Intein-Mediated Protein Engineering for Cell-Based Biosensors
Abstract
:1. Introduction
2. Bio-Recognition Elements and Monitoring of Cellular Signaling
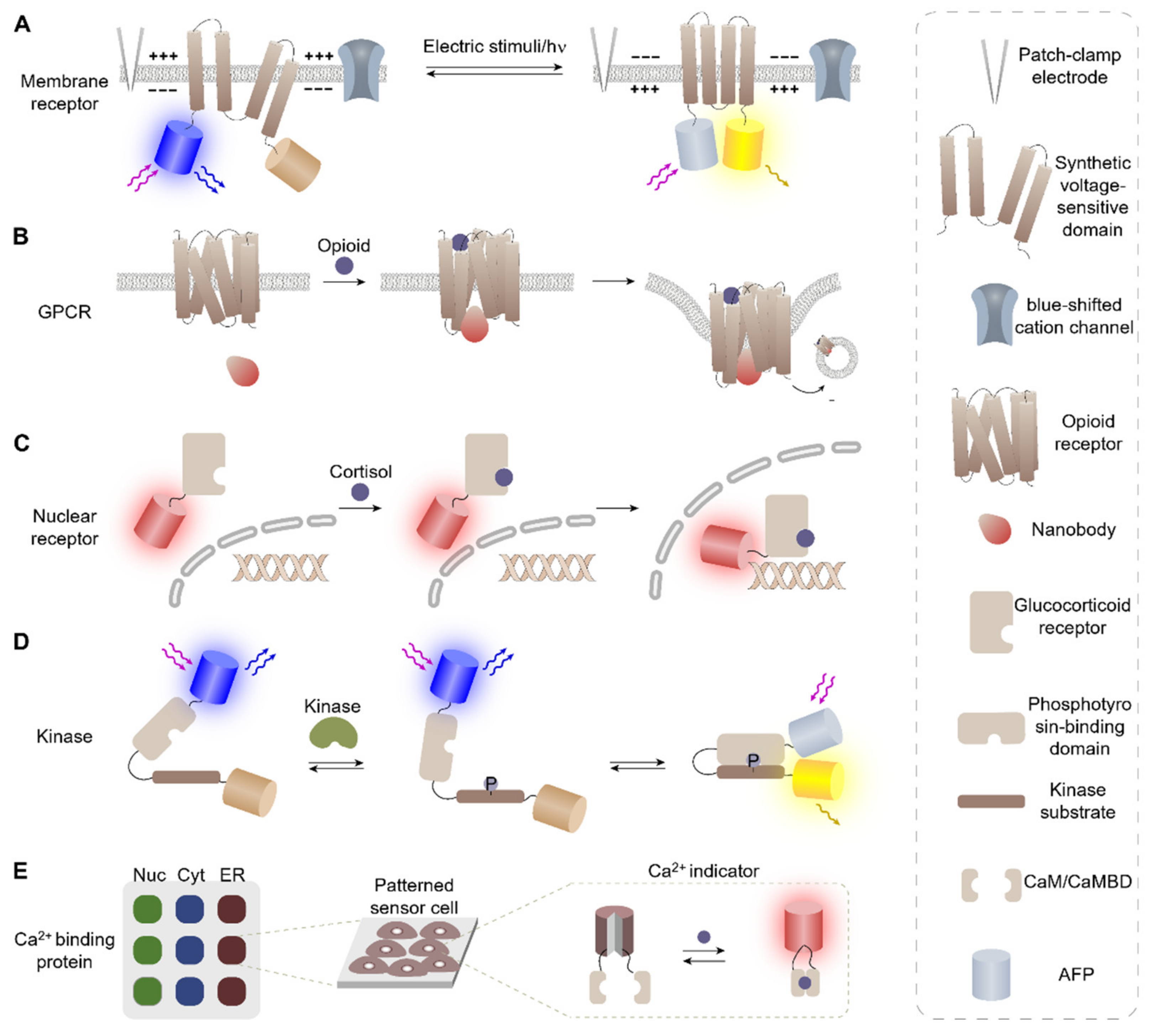
2.1. Membrane Receptors
2.2. Nuclear Receptors
2.3. Kinase Activity Assays
2.4. Genetically Encoded Ca2+ Indicators
3. Inteins and Intein-Mediated Protein Engineering
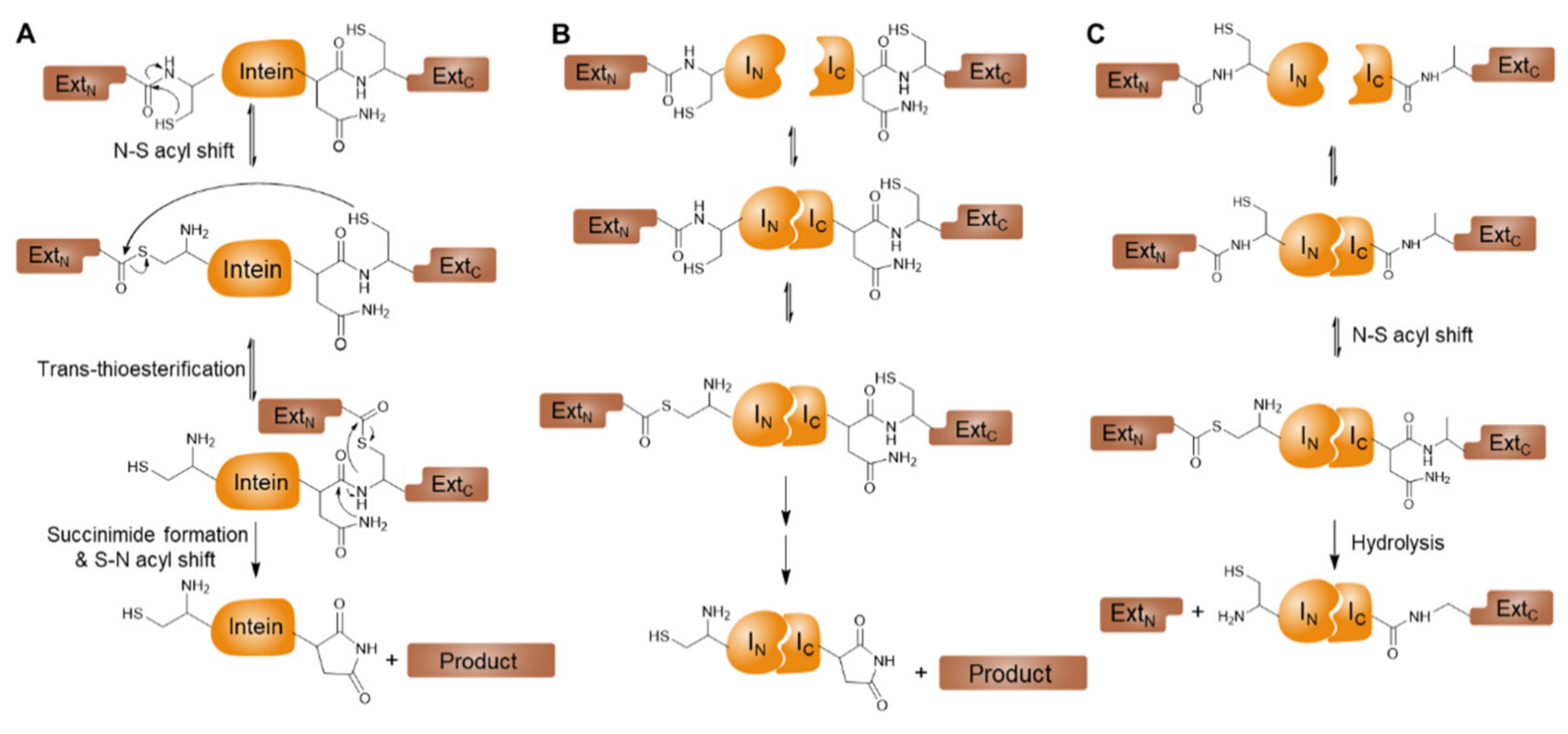
3.1. Inteins and Intein-Mediated Protein Splicing
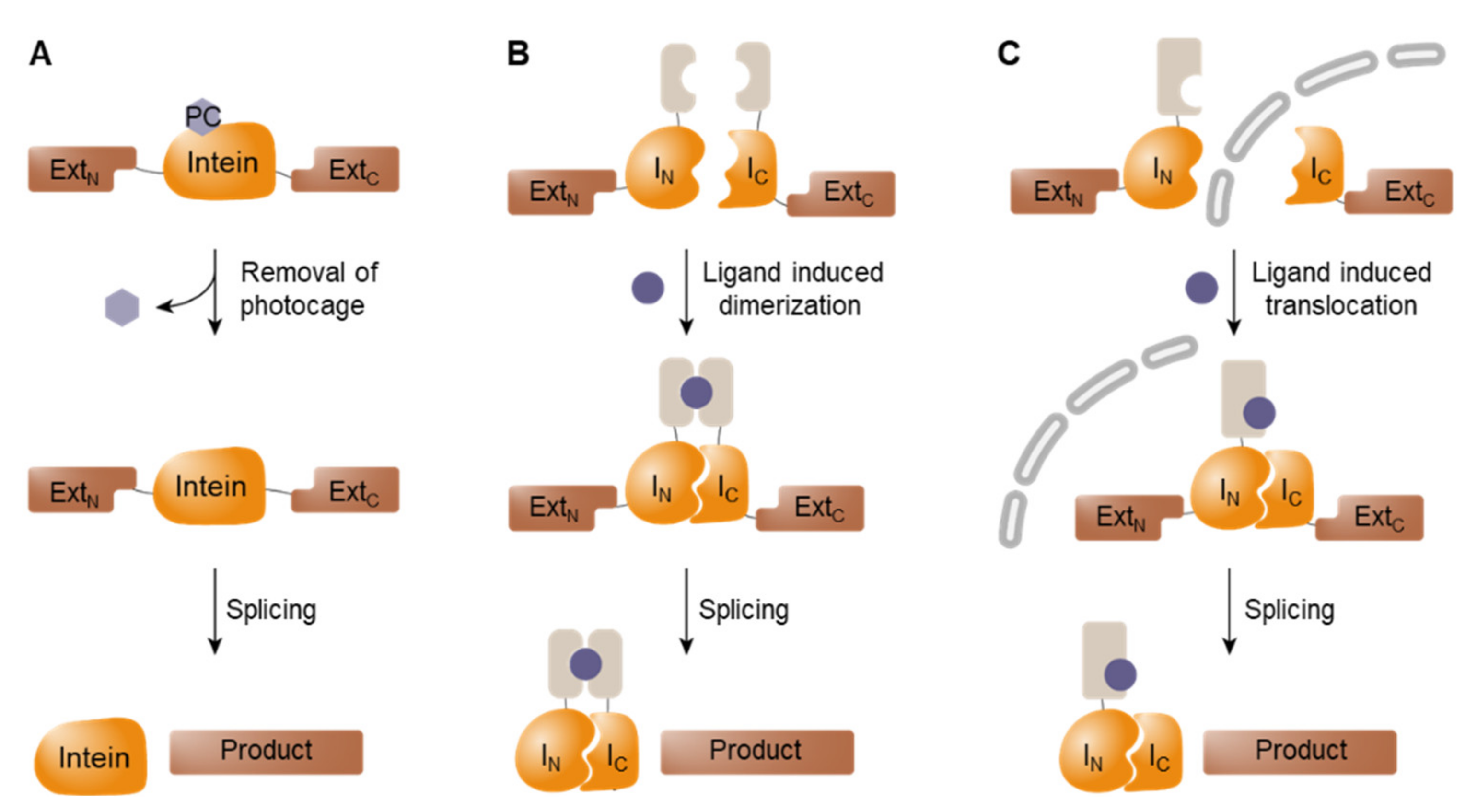
3.2. Conditional Protein Splicing (CPS)
4. Application of Intein-Mediated Reactions in Cell-Based Sensor Fabrication
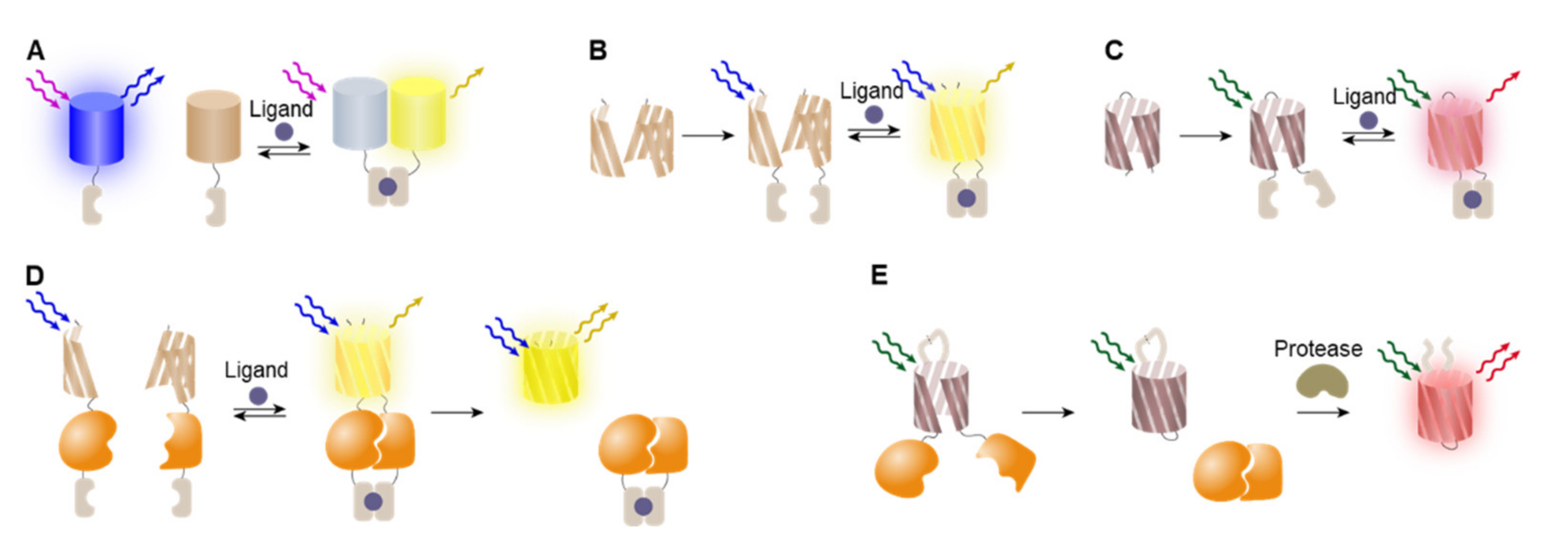
4.1. FRET-Based Detection System
4.2. BiFC-Based Detection System
4.3. Circularly Permuted Proteins for Biosensors
4.4. Cyclic Peptides as Biosensing Scaffolds
4.5. Intein Zymogens for Protease Activity Monitoring
4.6. Fluorescence Translocation Sensor via Signal Peptide Reconstitution
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Clark, L.C., Jr.; Lyons, C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann. N. Y. Acad. Sci. 1962, 102, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Gui, Q.; Lawson, T.; Shan, S.; Yan, L.; Liu, Y. The application of whole cell-based biosensors for use in environmental analysis and in medical diagnostics. Sensors 2017, 17, 1623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berti, F.; Turner, A.P. New micro-and nanotechnologies for electrochemical biosensor development. In Biosensor Nanomaterials; Li, S., Singh, J., Li, H., Banerjee, I.A., Eds.; Wiley-VCH: Weinheim, Germany, 2011; pp. 1–35. [Google Scholar]
- Lee, K.H.; Kim, D.M. In Vitro Use of Cellular Synthetic Machinery for Biosensing Applications. Front. Pharmacol. 2019, 10, 1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-C.E.; Lee, I.-C. The current trends of biosensors in tissue engineering. Biosensors 2020, 10, 88. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, A. Live cell-based sensor cells. Biomaterials 2010, 31, 5911–5915. [Google Scholar] [CrossRef]
- Zhou, Q.; Son, K.; Liu, Y.; Revzin, A. Biosensors for cell analysis. Annu. Rev. Biomed. Eng. 2015, 17, 165–190. [Google Scholar] [CrossRef] [Green Version]
- Li, Y. Split-inteins and their bioapplications. Biotechnol. Lett. 2015, 37, 2121–2137. [Google Scholar] [CrossRef]
- Kanno, A.; Ozawa, T.; Umezawa, Y. Intein-Mediated Reporter Gene Assay for Detecting Protein−Protein Interactions in Living Mammalian Cells. Anal. Chem. 2006, 78, 556–560. [Google Scholar] [CrossRef]
- Tian, L.; Hires, S.A.; Looger, L.L. Imaging neuronal activity with genetically encoded calcium indicators. Cold Spring Harb. Protoc. 2012, 2012, pdb.top069609. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Nakata, E.; Hamachi, I. Recent progress in strategies for the creation of protein-based fluorescent biosensors. ChemBioChem 2009, 10, 2560–2577. [Google Scholar] [CrossRef]
- Kim, Y.S.; Raston, N.H.; Gu, M.B. Aptamer-based nanobiosensors. Biosens. Bioelectron. 2016, 76, 2–19. [Google Scholar] [PubMed]
- Miller, K.E.; Kim, Y.; Huh, W.-K.; Park, H.-O. Bimolecular fluorescence complementation (BiFC) analysis: Advances and recent applications for genome-wide interaction studies. J. Mol. Biol. 2015, 427, 2039–2055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frommer, W.B.; Davidson, M.W.; Campbell, R.E. Genetically encoded biosensors based on engineered fluorescent proteins. Chem. Soc. Rev. 2009, 38, 2833–2841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozawa, T.; Nogami, S.; Sato, M.; Ohya, Y.; Umezawa, Y. A fluorescent indicator for detecting protein−protein interactions in vivo based on protein splicing. Anal. Chem. 2000, 72, 5151–5157. [Google Scholar] [CrossRef]
- Jung, D.; Sato, K.; Min, K.; Shigenaga, A.; Jung, J.; Otaka, A.; Kwon, Y. Photo-triggered fluorescent labelling of recombinant proteins in live cells. Chem. Commun. 2015, 51, 9670–9673. [Google Scholar] [CrossRef]
- Sonntag, T.; Mootz, H.D. An intein-cassette integration approach used for the generation of a split TEV protease activated by conditional protein splicing. Mol. Biosyst. 2011, 7, 2031–2039. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, X.; Cui, W.; Wang, W.; Zhang, H.; Liu, L.; Zhang, Z.; Li, Z.; Ying, G.; Zhang, N. Visualization of caspase-3-like activity in cells using a genetically encoded fluorescent biosensor activated by protein cleavage. Nat. Commun. 2013, 4, 2157. [Google Scholar] [CrossRef]
- Kim, S.B.; Ozawa, T.; Umezawa, Y. Genetically encoded stress indicator for noninvasively imaging endogenous corticosterone in living mice. Anal. Chem. 2005, 77, 6588–6593. [Google Scholar] [CrossRef]
- Jones, D.; Mistry, I.; Tavassoli, A. Post-translational control of protein function with light using a LOV-intein fusion protein. Mol. Biosyst. 2016, 12, 1388–1393. [Google Scholar] [CrossRef] [Green Version]
- Shah, N.H.; Muir, T.W. Inteins: Nature’s gift to protein chemists. Chem. Sci. 2014, 5, 446–461. [Google Scholar] [CrossRef] [Green Version]
- Yeagle, P.L. Membrane Receptors. In The Membranes of Cells, 3rd ed.; Academic Press: Boston, MA, USA, 2016; pp. 401–425. [Google Scholar]
- Misawa, N.; Osaki, T.; Takeuchi, S. Membrane protein-based biosensors. J. R. Soc. Interface 2018, 15, 20170952. [Google Scholar] [CrossRef] [PubMed]
- Denisov, I.G.; Sligar, S.G. Nanodiscs in Membrane Biochemistry and Biophysics. Chem. Rev. 2017, 117, 4669–4713. [Google Scholar] [CrossRef] [PubMed]
- Jing, M.; Zhang, P.; Wang, G.; Feng, J.; Mesik, L.; Zeng, J.; Jiang, H.; Wang, S.; Looby, J.C.; Guagliardo, N.A. A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nat. Biotechnol. 2018, 36, 726–737. [Google Scholar] [CrossRef] [PubMed]
- Patriarchi, T.; Cho, J.R.; Merten, K.; Howe, M.W.; Marley, A.; Xiong, W.H.; Folk, R.W.; Broussard, G.J.; Liang, R.; Jang, M.J.; et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 2018, 360, eaat4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, F.; Zeng, J.; Jing, M.; Zhou, J.; Feng, J.; Owen, S.F.; Luo, Y.; Li, F.; Wang, H.; Yamaguchi, T. A genetically encoded fluorescent sensor enables rapid and specific detection of dopamine in flies, fish, and mice. Cell 2018, 174, 481–496.e419. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, C.; Lischinsky, J.E.; Jing, M.; Zhou, J.; Wang, H.; Zhang, Y.; Dong, A.; Wu, Z.; Wu, H. A genetically encoded fluorescent sensor for rapid and specific in vivo detection of norepinephrine. Neuron 2019, 102, 745–761.e8. [Google Scholar] [CrossRef]
- Monakhov, M.V.; Matlashov, M.E.; Colavita, M.; Song, C.; Shcherbakova, D.M.; Antic, S.D.; Verkhusha, V.V.; Knöpfel, T. Screening and cellular characterization of genetically encoded voltage indicators based on near-infrared fluorescent proteins. ACS Chem. Neurosci. 2020, 11, 3523–3531. [Google Scholar] [CrossRef]
- Stoeber, M.; Jullié, D.; Lobingier, B.T.; Laeremans, T.; Steyaert, J.; Schiller, P.W.; Manglik, A.; von Zastrow, M. A genetically encoded biosensor reveals location bias of opioid drug action. Neuron 2018, 98, 963–976.e965. [Google Scholar] [CrossRef] [Green Version]
- Mazaira, G.I.; Zgajnar, N.R.; Lotufo, C.M.; Daneri-Becerra, C.; Sivils, J.C.; Soto, O.B.; Cox, M.B.; Galigniana, M.D. The nuclear receptor field: A historical overview and future challenges. Nucl. Recept. Res. 2018, 5, 101320. [Google Scholar] [CrossRef]
- Siegel, J.A.; Saukko, P.J. Pharmacology and Mechanism of Action of Drugs. In Encyclopedia of Forensic Sciences, 2nd ed.; Academic Press: London, UK, 2012; pp. 210–218. [Google Scholar]
- Burris, T.P.; Solt, L.A.; Wang, Y.; Crumbley, C.; Banerjee, S.; Griffett, K.; Lundasen, T.; Hughes, T.; Kojetin, D.J. Nuclear receptors and their selective pharmacologic modulators. Pharmacol. Rev. 2013, 65, 710–778. [Google Scholar] [CrossRef] [Green Version]
- Ripa, L.; Edman, K.; Dearman, M.; Edenro, G.; Hendrickx, R.; Ullah, V.; Chang, H.F.; Lepistö, M.; Chapman, D.; Geschwindner, S.; et al. Discovery of a Novel Oral Glucocorticoid Receptor Modulator (AZD9567) with Improved Side Effect Profile. J. Med. Chem. 2018, 61, 1785–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pang, J.-P.; Hu, X.-P.; Wang, Y.-X.; Liao, J.-N.; Chai, X.; Wang, X.-W.; Shen, C.; Wang, J.-J.; Zhang, L.-L.; Wang, X.-Y.; et al. Discovery of a novel nonsteroidal selective glucocorticoid receptor modulator by virtual screening and bioassays. Acta Pharmacol. Sin. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.-C.; Liu, Y.-B.; Lu, L.-S.; Lin, C.-W. Imaging reactive oxygen species dynamics in living cells and tissues. Front. Biosci. (Sch. Ed) 2009, 1, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.; Prack, M.; Zhu, Y.; Kolb, J.; Nowak, K.; Ryseck, R.; Shen, D.; Cvijic, M.E.; Somerville, J.; Nadler, S.; et al. A high-content glucocorticoid receptor translocation assay for compound mechanism-of-action evaluation. J. Biomol. Screen. 2007, 12, 1029–1041. [Google Scholar] [CrossRef] [Green Version]
- Ryu, J.; Lee, E.; Kang, C.; Lee, M.; Kim, S.; Park, S.; Lee, D.Y.; Kwon, Y. Rapid Screening of Glucocorticoid Receptor (GR) Effectors Using Cortisol-Detecting Sensor Cells. Int. J. Mol. Sci. 2021, 22, 4747. [Google Scholar] [CrossRef]
- Martinez-Val, A.; Bekker-Jensen, D.B.; Steigerwald, S.; Koenig, C.; Østergaard, O.; Mehta, A.; Tran, T.; Sikorski, K.; Torres-Vega, E.; Kwasniewicz, E. Spatial-proteomics reveals phospho-signaling dynamics at subcellular resolution. Nat. Commun. 2021, 12, 7113. [Google Scholar] [CrossRef]
- Li, F.; Long, Y.; Xie, J.; Ren, J.; Zhou, T.; Song, G.; Li, Q.; Cui, Z. Generation of GCaMP6s-Expressing Zebrafish to Monitor Spatiotemporal Dynamics of Calcium Signaling Elicited by Heat Stress. Int. J. Mol. Sci. 2021, 22, 5551. [Google Scholar] [CrossRef]
- Mehta, S.; Zhang, Y.; Roth, R.H.; Zhang, J.-F.; Mo, A.; Tenner, B.; Huganir, R.L.; Zhang, J. Single-fluorophore biosensors for sensitive and multiplexed detection of signalling activities. Nat. Cell Biol. 2018, 20, 1215–1225. [Google Scholar] [CrossRef]
- Zhang, J.-F.; Liu, B.; Hong, I.; Mo, A.; Roth, R.H.; Tenner, B.; Lin, W.; Zhang, J.Z.; Molina, R.S.; Drobizhev, M. An ultrasensitive biosensor for high-resolution kinase activity imaging in awake mice. Nat. Chem. Biol. 2021, 17, 39–46. [Google Scholar] [CrossRef]
- Prével, C.; Pellerano, M.; Van, T.N.; Morris, M.C. Fluorescent biosensors for high throughput screening of protein kinase inhibitors. Biotechnol. J. 2014, 9, 253–265. [Google Scholar] [CrossRef]
- Allen, M.D.; DiPilato, L.M.; Rahdar, M.; Ren, Y.R.; Chong, C.; Liu, J.O.; Zhang, J. Reading dynamic kinase activity in living cells for high-throughput screening. ACS Chem. Biol. 2006, 1, 371–376. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Kondo, T.; Darmanin, S.; Tsuda, M.; Tanaka, S.; Tobiume, M.; Asaka, M.; Ohba, Y. A novel FRET-based biosensor for the measurement of BCR-ABL activity and its response to drugs in living cells. Clin. Cancer. Res. 2010, 16, 3964–3975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Wang, Y. Fluorescence resonance energy transfer biosensors for cancer detection and evaluation of drug efficacy. Clin. Cancer Res. 2010, 16, 3822–3824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Mehta, S.; Zhang, J. Genetically encoded fluorescent biosensors illuminate kinase signaling in cancer. J. Biol. Chem. 2019, 294, 14814–14822. [Google Scholar] [CrossRef] [Green Version]
- Cohen, P.; Cross, D.; Jänne, P.A. Kinase drug discovery 20 years after imatinib: Progress and future directions. Nat. Rev. Drug Discov. 2021, 20, 551–569. [Google Scholar] [CrossRef]
- Greotti, E.; De Stefani, D. Biosensors for detection of calcium. In Methods in Cell Biology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 337–368. [Google Scholar]
- Van Kuyck, K.; Gabriëls, L.; Nuttin, B. Electrical Brain Stimulation in Treatment-Resistant Obsessive–Compulsive Disorder: Parcellation, and Cyto-and Chemoarchitecture of the Bed Nucleus of the Stria Terminalis–a Review. Neuromodulation 2009, 677–687. [Google Scholar] [CrossRef]
- Ast, C.; Foret, J.; Oltrogge, L.M.; De Michele, R.; Kleist, T.J.; Ho, C.-H.; Frommer, W.B. Ratiometric Matryoshka biosensors from a nested cassette of green-and orange-emitting fluorescent proteins. Nat. Commun. 2017, 8, 431. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Wildes, C.P.; Pattarabanjird, T.; Sanchez, M.I.; Glober, G.F.; Matthews, G.A.; Tye, K.M.; Ting, A.Y. A light-and calcium-gated transcription factor for imaging and manipulating activated neurons. Nat. Biotechnol. 2017, 35, 864–871. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Hyun, J.H.; Jung, K.; Hannan, P.; Kwon, H.-B. A calcium-and light-gated switch to induce gene expression in activated neurons. Nat. Biotechnol. 2017, 35, 858–863. [Google Scholar] [CrossRef]
- Kotlikoff, M.I. Genetically encoded Ca2+ indicators: Using genetics and molecular design to understand complex physiology. J. Physiol. 2007, 578, 55–67. [Google Scholar] [CrossRef]
- Vicario, M.; Calì, T. Measuring Ca2+ Levels in Subcellular Compartments with Genetically Encoded GFP-Based Indicators. In Calcium Signalling; Springer: New York, NY, USA, 2019; pp. 31–42. [Google Scholar]
- Park, J.G.; Palmer, A.E. Properties and use of genetically encoded FRET sensors for cytosolic and organellar Ca2+ measurements. Cold Spring Harb. Protoc. 2015, 2015, pdb.top066043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werley, C.A.; Boccardo, S.; Rigamonti, A.; Hansson, E.M.; Cohen, A.E. Multiplexed Optical Sensors in Arrayed Islands of Cells for multimodal recordings of cellular physiology. Nat. Commun. 2020, 11, 3881. [Google Scholar] [CrossRef] [PubMed]
- Jeon, H.; Lee, M.; Jang, W.; Kwon, Y. Intein-mediated protein engineering for biosensor fabrication. BioChip J. 2016, 10, 277–287. [Google Scholar] [CrossRef]
- Aryal, S.P.; Xia, M.; Adindu, E.; Davis, C.; Ortinski, P.I.; Richards, C.I. ER-GCaMP6f: An Endoplasmic Reticulum-Targeted Genetic Probe to Measure Calcium Activity in Astrocytic Processes. Anal. Chem. 2022, 94, 2099–2108. [Google Scholar] [CrossRef] [PubMed]
- Pavankumar, T.L. Inteins: Localized Distribution, Gene Regulation, and Protein Engineering for Biological Applications. Microorganisms 2018, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Topilina, N.I.; Mills, K.V. Recent advances in in vivo applications of intein-mediated protein splicing. Mob. DNA 2014, 5, 5. [Google Scholar] [CrossRef]
- Hirata, R.; Ohsumk, Y.; Nakano, A.; Kawasaki, H.; Suzuki, K.; Anraku, Y. Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae. J. Biol. Chem. 1990, 265, 6726–6733. [Google Scholar] [CrossRef]
- Elleuche, S.; Pöggeler, S. Inteins, valuable genetic elements in molecular biology and biotechnology. Appl. Microbiol. Biotechnol. 2010, 87, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Mills, K.V.; Johnson, M.A.; Perler, F.B. Protein splicing: How inteins escape from precursor proteins. J. Biol. Chem. 2014, 289, 14498–14505. [Google Scholar] [CrossRef] [Green Version]
- Frutos, S.; Goger, M.; Giovani, B.; Cowburn, D.; Muir, T.W. Branched intermediate formation stimulates peptide bond cleavage in protein splicing. Nat. Chem. Biol. 2010, 6, 527–533. [Google Scholar] [CrossRef] [Green Version]
- Mills, K.V.; Manning, J.S.; Garcia, A.M.; Wuerdeman, L.A. Protein splicing of a Pyrococcus abyssi intein with a C-terminal glutamine. J. Biol. Chem. 2004, 279, 20685–20691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topilina, N.I.; Green, C.M.; Jayachandran, P.; Kelley, D.S.; Stanger, M.J.; Piazza, C.L.; Nayak, S.; Belfort, M. SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses. Proc. Natl. Acad. Sci. USA 2015, 112, 10348–10353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, N.H.; Dann, G.P.; Vila-Perelló, M.; Liu, Z.; Muir, T.W. Ultrafast protein splicing is common among cyanobacterial split inteins: Implications for protein engineering. J. Am. Chem. Soc. 2012, 134, 11338–11341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenzel, S.; Kurpiers, T.; Mootz, H.D. Engineering artificially split inteins for applications in protein chemistry: Biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein. Biochemistry 2006, 45, 1571–1578. [Google Scholar] [CrossRef]
- Mills, K.V.; Lew, B.M.; Jiang, S.; Paulus, H. Protein splicing in trans by purified N- and C-terminal fragments of the Mycobacterium tuberculosis RecA intein. Proc. Natl. Acad. Sci. USA 1998, 95, 3543–3548. [Google Scholar] [CrossRef] [Green Version]
- Thiel, I.V.; Volkmann, G.; Pietrokovski, S.; Mootz, H.D. An atypical naturally split intein engineered for highly efficient protein labeling. Angew. Chem. Int. Ed. 2014, 53, 1306–1310. [Google Scholar] [CrossRef]
- Ludwig, C.; Pfeiff, M.; Linne, U.; Mootz, H.D. Ligation of a synthetic peptide to the N terminus of a recombinant protein using semisynthetic protein trans-splicing. Angew. Chem. Int. Ed. 2006, 45, 5218–5221. [Google Scholar] [CrossRef]
- Appleby, J.H.; Zhou, K.; Volkmann, G.; Liu, X.Q. Novel split intein for trans-splicing synthetic peptide onto C terminus of protein. J. Biol. Chem. 2009, 284, 6194–6199. [Google Scholar] [CrossRef] [Green Version]
- Klabunde, T.; Sharma, S.; Telenti, A.; Jacobs, W.R.; Sacchettini, J.C. Crystal structure of GyrA intein from Mycobacterium xenopi reveals structural basis of protein splicing. Nat. Struct. Biol. 1998, 5, 31–36. [Google Scholar] [CrossRef]
- Gogarten, J.P.; Senejani, A.G.; Zhaxybayeva, O.; Olendzenski, L.; Hilario, E. Inteins: Structure, function, and evolution. Annu. Rev. Microbiol. 2002, 56, 263–287. [Google Scholar] [CrossRef] [Green Version]
- Saleh, L.; Perler, F.B. Protein splicing in cis and in trans. Chem. Rec. 2006, 6, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Paulus, H. Protein splicing and related forms of protein autoprocessing. Annu. Rev. Biochem. 2000, 69, 447–496. [Google Scholar] [CrossRef] [PubMed]
- Iwai, H.; Züger, S.; Jin, J.; Tam, P.-H. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006, 580, 1853–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zettler, J.; Schütz, V.; Mootz, H.D. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009, 583, 909–914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.-E.; Cui, Z.; Wang, D. Sensing of biomolecular interactions using fluorescence complementing systems in living cells. Biosens. Bioelectron. 2016, 76, 243–250. [Google Scholar] [CrossRef]
- Gramespacher, J.A.; Stevens, A.J.; Nguyen, D.P.; Chin, J.W.; Muir, T.W. Intein zymogens: Conditional assembly and splicing of split inteins via targeted proteolysis. J. Am. Chem. Soc. 2017, 139, 8074–8077. [Google Scholar] [CrossRef] [Green Version]
- Leavesley, S.J.; Rich, T.C. Overcoming limitations of FRET measurements. Cytom. J. Int. Soc. Anal. Cytol. 2016, 89, 325. [Google Scholar] [CrossRef] [Green Version]
- Böcker, J.K.; Friedel, K.; Matern, J.C.; Bachmann, A.L.; Mootz, H.D. Generation of a genetically encoded, photoactivatable intein for the controlled production of cyclic peptides. Angew. Chem. Int. Ed. 2015, 54, 2116–2120. [Google Scholar] [CrossRef]
- Mootz, H.D.; Muir, T.W. Protein splicing triggered by a small molecule. J. Am. Chem. Soc. 2002, 124, 9044–9045. [Google Scholar] [CrossRef]
- Jeon, H.; Lee, E.; Kim, D.; Lee, M.; Ryu, J.; Kang, C.; Kim, S.; Kwon, Y. Cell-Based Biosensors Based on Intein-Mediated Protein Engineering for Detection of Biologically Active Signaling Molecules. Anal. Chem. 2018, 90, 9779–9786. [Google Scholar] [CrossRef]
- Xu, X.; Soutto, M.; Xie, Q.; Servick, S.; Subramanian, C.; von Arnim, A.G.; Johnson, C.H. Imaging protein interactions with bioluminescence resonance energy transfer (BRET) in plant and mammalian cells and tissues. Proc. Natl. Acad. Sci. USA 2007, 104, 10264–10269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piston, D.W.; Kremers, G.-J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem. Sci. 2007, 32, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.; Louis, J.M.; Gopich, I.V.; Chung, H.S. Three-color single-molecule FRET and fluorescence lifetime analysis of fast protein folding. J. Phys. Chem. B 2018, 122, 11702–11720. [Google Scholar] [CrossRef] [PubMed]
- Ai, H.-W.; Hazelwood, K.L.; Davidson, M.W.; Campbell, R.E. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods 2008, 5, 401–403. [Google Scholar] [CrossRef] [PubMed]
- Miyawaki, A.; Llopis, J.; Heim, R.; McCaffery, J.M.; Adams, J.A.; Ikura, M.; Tsien, R.Y. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997, 388, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, M.T.; Ni, Q.; Tsien, R.Y.; Zhang, J.; Newton, A.C. Spatio-temporal dynamics of protein kinase B/Akt signaling revealed by a genetically encoded fluorescent reporter. J. Biol. Chem. 2005, 280, 5581–5587. [Google Scholar] [CrossRef] [Green Version]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A guide to fluorescent protein FRET pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Borra, R.; Dong, D.; Elnagar, A.Y.; Woldemariam, G.A.; Camarero, J.A. In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. J. Am. Chem. Soc. 2012, 134, 6344–6353. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, S.J.; Shin, H.; Kim, Y.P. Collagen-Immobilized Extracellular FRET Reporter for Visualizing Protease Activity Secreted by Living Cells. ACS Sens. 2020, 5, 655–664. [Google Scholar] [CrossRef]
- Kodama, Y.; Hu, C.D. Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. BioTechniques 2012, 53, 285–298. [Google Scholar] [CrossRef]
- Hu, C.-D.; Chinenov, Y.; Kerppola, T.K. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 2002, 9, 789–798. [Google Scholar] [CrossRef]
- Ibraheem, A.; Campbell, R.E. Designs and applications of fluorescent protein-based biosensors. Curr. Opin. Chem. Biol. 2010, 14, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.W.; Camarero, J.A. Intein applications: From protein purification and labeling to metabolic control methods. J. Biol. Chem. 2014, 289, 14512–14519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Deng, M.; Yang, C.; Liu, F.; Guan, X.; Du, Y.; Wang, L.; Chu, J. Genetically encoded single circularly permuted fluorescent protein-based intensity indicators. J. Phys. D Appl. Phys. 2020, 53, 113001. [Google Scholar] [CrossRef]
- Nagai, T.; Yamada, S.; Tominaga, T.; Ichikawa, M.; Miyawaki, A. Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA 2004, 101, 10554–10559. [Google Scholar] [CrossRef] [Green Version]
- Nagai, T.; Sawano, A.; Park, E.S.; Miyawaki, A. Circularly permuted green fluorescent proteins engineered to sense Ca2+. Proc. Natl. Acad. Sci. USA 2001, 98, 3197–3202. [Google Scholar] [CrossRef] [Green Version]
- Kostyuk, A.I.; Demidovich, A.D.; Kotova, D.A.; Belousov, V.V.; Bilan, D.S. Circularly permuted fluorescent protein-based indicators: History, principles, and classification. Int. J. Mol. Sci. 2019, 20, 4200. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, M.R.; Freitas, D.F.; Alves, P.M.; Coroadinha, A.S. Detection and Quantification of Label-Free Infectious Adenovirus Using a Switch-On Cell-Based Fluorescent Biosensor. ACS Sens. 2019, 4, 1654–1661. [Google Scholar] [CrossRef]
- Townend, J.E.; Tavassoli, A. Traceless Production of Cyclic Peptide Libraries in E. coli. ACS Chem. Biol. 2016, 11, 1624–1630. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Xiong, S. Intein-mediated backbone cyclization of VP1 protein enhanced protection of CVB3-induced viral myocarditis. Sci. Rep. 2017, 7, 41485. [Google Scholar] [CrossRef] [Green Version]
- Sakamoto, S.; Terauchi, M.; Hugo, A.; Kim, T.; Araki, Y.; Wada, T. Creation of a caspase-3 sensing system using a combination of split-GFP and split-intein. Chem. Commun. 2013, 49, 10323–10325. [Google Scholar] [CrossRef] [PubMed]
- Kanno, A.; Yamanaka, Y.; Hirano, H.; Umezawa, Y.; Ozawa, T. Cyclic luciferase for real-time sensing of caspase-3 activities in living mammals. Angew. Chem. Int. Ed. 2007, 46, 7595–7599. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Jeon, H.; Ryu, J.; Kang, C.; Kim, S.; Park, S.; Kwon, Y. Genetically encoded biosensors for the detection of rapamycin: Toward the screening of agonists and antagonists. Analyst 2020, 145, 5571–5577. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Kim, S.; Lee, E.; Ryu, J.; Lee, M.; Kwon, Y. Genetically Encoded Sensor Cells for the Screening of Glucocorticoid Receptor (GR) Effectors in Herbal Extracts. Biosensors 2021, 11, 341. [Google Scholar] [CrossRef]
- Kapp, K.; Schrempf, S.; Lemberg, M.; Dobberstein, B. Post-Targeting Functions of Signal Peptides. In Protein Transport Into the Endoplasmic Reticulum; Landes Bioscience: Austin, TX, USA, 2009; pp. 1–16. [Google Scholar]
- Kojima, R.; Aubel, D.; Fussenegger, M. Building sophisticated sensors of extracellular cues that enable mammalian cells to work as “doctors” in the body. Cell. Mol. Life Sci. 2020, 77, 3567–3581. [Google Scholar] [CrossRef] [Green Version]

| Intein | Type | ksplice (s−1) | t1/2 | Yield (%) | Ref. |
|---|---|---|---|---|---|
| Mxe GyrA | Contiguous | 1.9 × 10−5 | 10 h | >90 | [65] |
| Pab PolIII | Contiguous | 1.6 × 10−5 | 12 h | 74 | [66] |
| Mtu SufB | Contiguous | [67] | |||
| Ssp DnaE | Naturally Split | 1.5 × 10−4 | 76 m | <50 | [68] |
| Npu DnaE | Naturally Split | 3.7 × 10−2 | 19 s | >90 | [68] |
| Ssp DnaB | Artificially Split | 9.9 × 10−4 | 12 m | 32–56 | [69] |
| Sce VMA 1 | Artificially Split | 1.2 × 10−3 | 10 m | 61–73 | [69] |
| Mtu RecA | Artificially Split | NA | 60–120 m | [70] | |
| AceL TerL | Naturally Split | 1.7 × 10−3 | 7.2 m | 90 | [71] |
| Ssp DnaB S1 | Artificially Split | 4.1 × 10−5 | 40–45 | [72] | |
| Ssp GyrB S11 | Artificially Split | 6.9 × 10−5 | 80 | [73] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C.; Shrestha, K.L.; Kwon, S.; Park, S.; Kim, J.; Kwon, Y. Intein-Mediated Protein Engineering for Cell-Based Biosensors. Biosensors 2022, 12, 283. https://doi.org/10.3390/bios12050283
Kang C, Shrestha KL, Kwon S, Park S, Kim J, Kwon Y. Intein-Mediated Protein Engineering for Cell-Based Biosensors. Biosensors. 2022; 12(5):283. https://doi.org/10.3390/bios12050283
Chicago/Turabian StyleKang, Chungwon, Keshab Lal Shrestha, San Kwon, Seungil Park, Jinsik Kim, and Youngeun Kwon. 2022. "Intein-Mediated Protein Engineering for Cell-Based Biosensors" Biosensors 12, no. 5: 283. https://doi.org/10.3390/bios12050283
APA StyleKang, C., Shrestha, K. L., Kwon, S., Park, S., Kim, J., & Kwon, Y. (2022). Intein-Mediated Protein Engineering for Cell-Based Biosensors. Biosensors, 12(5), 283. https://doi.org/10.3390/bios12050283





