Baba, M.; Akashi, M. Biodegradable nanoparticles as vaccine adjuvants and delivery systems: Regulation of immune responses by nanoparticle-based vaccine. In Polymers in Nanomedicine; Springer: Berlin/Heidelberg, Germany, 2011; pp. 31–64. [Google Scholar]
- Vilar, G.; Tulla-Puche, J.; Albericio, F. Polymers and drug delivery systems. Curr. Drug Deliv. 2012, 9, 367–394. [Google Scholar] [CrossRef] [PubMed]
- Arefi, F.; Andre, V.; Montazer-Rahmati, P.; Amouroux, J. Plasma polymerization and surface treatment of polymers. Pure Appl. Chem. 1992, 64, 715–723. [Google Scholar] [CrossRef]
- Bazaka, K.; Jacob, M.V.; Crawford, R.J.; Ivanova, E.P. Plasma-assisted surface modification of organic biopolymers to prevent bacterial attachment. Acta Biomater. 2011, 7, 2015–2028. [Google Scholar] [CrossRef] [PubMed]
- Mousavi, S.M.; Hashemi, S.A.; Salahi, S.; Hosseini, M.; Amani, A.M.; Babapoor, A. Development of Clay Nanoparticles toward Bio and Medical Applications; IntechOpen: London, UK, 2018. [Google Scholar]
- Meichsner, J.; Schmidt, M.; Schneider, R.; Wagner, H.-E. Nonthermal Plasma Chemistry and Physics; CRC Press: Boca Raton, FL, USA, 2013. [Google Scholar]
- Vossen, J.L.; Kern, W.; Kern, W. Thin Film Processes II; Gulf Professional Publishing: Houston, TX, USA, 1991; Volume 2. [Google Scholar]
- Biederman, H.; Kudrna, P.; Slavinska, D. Hard plasma polymers, composites and plasma polymer films prepared by rf sputtering of conventional polymers. In Plasma Polymer Films; World Scientific: Singapore, 2004; pp. 289–324. [Google Scholar]
- Mousavi, S.M.; Low, F.W.; Hashemi, S.A.; Samsudin, N.A.; Shakeri, M.; Yusoff, Y.; Rahsepar, M.; Lai, C.W.; Babapoor, A.; Soroshnia, S. Development of hydrophobic reduced graphene oxide as a new efficient approach for photochemotherapy. RSC Adv. 2020, 10, 12851–12863. [Google Scholar] [CrossRef]
- Kalashgarani, M.Y.; Babapoor, A. Application of nano-antibiotics in the diagnosis and treatment of infectious diseases. Adv. Appl. NanoBio-Technol. 2022, 3, 22–35. [Google Scholar]
- Ao, W.; Lim, J.-S.; Shin, P.-K. Preparation and characterization of plasma polymerized methyl methacrylate thin films as gate dielectric for organic thin film transistor. J. Electr. Eng. Technol. 2011, 6, 836–841. [Google Scholar] [CrossRef]
- Krishnapura, P.R.; Belur, P.D.; Subramanya, S. A critical review on properties and applications of microbial l-asparaginases. Crit. Rev. Microbiol. 2016, 42, 720–737. [Google Scholar]
- Nagpal, K.; Singh, S.K.; Mishra, D.N. Chitosan nanoparticles: A promising system in novel drug delivery. Chem. Pharm. Bull. 2010, 58, 1423–1430. [Google Scholar] [CrossRef]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M.M. A comprehensive review on L-asparaginase and its applications. Appl. Biochem. Biotechnol. 2016, 178, 900–923. [Google Scholar] [CrossRef]
- Manrich, A.; Galvão, C.M.; Jesus, C.D.; Giordano, R.C.; Giordano, R.L. Immobilization of trypsin on chitosan gels: Use of different activation protocols and comparison with other supports. Int. J. Biol. Macromol. 2008, 43, 54–61. [Google Scholar] [CrossRef]
- Huang, K.-S.; Sheu, Y.-R.; Chao, I.-C. Preparation and properties of nanochitosan. Polym. Plast. Technol. Eng. 2009, 48, 1239–1243. [Google Scholar] [CrossRef]
- Borcia, G.; Anderson, C.; Brown, N. Dielectric barrier discharge for surface treatment: Application to selected polymers in film and fibre form. Plasma Sources Sci. Technol. 2003, 12, 335. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Ramakrishna, S.; Esmaeili, H.; Bahrani, S.; Koosha, M.; Babapoor, A. Green synthesis of supermagnetic Fe3O4–MgO nanoparticles via Nutmeg essential oil toward superior anti-bacterial and anti-fungal performance. J. Drug Deliv. Sci. Technol. 2019, 54, 101352. [Google Scholar] [CrossRef]
- Holubka, J.W.; Haack, L.P.; Straccia, A. Environmentally Friendly Reactive Fixture to Allow Localized Surface Engineering for Improved Adhesion to Coated and Non-Coated Substrates. U.S. Patent 8,586,149 B2, 19 November 2013. [Google Scholar]
- Fridman, A. Plasma Chemistry; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Dorranian, D.; Abedini, Z.; Hojabri, A.; Ghoranneviss, M. Structural and optical characterization of PMMA surface treated in low power nitrogen and oxygen RF plasmas. J. Non-Oxide Glasses 2009, 1, 217–229. [Google Scholar]
- Yamamoto, T.; Okubo, M.; Hayakawa, K.; Kitaura, K. Towards ideal NO/sub x/control technology using a plasma-chemical hybrid process. IEEE Trans. Ind. Appl. 2001, 37, 1492–1498. [Google Scholar] [CrossRef]
- Dorranian, D.; Golian, Y.; Hojabri, A. Investigation of nitrogen plasma effect on the nonlinear optical properties of PMMA. J. Theor. Appl. Phys. 2012, 6, 1. [Google Scholar] [CrossRef]
- Inagaki, N. Plasma Surface Modification and Plasma Polymerization; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Ahmadi, S.; Fazilati, M.; Mousavi, S.M.; Nazem, H. Anti-bacterial/fungal and anti-cancer performance of green synthesized Ag nanoparticles using summer savory extract. J. Exp. Nanosci. 2020, 15, 363–380. [Google Scholar] [CrossRef]
- Garbassi, F.; Morra, M.; Ochiello, E. From Physics to Technology. In Polymer Surfaces; Wiley: Hoboken, NJ, USA, 1994. [Google Scholar]
- Shishoo, R. Plasma Technologies for Textiles; Elsevier: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Mousavi, S.M.; Low, F.W.; Hashemi, S.A.; Lai, C.W.; Ghasemi, Y.; Soroshnia, S.; Savardashtaki, A.; Babapoor, A.; Pynadathu Rumjit, N.; Goh, S.M. Development of graphene based nanocomposites towards medical and biological applications. Artif. Cells Nanomed. Biotechnol. 2020, 48, 1189–1205. [Google Scholar] [CrossRef]
- Chu, P.K.; Chen, J.; Wang, L.; Huang, N. Plasma-surface modification of biomaterials. Mater. Sci. Eng. R Rep. 2002, 36, 143–206. [Google Scholar] [CrossRef]
- Surmenev, R.; Chernozem, R.; Syromotina, D.; Oehr, C.; Baumbach, T.; Krause, B.; Boyandin, A.; Dvoinina, L.; Volova, T.; Surmeneva, M. Low-temperature argon and ammonia plasma treatment of poly-3-hydroxybutyrate films: Surface topography and chemistry changes affect fibroblast cells in vitro. Eur. Polym. J. 2019, 112, 137–145. [Google Scholar] [CrossRef]
- Kalia, S. Biodegradable Green Composites; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Yang, P.; Moloney, M.G.; Zhang, F.; Ji, W. Surface hydrophobic modification of polymers with fluorodiazomethanes. Mater. Lett. 2018, 210, 295–297. [Google Scholar] [CrossRef]
- Ahmadi, S.; Fazilati, M.; Nazem, H.; Mousavi, S.M. Green synthesis of magnetic nanoparticles using Satureja hortensis essential oil toward superior antibacterial/fungal and anticancer performance. BioMed Res. Int. 2021, 2021, 8822645. [Google Scholar] [CrossRef] [PubMed]
- Magliulo, M.; Pistillo, B.R.; Mulla, M.Y.; Cotrone, S.; Ditaranto, N.; Cioffi, N.; Favia, P.; Torsi, L. PE-CVD of Hydrophilic-COOH Functionalized Coatings on Electrolyte Gated Field-Effect Transistor Electronic Layers. Plasma Processes Polym. 2013, 10, 102–109. [Google Scholar] [CrossRef]
- Group, B.D.W.; Atkinson, A.J., Jr.; Colburn, W.A.; DeGruttola, V.G.; DeMets, D.L.; Downing, G.J.; Hoth, D.F.; Oates, J.A.; Peck, C.C.; Schooley, R.T. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 2001, 69, 89–95. [Google Scholar]
- Wu, S.; Liu, L.; Li, G.; Jing, F.; Mao, H.; Jin, Q.; Zhai, W.; Zhang, H.; Zhao, J.; Jia, C. Multiplexed detection of lung cancer biomarkers based on quantum dots and microbeads. Talanta 2016, 156, 48–54. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, C.; Bai, S.; Gao, Y.; Metcalfe, G.; Cheng, W.; Zhu, Y. Multiplexed detection of cancer biomarkers using a microfluidic platform integrating single bead trapping and acoustic mixing techniques. Nanoscale 2018, 10, 20196–20206. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Mazraedoost, S.; Yousefi, K.; Gholami, A.; Behbudi, G.; Ramakrishna, S.; Omidifar, N.; Alizadeh, A.; Chiang, W.-H. Multifunctional gold nanorod for therapeutic applications and pharmaceutical delivery considering cellular metabolic responses, oxidative stress and cellular longevity. Nanomaterials 2021, 11, 1868. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, J.; Liu, J.; Sun, S.; Xiong, Y.; Ma, Y.; Yan, S.; Yang, Y.; Yin, H.; Cai, X. Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers. Biosens. Bioelectron. 2019, 136, 84–90. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Luan, C.; Wang, H.; Chen, B.; Zhao, Y. Hybrid hydrogel photonic barcodes for multiplex detection of tumor markers. Biosens. Bioelectron. 2017, 87, 264–270. [Google Scholar] [CrossRef]
- Zou, Z.; Yang, H.; Yan, Q.; Qi, P.; Qing, Z.; Zheng, J.; Xu, X.; Zhang, L.; Feng, F.; Yang, R. Synchronous screening of multiplexed biomarkers of Alzheimer’s disease by a length-encoded aerolysin nanopore-integrated triple-helix molecular switch. Chem. Commun. 2019, 55, 6433–6436. [Google Scholar] [CrossRef]
- Fabri-Faja, N.; Calvo-Lozano, O.; Dey, P.; Terborg, R.A.; Estevez, M.-C.; Belushkin, A.; Yesilköy, F.; Duempelmann, L.; Altug, H.; Pruneri, V. Early sepsis diagnosis via protein and miRNA biomarkers using a novel point-of-care photonic biosensor. Anal. Chim. Acta 2019, 1077, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wang, W.; Yao, S.; Wu, S.; Zhang, H.; Zhang, J.; Jing, F.; Mao, H.; Jin, Q.; Cong, H. Highly sensitive detection of multiple tumor markers for lung cancer using gold nanoparticle probes and microarrays. Anal. Chim. Acta 2017, 958, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Abootalebi, S.N.; Mousavi, S.M.; Hashemi, S.A.; Shorafa, E.; Omidifar, N.; Gholami, A. Antibacterial effects of green-synthesized silver nanoparticles using Ferula asafoetida against Acinetobacter baumannii isolated from the hospital environment and assessment of their cytotoxicity on the human cell lines. J. Nanomater. 2021, 2021, 6676555. [Google Scholar] [CrossRef]
- Ngo, H.T.; Wang, H.-N.; Burke, T.; Ginsburg, G.S.; Vo-Dinh, T. Multiplex detection of disease biomarkers using SERS molecular sentinel-on-chip. Anal. Bioanal. Chem. 2014, 406, 3335–3344. [Google Scholar] [CrossRef]
- Guo, J.; Huang, X.; Ma, X. Clinical identification of diabetic ketosis/diabetic ketoacidosis acid by electrochemical dual channel test strip with medical smartphone. Sens. Actuators B Chem. 2018, 275, 446–450. [Google Scholar] [CrossRef]
- Guo, J.; Ma, X. Simultaneous monitoring of glucose and uric acid on a single test strip with dual channels. Biosens. Bioelectron. 2017, 94, 415–419. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Gholami, A.; Omidifar, N.; Zarei, M.; Bahrani, S.; Yousefi, K.; Chiang, W.-H.; Babapoor, A. Bioinorganic synthesis of polyrhodanine stabilized Fe3O4/Graphene oxide in microbial supernatant media for anticancer and antibacterial applications. Bioinorg. Chem. Appl. 2021, 2021, 9972664. [Google Scholar] [CrossRef]
- Elnifro, E.M.; Ashshi, A.M.; Cooper, R.J.; Klapper, P.E. Multiplex PCR: Optimization and application in diagnostic virology. Clin. Microbiol. Rev. 2000, 13, 559–570. [Google Scholar] [CrossRef]
- Jackson, B.; Busch, M.; Stramer, S.; AuBuchon, J. The cost-effectiveness of NAT for HIV, HCV, and HBV in whole-blood donations. Transfusion 2003, 43, 721–729. [Google Scholar] [CrossRef]
- Masoumzadeh, R. Polyethyleneimine-based materials for gene therapy, bioimaging and drug delivery systems applications. Adv. Appl. NanoBio-Technol. 2021, 2, 13–16. [Google Scholar]
- Candotti, D.; Temple, J.; Owusu-Ofori, S.; Allain, J.-P. Multiplex real-time quantitative RT-PCR assay for hepatitis B virus, hepatitis C virus, and human immunodeficiency virus type 1. J. Virol. Methods 2004, 118, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Volk, W.A.; Gebhardt, B.; Hammaskjold, M.; Kaomer, R. Medical Microbiology; Lippincott-Raven: Philadelphia, PA, USA, 1995. [Google Scholar]
- Poritz, M.A.; Lingenfelter, B. Multiplex PCR for Detection and identification of microbial pathogens. In Advanced Techniques in Diagnostic Microbiology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 475–493. [Google Scholar]
- Wong, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. Biotechniques 2005, 39, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Dobaño, C.; Vidal, M.; Santano, R.; Jiménez, A.; Chi, J.; Barrios, D.; Ruiz-Olalla, G.; Rodrigo Melero, N.; Carolis, C.; Parras, D. Highly sensitive and specific multiplex antibody assays to quantify immunoglobulins M, A, and G against SARS-CoV-2 antigens. J. Clin. Microbiol. 2020, 59, e01731-20. [Google Scholar] [CrossRef]
- Hoste, A.C.; Ruiz, T.; Fernández-Pacheco, P.; Jiménez-Clavero, M.Á.; Djadjovski, I.; Moreno, S.; Brun, A.; Edwards, T.A.; Barr, J.N.; Rueda, P. Development of a multiplex assay for antibody detection in serum against pathogens affecting ruminants. Transbound. Emerg. Dis. 2021, 68, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.A.; Kullo, I.J.; Bailey, K.R.; Klee, G.G. Antibody-based protein multiplex platforms: Technical and operational challenges. Clin. Chem. 2010, 56, 186–193. [Google Scholar] [CrossRef]
- Alipour, A.; Kalashgarani, M.Y. Nano Protein and Peptides for Drug Delivery and Anticancer Agents. Adv. Appl. NanoBio-Technol. 2022, 3, 60–64. [Google Scholar]
- Munro, S.B.; Kuypers, J.; Jerome, K.R. Comparison of a multiplex real-time PCR assay with a multiplex Luminex assay for influenza virus detection. J. Clin. Microbiol. 2013, 51, 1124–1129. [Google Scholar] [CrossRef][Green Version]
- Cordray, M.S.; Richards-Kortum, R.R. Emerging nucleic acid–based tests for point-of-care detection of malaria. Am. J. Trop. Med. Hyg. 2012, 87, 223. [Google Scholar] [CrossRef]
- Dudak, F.C.; Boyaci, I.H. Multiplex Detection of Escherichia Coli and Salmonella Enteritidis by Using Quantum Dot-Labeled Antibodies. J. Rapid Methods Autom. Microbiol. 2009, 17, 315–327. [Google Scholar] [CrossRef]
- Bilan, R.; Ametzazurra, A.; Brazhnik, K.; Escorza, S.; Fernández, D.; Uríbarri, M.; Nabiev, I.; Sukhanova, A. Quantum-dot-based suspension microarray for multiplex detection of lung cancer markers: Preclinical validation and comparison with the Luminex xMAP® system. Sci. Rep. 2017, 7, 44668. [Google Scholar] [CrossRef]
- Allain, J.-P.; Stramer, S.L.; Carneiro-Proietti, A.; Martins, M.; Da Silva, S.L.; Ribeiro, M.; Proietti, F.; Reesink, H.W. Transfusion-transmitted infectious diseases. Biologicals 2009, 37, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Traineau, R.; Elghouzzi, M.-H.; Bierling, P. Update on infectious risks associated with blood products. Rev. Prat. 2009, 59, 86–89. [Google Scholar] [PubMed]
- Thomas, D.L. The challenge of hepatitis C in the HIV-infected person. Annu. Rev. Med. 2008, 59, 473–485. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hosseini, M.; Seyed, A.N.S.; Kheyr, A.P.; Esmaeili, J.G.R.; Shirzad, H.; Karami, N.; Jahani, M.; Seyed, A.M.; Paivarmehr, F.; Mohraz, M. Prevalence and correlates of co-infection with human immunodeficiency virus and hepatitis C virus in male injection drug users in Iran. Arch. Iran. Med. 2010, 13, 318–323. [Google Scholar] [PubMed]
- Mohammadi, M.; Talei, G.; Sheikhian, A.; Ebrahimzade, F.; Pournia, Y.; Ghasemi, E.; Boroun, H. Survey of both hepatitis B virus (HBsAg) and hepatitis C virus (HCV-Ab) coinfection among HIV positive patients. Virol. J. 2009, 6, 202. [Google Scholar] [CrossRef]
- Nelson, K.E.; Thomas, D.L. Reciprocal interaction of human immunodeficiency virus and hepatitis C virus infections. Clin. Diagn. Lab. Immunol. 2001, 8, 867–870. [Google Scholar] [CrossRef][Green Version]
- Takmil, F.; Esmaeili, H.; Mousavi, S.M.; Hashemi, S.A. Nano-magnetically modified activated carbon prepared by oak shell for treatment of wastewater containing fluoride ion. Adv. Powder Technol. 2020, 31, 3236–3245. [Google Scholar] [CrossRef]
- Busch, M.; Kleinman, S.; Jackson, B.; Stramer, S.; Hewlett, I.; Preston, S. Committee report. Nucleic acid amplification testing of blood donors for transfusion-transmitted infectious diseases: Report of the Interorganizational Task Force on Nucleic Acid Amplification Testing of Blood Donors. Transfusion 2000, 40, 143–159. [Google Scholar]
- Bierbaum, S.; Forster, J.; Berner, R.; Rücker, G.; Rohde, G.; Neumann-Haefelin, D.; Panning, M. Detection of respiratory viruses using a multiplex real-time PCR assay in Germany, 2009/10. Arch. Virol. 2014, 159, 669–676. [Google Scholar] [CrossRef]
- Cecilia, D.; Kakade, M.; Alagarasu, K.; Patil, J.; Salunke, A.; Parashar, D.; Shah, P. Development of a multiplex real-time RT-PCR assay for simultaneous detection of dengue and chikungunya viruses. Arch. Virol. 2015, 160, 323–327. [Google Scholar] [CrossRef]
- Xu, X.; Bao, H.; Ma, Y.; Sun, J.; Zhao, Y.; Wang, Y.; Shi, J.; Zeng, X.; Li, Y.; Wang, X. Simultaneous detection of novel H7N9 and other influenza A viruses in poultry by multiplex real-time RT-PCR. Virol. J. 2015, 12, 69. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santiago, G.A.; Vergne, E.; Quiles, Y.; Cosme, J.; Vazquez, J.; Medina, J.F.; Medina, F.; Colón, C.; Margolis, H.; Muñoz-Jordán, J.L. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl. Trop. Dis. 2013, 7, e2311. [Google Scholar] [CrossRef]
- Mousavi, S.M.; Hashemi, S.A.; Amani, A.M.; Saed, H.; Jahandideh, S.; Mojoudi, F. Polyethylene terephthalate/acryl butadiene styrene copolymer incorporated with oak shell, potassium sorbate and egg shell nanoparticles for food packaging applications: Control of bacteria growth, physical and mechanical properties. Polym. Renew. Resour. 2017, 8, 177–196. [Google Scholar] [CrossRef]
- Höfler, D.; Nicklas, W.; Mauter, P.; Pawlita, M.; Schmitt, M. A bead-based multiplex assay for the detection of DNA viruses infecting laboratory rodents. PLoS ONE 2014, 9, e97525. [Google Scholar] [CrossRef] [PubMed]
- Barbara, J.A.; Garson, J.A. Polymerase chain reaction and transfusion microbiology. Vox Sanguinis. 1993, 64, 73–81. [Google Scholar] [CrossRef]
- Kalland, K.-H.; Myrmel, H.; Nordbø, S.A. Nukleinsyrediagnostikk i medisinsk mikrobiologi. Tidsskr. Den Nor. Legeforening 2005. Available online: https://tidsskriftet.no/2005/11/medisin-og-vitenskap/nukleinsyrediagnostikk-i-medisinsk-mikrobiologi (accessed on 29 March 2022).
- Mackay, I.M.; Arden, K.E.; Nitsche, A. Real-time PCR in virology. Nucleic Acids Res. 2002, 30, 1292–1305. [Google Scholar] [CrossRef]
- Hashemi, S.A.; Mousavi, S.M.; Bahrani, S.; Ramakrishna, S. Integrated polyaniline with graphene oxide-iron tungsten nitride nanoflakes as ultrasensitive electrochemical sensor for precise detection of 4-nitrophenol within aquatic media. J. Electroanal. Chem. 2020, 873, 114406. [Google Scholar] [CrossRef]
- Fu, X.; Cheng, Z.; Yu, J.; Choo, P.; Chen, L.; Choo, J. A SERS-based lateral flow assay biosensor for highly sensitive detection of HIV-1 DNA. Biosens. Bioelectron. 2016, 78, 530–537. [Google Scholar] [CrossRef]
- Rahimian, P.; He, J.J. HIV/neuroAIDS biomarkers. Prog. Neurobiol. 2017, 157, 117–132. [Google Scholar] [CrossRef]
- Azhdari, R.; Mousavi, S.M.; Hashemi, S.A.; Bahrani, S.; Ramakrishna, S. Decorated graphene with aluminum fumarate metal organic framework as a superior non-toxic agent for efficient removal of Congo Red dye from wastewater. J. Environ. Chem. Eng. 2019, 7, 103437. [Google Scholar] [CrossRef]
- Perlman, S. Another decade, another coronavirus. Mass Med. Soc. 2020, 382, 760–762. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. COVID-19: A drug repurposing and biomarker identification by using comprehensive gene-disease associations through protein-protein interaction network analysis. Preprints 2020. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Fadaka, A.O.; Sibuyi, N.R.S.; Adewale, O.B.; Bakare, O.O.; Akanbi, M.O.; Klein, A.; Madiehe, A.M.; Meyer, M. Understanding the epidemiology, pathophysiology, diagnosis and management of SARS-CoV-2. J. Int. Med. Res. 2020, 48, 0300060520949077. [Google Scholar] [CrossRef]
- Campos, C.; Bernuci, M.; Vireque, A.; Campos, J.; Silva-de-Sá, M.; Jamur, M.; Rosa-e-Silva, A. Preventing microbial contamination during long-term in vitro culture of human granulosa-lutein cells: An ultrastructural analysis. Int. Sch. Res. Not. 2012, 2012, 152781. [Google Scholar] [CrossRef]
- Sanyaolu, A.; Okorie, C.; Marinkovic, A.; Ayodele, O.; Abbasi, A.F.; Prakash, S.; Ahmed, M.; Kayode, D.; Jaferi, U.; Haider, N. Navigating the Diagnostics of COVID-19. SN Compr. Clin. Med. 2020, 2, 1393–1400. [Google Scholar] [CrossRef]
- Samson, R.; Navale, G.R.; Dharne, M.S. Biosensors: Frontiers in rapid detection of COVID-19. 3 Biotech 2020, 10, 385. [Google Scholar] [CrossRef]
- Zhu, X.; Wang, X.; Han, L.; Chen, T.; Wang, L.; Li, H.; Li, S.; He, L.; Fu, X.; Chen, S. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020, 166, 112437. [Google Scholar] [CrossRef]
- Gholami, A.; Hashemi, S.A.; Yousefi, K.; Mousavi, S.M.; Chiang, W.-H.; Ramakrishna, S.; Mazraedoost, S.; Alizadeh, A.; Omidifar, N.; Behbudi, G. 3D nanostructures for tissue engineering, cancer therapy, and gene delivery. J. Nanomater. 2020, 2020, 1852946. [Google Scholar] [CrossRef]
- Khan, M.; Hasan, M.; Hossain, S.; Ahommed, M.; Daizy, M. Ultrasensitive detection of pathogenic viruses with electrochemical biosensor: State of the art. Biosens. Bioelectron. 2020, 166, 112431. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.-H.; Chang, T.-J.; Wang, M.-L.; Tsai, P.-H.; Lin, T.-H.; Wang, C.-T.; Yang, D.-M. Novel biosensor platforms for the detection of coronavirus infection and severe acute respiratory syndrome coronavirus 2. J. Chin. Med. Assoc. 2020, 83, 701. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.S.; Toh, C.-S. Novel biosensing methodologies for ultrasensitive detection of viruses. Analyst 2013, 138, 6219–6229. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Rawal, G.; Baxi, M. Zika virus: An emergence of a new arbovirus. J. Clin. Diagn. Res. JCDR 2016, 10, DM01. [Google Scholar] [CrossRef]
- Tavakoli, A.; Karbalaie Niya, M.H.; Keshavarz, M.; Safarnezhad Tameshke, F.; Monavari, S.H. Middle east respiratory syndrome coronavirus (MERS-CoV). Iran J. Med. Microbiol. 2017, 11, 1–8. [Google Scholar]
- Mortazavi, H.S.; Monavari, S.H.; Ataei Pirkooh, A.; Tavakoli, A. Middle east respiratory syndrome coronavirus (MERS-CoV): A review article. Iran. J. Virol. 2014, 8, 59–68. [Google Scholar] [CrossRef]
- Slenczka, W. Zika virus disease. In Emerging Infections 10; Wiley: Hoboken, NJ, USA, 2016; pp. 163–173. [Google Scholar]
- Guo, D.; Wang, Y.-W.; Ma, J.; Yan, L.; Li, T.-F.; Han, X.-W.; Shui, S.-F. Study on the role of Cathepsin B and JNK signaling pathway in the development of cerebral aneurysm. Asian Pac. J. Trop. Med. 2016, 9, 499–502. [Google Scholar] [CrossRef]
- Younger, D.S. Epidemiology of Zika virus. Neurol. Clin. 2016, 34, 1049–1056. [Google Scholar] [CrossRef]
- Cabral-Miranda, G.; de Jesus, J.; Oliveira, P.S.; Britto, G.G.; Pontes-de-Carvalho, L.; Dutra, R.; Alcântara-Neves, N. Detection of parasite antigens in Leishmania infantum–infected spleen tissue by monoclonal antibody-, piezoelectric-based Immunosensors. J. Parasitol. 2014, 100, 73–78. [Google Scholar] [CrossRef]
- Cabral-Miranda, G.; Yamashiro-Kanashiro, E.; Gidlund, M.; Sales, M.G.F. Specific label-free and real-time detection of oxidized low density lipoprotein (oxLDL) using an immunosensor with three monoclonal antibodies. J. Mater. Chem. B 2014, 2, 477–484. [Google Scholar] [CrossRef]
- Cardoso, A.R.; Cabral-Miranda, G.; Reyes-Sandoval, A.; Bachmann, M.F.; Sales, M.G.F. Detecting circulating antibodies by controlled surface modification with specific target proteins: Application to malaria. Biosens. Bioelectron. 2017, 91, 833–841. [Google Scholar] [CrossRef]
- Afsahi, S.; Lerner, M.B.; Goldstein, J.M.; Lee, J.; Tang, X.; Bagarozzi, D.A., Jr.; Pan, D.; Locascio, L.; Walker, A.; Barron, F. Novel graphene-based biosensor for early detection of Zika virus infection. Biosens. Bioelectron. 2018, 100, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Francis, M.E.; King, M.L.; Kelvin, A.A. Back to the future for influenza preimmunity—Looking back at influenza virus history to infer the outcome of future infections. Viruses 2019, 11, 122. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.; Palese, P. Orthomyxoviridae. Fields Virol. 2013, 1, 1151–1185. [Google Scholar]
- Auladell, M.; Phuong, H.V.M.; Mai, L.T.Q.; Tseng, Y.-Y.; Carolan, L.; Wilks, S.; Thai, P.Q.; Price, D.; Duong, N.T.; Hang, N.L.K. Influenza virus infection history shapes antibody responses to influenza vaccination. Nat. Med. 2022, 28, 363–372. [Google Scholar] [CrossRef]
- Javid Khojasteh, V.; Mojtahedi, A.; Hosseini, S.; Joukar, F. Relative frequency of influenza A/H1N1 virus in Guilan province, Iran. Iran. J. Med. Microbiol. 2017, 11, 83–89. [Google Scholar]
- Kavunga-Membo, H.; Nkwembe, E.; Simulundu, E.; Karhemere, S.; Babakazo, P.; Manya, L.; Kabamba, J.; Okitolonda, E.; Ahuka-Mundeke, S.; Muyembe, J.J. Epidemiology of circulating human influenza viruses from the Democratic Republic of Congo, 2015. PLoS ONE 2018, 13, e0203995. [Google Scholar] [CrossRef]
- Van Kerkhove, M.D. Brief literature review for the WHO global influenza research agenda–highly pathogenic avian influenza H5N1 risk in humans. Influenza Other Respir. Viruses 2013, 7, 26–33. [Google Scholar] [CrossRef]
- Mahardika, G.N.; Suartha, N.I.; Kencana, G.A.; Suardana, I.B.; Mahardika, W.W.; Budayanti, N.S. Biochemistry and computer-generated graph comparison of the structural and nonstructural proteins of Spanish-1918 Influenza, pandemic-2009, and bird flu viruses. Acta Biochim. Pol. 2019, 66, 329–336. [Google Scholar] [CrossRef]
- Myers, K.P.; Olsen, C.W.; Gray, G.C. Cases of swine influenza in humans: A review of the literature. Clin. Infect. Dis. 2007, 44, 1084–1088. [Google Scholar] [CrossRef]
- Donatelli, I.; Castrucci, M.R.; Marco, M.A.D.; Delogu, M.; Webster, R.G. Human–animal interface: The case for influenza interspecies transmission. In Emerging and Re-Emerging Viral Infections; Springer: Berlin/Heidelberg, Germany, 2016; pp. 17–33. [Google Scholar]
- Xie, Z.; Pang, Y.-s.; Liu, J.; Deng, X.; Tang, X.; Sun, J.; Khan, M.I. A multiplex RT-PCR for detection of type A influenza virus and differentiation of avian H5, H7, and H9 hemagglutinin subtypes. Mol. Cell. Probes 2006, 20, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.-S.; Chang, P.-C.; Shien, J.-H.; Cheng, M.-C.; Shieh, H.K. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J. Virol. Methods 2001, 97, 13–22. [Google Scholar] [CrossRef]
- Wang, R.; Yan, W.; Du, M.; Tao, L.; Liu, J. The effect of influenza virus infection on pregnancy outcomes: A systematic review and meta-analysis of cohort studies. Int. J. Infect. Dis. 2021, 105, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.; Gamblin, S.; Haire, L.; Stevens, D.; Xiao, B.; Ha, Y.; Skehel, J. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 2004, 325, 287–296. [Google Scholar] [CrossRef]
- Webster, R.; Shortridge, K.; Kawaoka, Y. Influenza: Interspecies transmission and emergence of new pandemics. FEMS Immunol. Med. Microbiol. 1997, 18, 275. [Google Scholar] [CrossRef]
- Krammer, F. The human antibody response to influenza A virus infection and vaccination. Nat. Rev. Immunol. 2019, 19, 383–397. [Google Scholar] [CrossRef]
- Murray, P.R.; Rosenthal, K.S.; Pfaller, M.A. Medical Microbiology E-Book; Elsevier Health Sciences: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Octaviani, C.P.; Goto, H.; Kawaoka, Y. Reassortment between seasonal H1N1 and pandemic (H1N1) 2009 influenza viruses is restricted by limited compatibility among polymerase subunits. J. Virol. 2011, 85, 8449–8452. [Google Scholar] [CrossRef]
- Tumer, A. Biosensors: Sense and sensitivity. Science 2000, 290, 1315–1317. [Google Scholar]
- Amano, Y.; Cheng, Q. Detection of influenza virus: Traditional approaches and development of biosensors. Anal. Bioanal. Chem. 2005, 381, 156–164. [Google Scholar] [CrossRef]
- Pejcic, B.; De Marco, R.; Parkinson, G. The role of biosensors in the detection of emerging infectious diseases. Analyst 2006, 131, 1079–1090. [Google Scholar] [CrossRef]
- Koncki, R. Recent developments in potentiometric biosensors for biomedical analysis. Anal. Chim. Acta 2007, 599, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, Y. Biosensors for rapid detection of Avian Influenza. In Steps Forwards in Diagnosing and Controlling Influenza; BoD—Books on Demand: Norderstedt, Germany, 2016; p. 61. [Google Scholar]



| Infectious Disease | Infectious Biomarker | Detection Techniques | Ref. |
|---|---|---|---|
| Hepatitis B | AFP | ELISA | [25] |
| Hepatitis B | DCP | Electrochemiluminescence immunoassay | [26] |
| Hepatitis B | miRNA-21 | qRT-PCR | [27] |
| HIV | microRNA | RT-qPCR | [28] |
| HIV | HIV RNA | Cerebrospinal Fluid (CSF) | [29] |
| HIV | HIV antibodies | Plasma | [30] |
| COVID-19 | Viral RNA Genome | PCR Point-of-care detection | [31] |
| COVID-19 | Spike Protein | ELISA Laboratory testing | [32] |
| ZIKV | IgM antibodies | RT-PCR | [33] |
| Influenza | miRNAs | RT-qPCR | [34] |
| Influenza | 719 DEGs | Weighted gene co-expression network analysis (WGCNA) | [35] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mousavi, S.M.; Hashemi, S.A.; Kalashgrani, M.Y.; Gholami, A.; Omidifar, N.; Babapoor, A.; Vijayakameswara Rao, N.; Chiang, W.-H. Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors 2022, 12, 286. https://doi.org/10.3390/bios12050286
Mousavi SM, Hashemi SA, Kalashgrani MY, Gholami A, Omidifar N, Babapoor A, Vijayakameswara Rao N, Chiang W-H. Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors. 2022; 12(5):286. https://doi.org/10.3390/bios12050286
Chicago/Turabian StyleMousavi, Seyyed Mojtaba, Seyyed Alireza Hashemi, Masoomeh Yari Kalashgrani, Ahmad Gholami, Navid Omidifar, Aziz Babapoor, Neralla Vijayakameswara Rao, and Wei-Hung Chiang. 2022. "Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis" Biosensors 12, no. 5: 286. https://doi.org/10.3390/bios12050286
APA StyleMousavi, S. M., Hashemi, S. A., Kalashgrani, M. Y., Gholami, A., Omidifar, N., Babapoor, A., Vijayakameswara Rao, N., & Chiang, W.-H. (2022). Recent Advances in Plasma-Engineered Polymers for Biomarker-Based Viral Detection and Highly Multiplexed Analysis. Biosensors, 12(5), 286. https://doi.org/10.3390/bios12050286









