DNA-Based Biosensors for the Biochemical Analysis: A Review
Abstract
1. Introduction
2. Functional DNA Strands-Based Biosensors
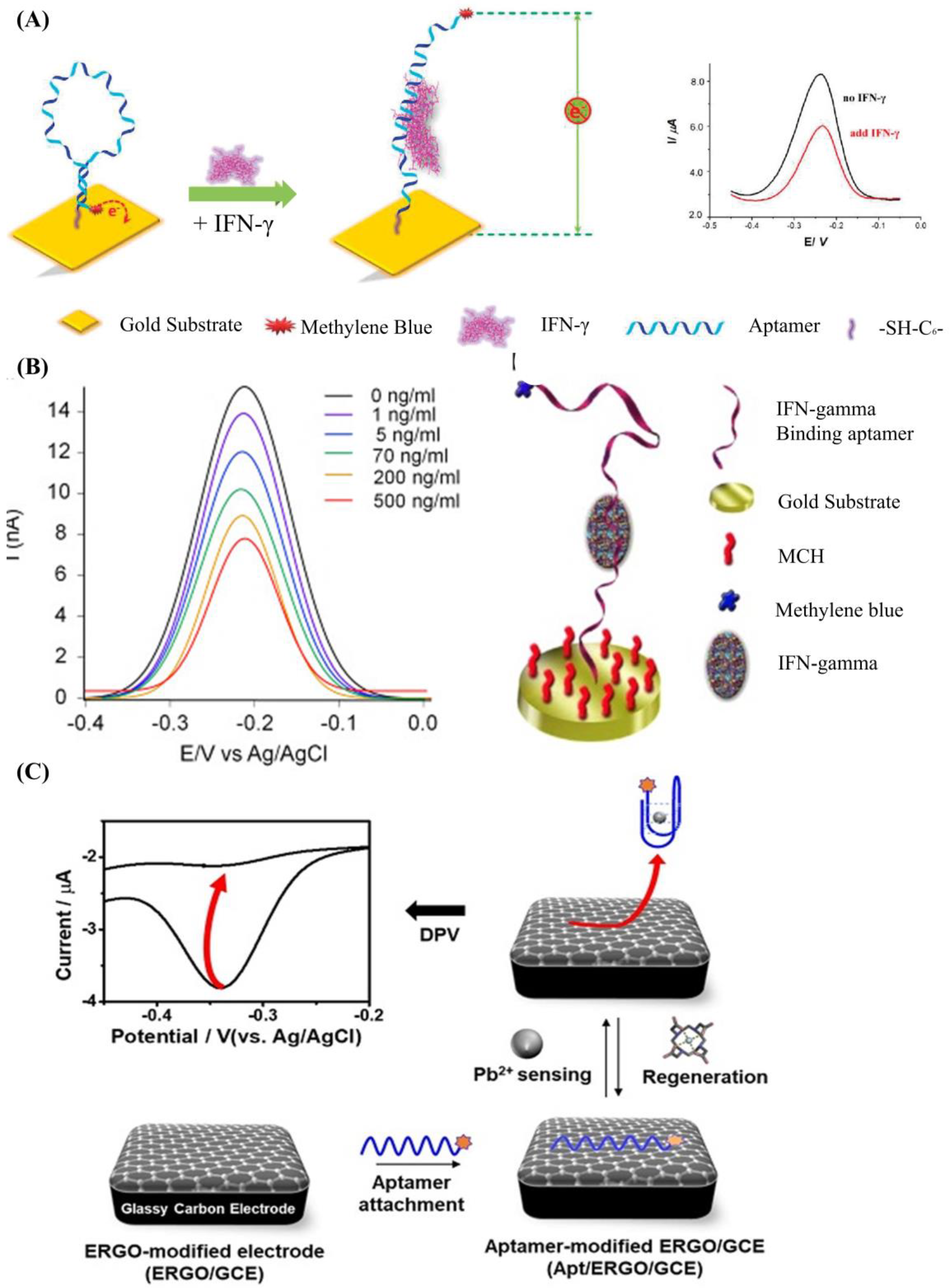
2.1. DNA Aptamer Biosensors
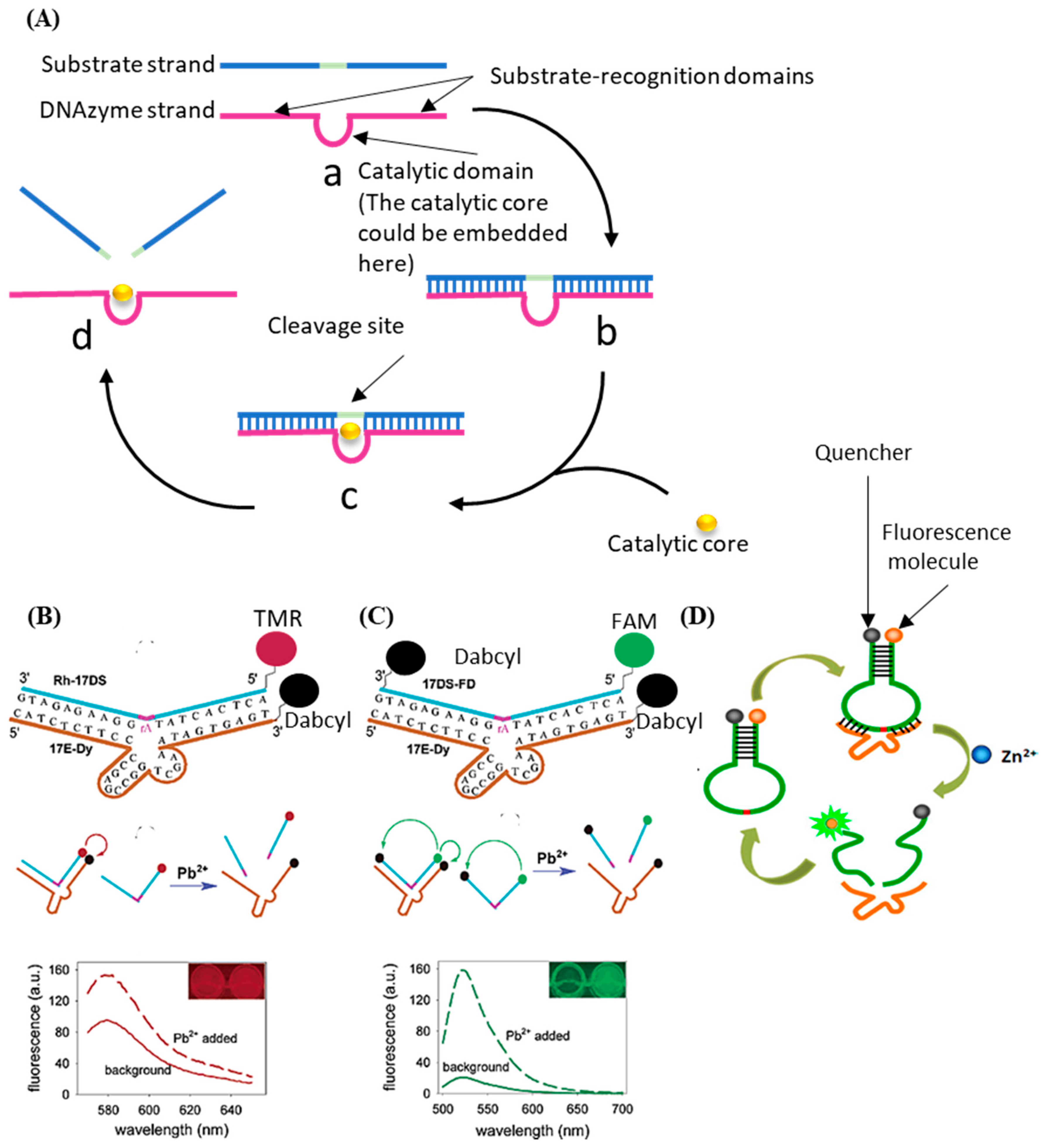
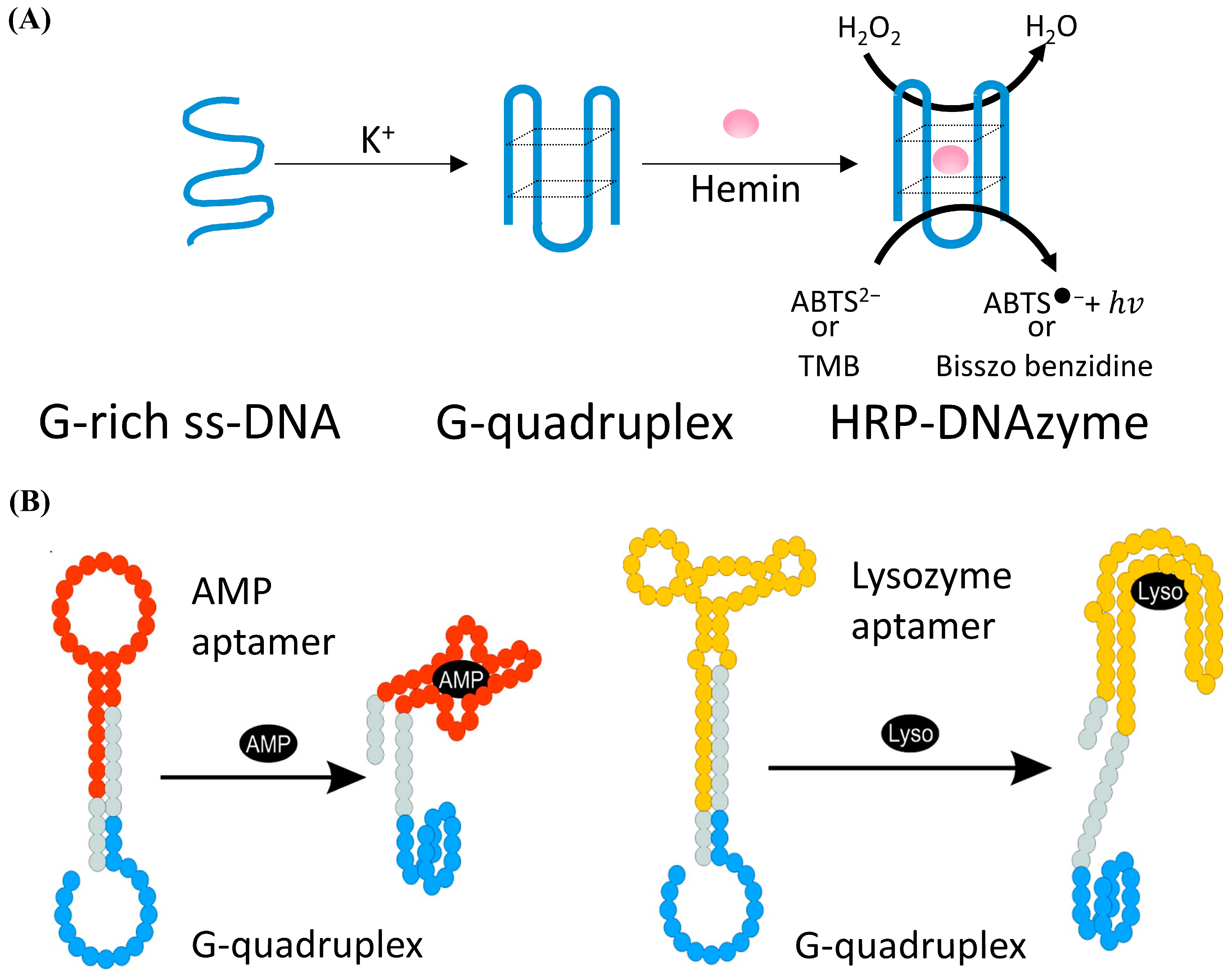
2.2. DNAzyme Biosensors
3. DNA Hybridization-Based Biosensors
3.1. Biosensor Probes Based on DNA Hairpin
3.2. Signal-Enhanced Biosensors Based on DNA Hybridization
4. DNA Templated Biosensors
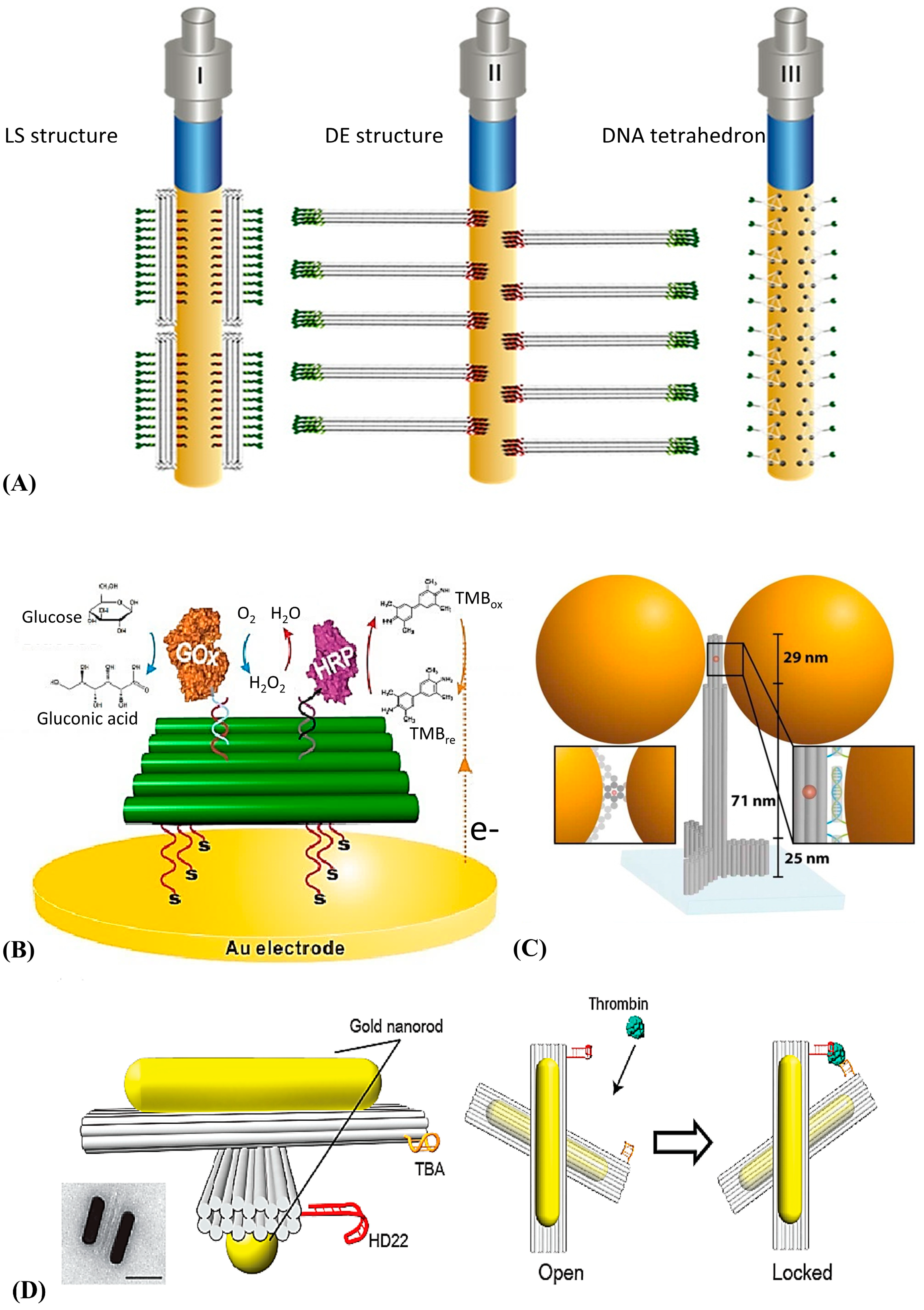
4.1. DNA Tile Assembly and Its Application
4.2. DNA Origami Assembly and Its Application
5. Summary and Conclusions
6. Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Morán, M.C.; Nogueira, D.R.; Vinardell, M.P.; Miguel, M.G.; Lindman, B. Mixed protein–DNA gel particles for DNA delivery: Role of protein composition and preparation method on biocompatibility. Int. J. Pharm. 2013, 454, 192–203. [Google Scholar] [CrossRef]
- Hutton, J.R. Renaturation Kinetics and thermal stability of DNA in aqueous solutions of formamide and urea. Nucleic Acids Res. 1977, 4, 3537–3555. [Google Scholar] [CrossRef]
- Blake, R.D.; Delcourt, S.G. Thermal stability of DNA. Nucleic Acids Res. 1998, 26, 3323–3332. [Google Scholar] [CrossRef]
- Giesen, U.; Kleider, W.; Berding, C.; Geiger, A.; Ørum, H.; Nielsen, P.E. A formula for thermal stability (Tm) prediction of PNA/DNA duplexes. Nucleic Acids Res. 1998, 26, 5004–5006. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Dever, B.; Li, X.-F.; Le, X.C. Thermal Stability of DNA Functionalized Gold Nanoparticles. Bioconjug. Chem. 2013, 24, 1790–1797. [Google Scholar] [CrossRef]
- Jäger, S.; Rasched, G.; Kornreich-Leshem, H.; Engeser, M.; Thum, O.; Famulok, M. A Versatile Toolbox for Variable DNA Functionalization at High Density. J. Am. Chem. Soc. 2005, 127, 15071–15082. [Google Scholar] [CrossRef]
- Saccà, B.; Niemeyer, C.M. Functionalization of DNA nanostructures with proteins. Chem. Soc. Rev. 2011, 40, 5910–5921. [Google Scholar] [CrossRef]
- Wijaya, A.; Hamad-Schifferli, K. Ligand Customization and DNA Functionalization of Gold Nanorods via Round-Trip Phase Transfer Ligand Exchange. Langmuir 2008, 24, 9966–9969. [Google Scholar] [CrossRef] [PubMed]
- Kerman, K.; Kobayashi, M.; Tamiya, E. Recent trends in electrochemical DNA biosensor technology. Meas. Sci. Technol. 2003, 15, 1–11. [Google Scholar] [CrossRef]
- He, P.; Xu, Y.; Fang, Y. A Review: Electrochemical DNA Biosensors for Sequence Recognition. Anal. Lett. 2005, 38, 2597–2623. [Google Scholar] [CrossRef]
- Saidur, M.R.; Aziz, A.R.A.; Basirun, W.J. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017, 90, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Liang, G.; Man, Y.; Li, A.; Jin, X.; Liu, X.; Pan, L. DNAzyme-based biosensor for detection of lead ion: A review. Microchem. J. 2017, 131, 145–153. [Google Scholar] [CrossRef]
- Reder-Christ, K.; Bendas, G. Biosensor Applications in the Field of Antibiotic Research—A Review of Recent Developments. Sensors 2011, 11, 9450–9466. [Google Scholar] [CrossRef] [PubMed]
- Dehghani, S.; Nosrati, R.; Yousefi, M.; Nezami, A.; Soltani, F.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosens. Bioelectron. 2018, 110, 23–37. [Google Scholar] [CrossRef]
- Mehlhorn, A.; Rahimi, P.; Joseph, Y. Aptamer-Based Biosensors for Antibiotic Detection: A Review. Biosensors 2018, 8, 54. [Google Scholar] [CrossRef]
- Bishop, G.R.; Ren, J.; Polander, B.C.; Jeanfreau, B.D.; Trent, J.O.; Chaires, J.B. Energetic basis of molecular recognition in a DNA aptamer. Biophys. Chem. 2007, 126, 165–175. [Google Scholar] [CrossRef]
- Smirnov, I.; Shafer, R.H. Effect of Loop Sequence and Size on DNA Aptamer Stability. Biochemistry 2000, 39, 1462–1468. [Google Scholar] [CrossRef]
- Xia, T.; Yuan, J.; Fang, X. Conformational Dynamics of an ATP-Binding DNA Aptamer: A Single-Molecule Study. J. Phys. Chem. B 2013, 117, 14994–15003. [Google Scholar] [CrossRef]
- Minagawa, H.; Kataoka, Y.; Kuwahara, M.; Horii, K.; Shiratori, I.; Waga, I. A high affinity modified DNA aptamer containing base-appended bases for human β-defensin. Anal. Biochem. 2020, 594, 113627. [Google Scholar] [CrossRef]
- Tan, L.; Neoh, K.G.; Kang, E.-T.; Choe, W.-S.; Su, X. Affinity analysis of DNA aptamer—Peptide interactions using gold nanoparticles. Anal. Biochem. 2012, 421, 725–731. [Google Scholar] [CrossRef]
- Lai, J.-C.; Hong, C.-Y. Magnetic-Assisted Rapid Aptamer Selection (MARAS) for Generating High-Affinity DNA Aptamer Using Rotating Magnetic Fields. ACS Comb. Sci. 2014, 16, 321–327. [Google Scholar] [CrossRef]
- Keum, J.-W.; Bermudez, H. Enhanced resistance of DNA nanostructures to enzymatic digestion. Chem. Commun. 2009, 7036–7038. [Google Scholar] [CrossRef]
- Xue, C.; Zhang, S.; Yu, X.; Hu, S.; Lu, Y.; Wu, Z.-S. Periodically Ordered, Nuclease-Resistant DNA Nanowires Decorated with Cell-Specific Aptamers as Selective Theranostic Agents. Angew. Chem. Int. Ed. 2020, 59, 17540–17547. [Google Scholar] [CrossRef]
- Ma, Y.; Ali, S.R.; Dodoo, A.S.; He, H. Enhanced Sensitivity for Biosensors: Multiple Functions of DNA-Wrapped Single-Walled Carbon Nanotubes in Self-Doped Polyaniline Nanocomposites. J. Phys. Chem. B 2006, 110, 16359–16365. [Google Scholar] [CrossRef]
- Li, J.; Fu, W.; Bao, J.; Wang, Z.; Dai, Z. Fluorescence Regulation of Copper Nanoclusters via DNA Template Manipulation toward Design of a High Signal-to-Noise Ratio Biosensor. ACS Appl. Mater. Interfaces 2018, 10, 6965–6971. [Google Scholar] [CrossRef]
- Liu, J. DNA-stabilized, fluorescent, metal nanoclusters for biosensor development. TrAC Trends Anal. Chem. 2014, 58, 99–111. [Google Scholar] [CrossRef]
- Blackwell, T.K.; Kretzner, L.; Blackwood, E.M.; Eisenman, R.N.; Weintraub, H. Sequence-Specific DNA Binding by the c-Myc Protein. Science 1990, 250, 1149–1151. [Google Scholar] [CrossRef]
- Paborsky, L.R.; McCurdy, S.N.; Griffin, L.C.; Toole, J.J.; Leung, L.L. The single-stranded DNA aptamer-binding site of human thrombin. J. Biol. Chem. 1993, 268, 20808–20811. [Google Scholar] [CrossRef]
- Muhammad, M.; Huang, Q. A review of aptamer-based SERS biosensors: Design strategies and applications. Talanta 2021, 227, 122188. [Google Scholar] [CrossRef]
- He, L.; Huang, R.; Xiao, P.; Liu, Y.; Jin, L.; Liu, H.; Li, S.; Deng, Y.; Chen, Z.; Li, Z.; et al. Current signal amplification strategies in aptamer-based electrochemical biosensor: A review. Chin. Chem. Lett. 2021, 32, 1593–1602. [Google Scholar] [CrossRef]
- Ekrami, E.; Pouresmaieli, M.; Shariati, P.; Mahmoudifard, M. A review on designing biosensors for the detection of trace metals. Appl. Geochem. 2021, 127, 104902. [Google Scholar] [CrossRef]
- Breaker, R.R.; Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994, 1, 223–229. [Google Scholar] [CrossRef]
- Yang, H.; Zhou, Y.; Liu, J. G-quadruplex DNA for construction of biosensors. TrAC Trends Anal. Chem. 2020, 132, 116060. [Google Scholar] [CrossRef]
- Wang, S. Construction of DNA Biosensors for Mercury (II) Ion Detection Based on Enzyme-Driven Signal Amplification Strategy. Biomolecules 2021, 11, 399. [Google Scholar] [CrossRef]
- Wang, F.; Zhang, Y.; Lu, M.; Du, Y.; Chen, M.; Meng, S.; Ji, W.; Sun, C.; Peng, W. Near-infrared band Gold nanoparticles-Au film “hot spot” model based label-free ultratrace lead (II) ions detection via fiber SPR DNAzyme biosensor. Sens. Actuators B Chem. 2021, 337, 129816. [Google Scholar] [CrossRef]
- Glick, G.D. Synthesis of a conformationally restricted DNA hairpin. J. Org. Chem. 1991, 56, 6746–6747. [Google Scholar] [CrossRef]
- Du, H.; Disney, M.D.; Miller, B.L.; Krauss, T.D. Hybridization-Based Unquenching of DNA Hairpins on Au Surfaces: Prototypical “Molecular Beacon” Biosensors. J. Am. Chem. Soc. 2003, 125, 4012–4013. [Google Scholar] [CrossRef]
- Du, H.; Strohsahl, C.M.; Camera, J.; Miller, B.L.; Krauss, T.D. Sensitivity and Specificity of Metal Surface-Immobilized “Molecular Beacon” Biosensors. J. Am. Chem. Soc. 2005, 127, 7932–7940. [Google Scholar] [CrossRef]
- Liu, A.; Wang, K.; Weng, S.; Lei, Y.; Lin, L.; Chen, W.; Lin, X.; Chen, Y. Development of electrochemical DNA biosensors. TrAC Trends Anal. Chem. 2012, 37, 101–111. [Google Scholar] [CrossRef]
- Seeman, N.C. Nucleic acid junctions and lattices. J. Theor. Biol. 1982, 99, 237–247. [Google Scholar] [CrossRef]
- Winfree, E.; Liu, F.; Wenzler, L.A.; Seeman, N.C. Design and self-assembly of two-dimensional DNA crystals. Nature 1998, 394, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Nangreave, J.K.; Li, Z.; Liu, Y.; Yan, H. Signal amplification on a DNA-tile-based biosensor with enhanced sensitivity. Nanomed. 2008, 3, 521–528. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Design of DNA origami. In Proceedings of the ICCAD-2005, IEEE/ACM International Conference on Computer-Aided Design, San Jose, CA, USA, 6–10 November 2005; pp. 471–478. [Google Scholar] [CrossRef]
- Douglas, S.M.; Dietz, H.; Liedl, T.; Högberg, B.; Graf, F.; Shih, W.M. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 2009, 459, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Zuo, X.; Pan, D.; Shi, J.; Huang, Q.; Fan, C. Scaffolded biosensors with designed DNA nanostructures. NPG Asia Mater. 2013, 5, 51. [Google Scholar] [CrossRef]
- Han, S.; Liu, W.; Yang, S.; Wang, R. Facile and Label-Free Electrochemical Biosensors for MicroRNA Detection Based on DNA Origami Nanostructures. ACS Omega 2019, 4, 11025–11031. [Google Scholar] [CrossRef] [PubMed]
- Sameiyan, E.; Bagheri, E.; Ramezani, M.; Alibolandi, M.; Abnous, K.; Taghdisi, S.M. DNA origami-based aptasensors. Biosens. Bioelectron. 2019, 143, 111662. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. TrAC Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Gong, L.; Zhao, Z.; Lv, Y.-F.; Huan, S.-Y.; Fu, T.; Zhang, X.-B.; Shen, G.-L.; Yu, R.-Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. 2014, 51, 979–995. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, J.; Sun, R.; Huang, Z.; Luo, Z.; Zhou, C.; Wu, M.; Duan, Y.; Li, Y. The Recent Development of Hybridization Chain Reaction Strategies in Biosensors. ACS Sens. 2020, 5, 2977–3000. [Google Scholar] [CrossRef]
- Li, N.; Du, M.; Liu, Y.; Ji, X.; He, Z. Multipedal DNA Walker Biosensors Based on Catalyzed Hairpin Assembly and Isothermal Strand-Displacement Polymerase Reaction for the Chemiluminescent Detection of Proteins. ACS Sens. 2018, 3, 1283–1290. [Google Scholar] [CrossRef]
- Xie, N.; Liu, S.; Yang, X.; He, X.; Huang, J.; Wang, K. DNA tetrahedron nanostructures for biological applications: Biosensors and drug delivery. Analyst 2017, 142, 3322–3332. [Google Scholar] [CrossRef]
- Loretan, M.; Domljanovic, I.; Lakatos, M.; Rüegg, C.; Acuna, G.P. DNA Origami as Emerging Technology for the Engineering of Fluorescent and Plasmonic-Based Biosensors. Materials 2020, 13, 2185. [Google Scholar] [CrossRef]
- Murphy, L. Biosensors and bioelectrochemistry. Curr. Opin. Chem. Biol. 2006, 10, 177–184. [Google Scholar] [CrossRef]
- Songa, E.A.; Okonkwo, J.O. Recent approaches to improving selectivity and sensitivity of enzyme-based biosensors for organophosphorus pesticides: A review. Talanta 2016, 155, 289–304. [Google Scholar] [CrossRef]
- Long, F.; Zhu, A.; Shi, H. Recent Advances in Optical Biosensors for Environmental Monitoring and Early Warning. Sensors 2013, 13, 13928–13948. [Google Scholar] [CrossRef] [PubMed]
- Metkar, S.K.; Girigoswami, K. Diagnostic biosensors in medicine—A review. Biocatal. Agric. Biotechnol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- Hu, Q.; Wu, J.; Chen, L.; Lou, X.; Xia, F. Recent Development of DNA-modified AIEgen Probes for Biomedical Application. Chem. Res. Chin. Univ. 2021, 37, 66–72. [Google Scholar] [CrossRef]
- Mannelli, I.; Minunni, M.; Tombelli, S.; Wang, R.; Michela Spiriti, M.; Mascini, M. Direct immobilisation of DNA probes for the development of affinity biosensors. Bioelectrochemistry 2005, 66, 129–138. [Google Scholar] [CrossRef]
- Wang, J. Survey and summary: From DNA biosensors to gene chips. Nucleic Acids Res. 2000, 28, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Canoura, J.; Yu, H.; Alkhamis, O.; Roncancio, D.; Farhana, R.; Xiao, Y. Accelerating Post-SELEX Aptamer Engineering Using Exonuclease Digestion. J. Am. Chem. Soc. 2021, 143, 805–816. [Google Scholar] [CrossRef]
- Zhuo, Z.; Yu, Y.; Wang, M.; Li, J.; Zhang, Z.; Liu, J.; Wu, X.; Lu, A.; Zhang, G.; Zhang, B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017, 18, 2142. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tuleouva, N.; Ramanculov, E.; Revzin, A. Aptamer-based electrochemical biosensor for interferon gamma detection. Anal. Chem. 2010, 82, 8131–8136. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Pui, T.S.; Kongsuphol, P.; Tang, K.C.; Arya, S.K. Aptamer-based array electrodes for quantitative interferon-γ detection. Biosens. Bioelectron. 2014, 53, 257–262. [Google Scholar] [CrossRef]
- Pan, H.M.; Gonuguntla, S.; Li, S.; Trau, D. 3.33 Conjugated Polymers for Biosensor Devices. In Comprehensive Biomaterials II; Ducheyne, P., Ed.; Elsevier: Oxford, UK, 2017; pp. 716–754. ISBN 978-0-08-100692-4. [Google Scholar] [CrossRef]
- Kabanov, A.V.; Felgner, P.L.; Seymour, L.W. Self-assembling complexes for gene delivery. Lab. Clin. Trial 1998, 197–218. [Google Scholar] [CrossRef]
- Ho, H.-A.; Boissinot, M.; Bergeron, M.G.; Corbeil, G.; Doré, K.; Boudreau, D.; Leclerc, M. Colorimetric and Fluorometric Detection of Nucleic Acids Using Cationic Polythiophene Derivatives. Angew. Chem. Int. Ed. 2002, 41, 1548–1551. [Google Scholar] [CrossRef]
- Ho, H.-A.; Leclerc, M. Optical Sensors Based on Hybrid Aptamer/Conjugated Polymer Complexes. J. Am. Chem. Soc. 2004, 126, 1384–1387. [Google Scholar] [CrossRef]
- Geim, A.K. Graphene: Status and Prospects. Science 2009, 324, 1530–1534. [Google Scholar] [CrossRef]
- Nag, A.; Mitra, A.; Mukhopadhyay, S.C. Graphene and Its Sensor-Based Applications: A Review. Sens. Actuators Phys. 2018, 270, 177–194. [Google Scholar] [CrossRef]
- Suvarnaphaet, P.; Pechprasarn, S. Graphene-Based Materials for Biosensors: A Review. Sensors 2017, 17, 2161. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Park, S.-J.; Min, D.-H. Emerging Approaches for Graphene Oxide Biosensor. Anal. Chem. 2017, 89, 232–248. [Google Scholar] [CrossRef]
- Mukherjee, S.; Meshik, X.; Choi, M.; Farid, S.; Datta, D.; Lan, Y.; Poduri, S.; Sarkar, K.; Baterdene, U.; Huang, C.-E. A Graphene and Aptamer Based Liquid Gated FET-like Electrochemical Biosensor to Detect Adenosine Triphosphate. IEEE Trans. Nanobioscience 2015, 14, 967–972. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Z.; Wang, J.; Li, J.; Lin, Y. Graphene and Graphene Oxide: Biofunctionalization and Applications in Biotechnology. Trends Biotechnol. 2011, 29, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. Adsorption of DNA onto Gold Nanoparticles and Graphene Oxide: Surface Science and Applications. Phys. Chem. Chem. Phys. 2012, 14, 10485–10496. [Google Scholar] [CrossRef]
- Yu, S.H.; Lee, C.-S.; Kim, T.H. Electrochemical Detection of Ultratrace Lead Ion through Attaching and Detaching DNA Aptamer from Electrochemically Reduced Graphene Oxide Electrode. Nanomaterials 2019, 9, 817. [Google Scholar] [CrossRef] [PubMed]
- Koshland, D.E. 78. Koshland, D.E. 7 The Molecular Basis for Enzyme Regulation. In The Enzymes; Boyer, P.D., Ed.; Academic Press: Cambridge, MA, USA, 1970; Volume 1, pp. 341–396. [Google Scholar] [CrossRef]
- Hasegawa, H.; Savory, N.; Abe, K.; Ikebukuro, K. Methods for Improving Aptamer Binding Affinity. Molecules 2016, 21, 421. [Google Scholar] [CrossRef] [PubMed]
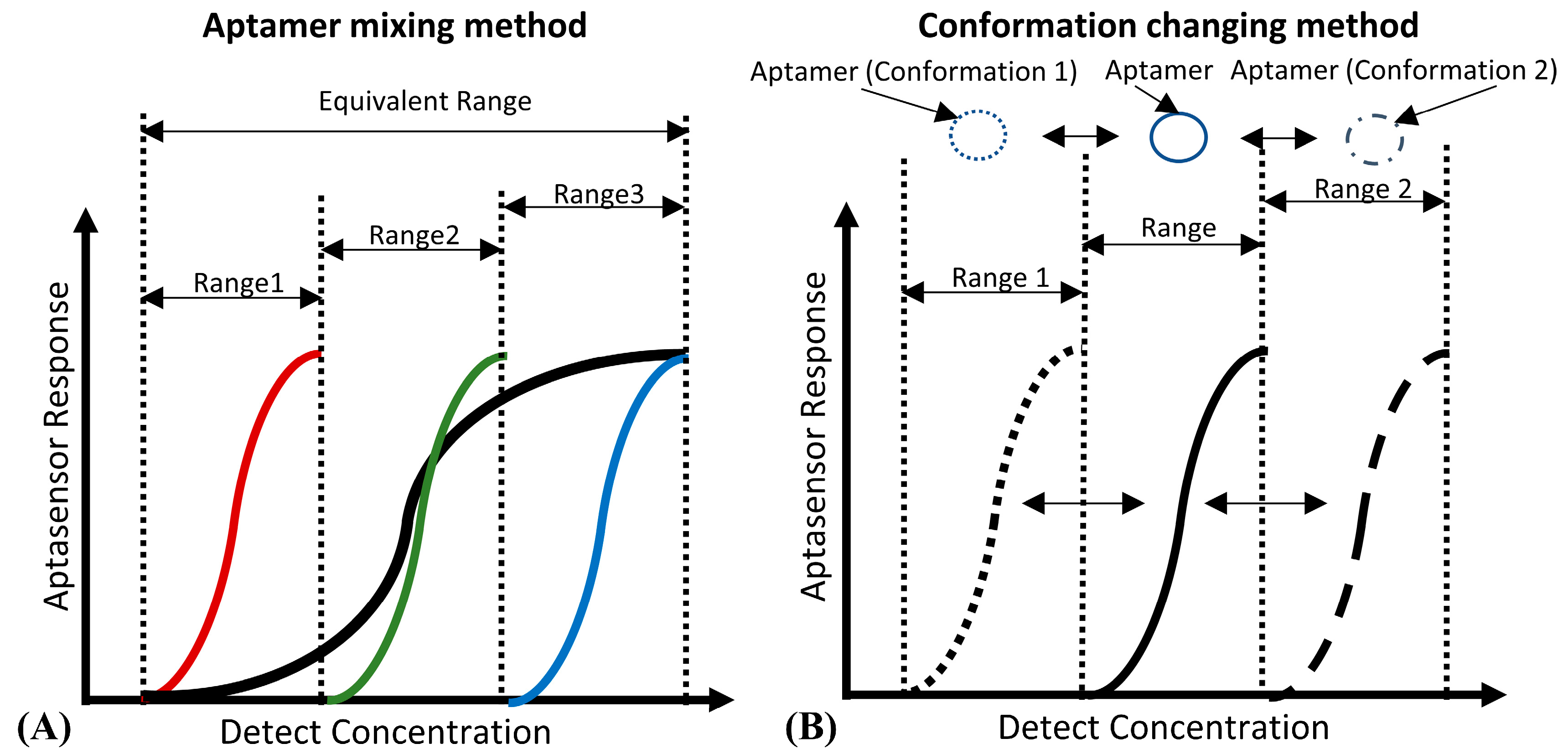
- Kang, D.; Vallée-Bélisle, A.; Porchetta, A.; Plaxco, K.W.; Ricci, F. Re-engineering Electrochemical Biosensors To Narrow or Extend Their Useful Dynamic Range. Angew. Chem. Int. Ed. 2012, 51, 6717–6721. [Google Scholar] [CrossRef] [PubMed]
- Satış, H.; Özger, H.S.; Aysert Yıldız, P.; Hızel, K.; Gulbahar, Ö.; Erbaş, G.; Aygencel, G.; Guzel Tunccan, O.; Öztürk, M.A.; Dizbay, M.; et al. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine 2021, 137, 155302. [Google Scholar] [CrossRef]
- Pepe, M.S.; Etzioni, R.; Feng, Z.; Potter, J.D.; Thompson, M.L.; Thornquist, M.; Winget, M.; Yasui, Y. Phases of Biomarker Development for Early Detection of Cancer. JNCI J. Natl. Cancer Inst. 2001, 93, 1054–1061. [Google Scholar] [CrossRef]
- Lu, Y.; Li, X.; Zhang, L.; Yu, P.; Su, L.; Mao, L. Aptamer-Based Electrochemical Sensors with Aptamer−Complementary DNA Oligonucleotides as Probe. Anal. Chem. 2008, 80, 1883–1890. [Google Scholar] [CrossRef]
- Mena, M.L.; Yanez-Sedeno, P.; Pingarron, J.M. A comparison of different strategies for the construction of amperometric enzyme biosensors using gold nanoparticle-modified electrodes. Anal. Biochem. 2005, 336, 20–27. [Google Scholar] [CrossRef]
- Zhao, Z.; Lei, W.; Zhang, X.; Wang, B.; Jiang, H. ZnO-Based Amperometric Enzyme Biosensors. Sensors 2010, 10, 1216–1231. [Google Scholar] [CrossRef] [PubMed]
- Delvaux, M.; Demoustier-Champagne, S. Immobilisation of glucose oxidase within metallic nanotubes arrays for application to enzyme biosensors. Biosens. Bioelectron. 2003, 18, 943–951. [Google Scholar] [CrossRef]
- Wilson, G.S.; Hu, Y.B. Enzyme based biosensors for in vivo measurements. Chem. Rev. 2000, 100, 2693–2704. [Google Scholar] [CrossRef]
- Secundo, F. Conformational changes of enzymes upon immobilisation. Chem. Soc. Rev. 2013, 42, 6250–6261. [Google Scholar] [CrossRef] [PubMed]
- Jesionowski, T.; Zdarta, J.; Krajewska, B. Enzyme immobilization by adsorption: A review. Adsorpt.-J. Int. Adsorpt. Soc. 2014, 20, 801–821. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Santoro, S.W.; Joyce, G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. USA 1997, 94, 4262–4266. [Google Scholar] [CrossRef] [PubMed]
- Schildkraut, C.; Lifson, S. Dependence of the melting temperature of DNA on salt concentration. Biopolymers 1965, 3, 195–208. [Google Scholar] [CrossRef] [PubMed]
- Panjkovich, A.; Melo, F. Comparison of different melting temperature calculation methods for short DNA sequences. Bioinformatics 2005, 21, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Nakano, S.; Sugimoto, N. Temperature dependence of thermodynamic properties for DNA/DNA and RNA/DNA duplex formation. Eur. J. Biochem. 2002, 269, 2821–2830. [Google Scholar] [CrossRef]
- Weber, G. Mesoscopic model parametrization of hydrogen bonds and stacking interactions of RNA from melting temperatures. Nucleic Acids Res. 2013, 41, 30. [Google Scholar] [CrossRef][Green Version]
- Brunet, A.; Salomé, L.; Rousseau, P.; Destainville, N.; Manghi, M.; Tardin, C. How does temperature impact the conformation of single DNA molecules below melting temperature? Nucleic Acids Res. 2018, 46, 2074–2081. [Google Scholar] [CrossRef]
- Kim, H.-K.; Rasnik, I.; Liu, J.; Ha, T.; Lu, Y. Dissecting metal ion–dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007, 3, 763–768. [Google Scholar] [CrossRef]
- Zhang, X.-B.; Kong, R.-M.; Lu, Y. Metal Ion Sensors Based on DNAzymes and Related DNA Molecules. Annu. Rev. Anal. Chem. 2011, 4, 105–128. [Google Scholar] [CrossRef]
- Hwang, K.; Hosseinzadeh, P.; Lu, Y. Biochemical and biophysical understanding of metal ion selectivity of DNAzymes. Inorganica Chim. Acta 2016, 452, 12–24. [Google Scholar] [CrossRef]
- Roth, A.; Breaker, R.R. An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. USA 1998, 95, 6027–6031. [Google Scholar] [CrossRef]
- Huang, K.; Martí, A.A. Recent trends in molecular beacon design and applications. Anal. Bioanal. Chem. 2012, 402, 3091–3102. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y. A Highly Sensitive and Selective Catalytic DNA Biosensor for Lead Ions. J. Am. Chem. Soc. 2000, 122, 10466–10467. [Google Scholar] [CrossRef]
- Liu, J.; Lu, Y. Improving Fluorescent DNAzyme Biosensors by Combining Inter- and Intramolecular Quenchers. Anal. Chem. 2003, 75, 6666–6672. [Google Scholar] [CrossRef]
- Zhao, X.-H.; Gong, L.; Zhang, X.-B.; Yang, B.; Fu, T.; Hu, R.; Tan, W.; Yu, R. Versatile DNAzyme-Based Amplified Biosensing Platforms for Nucleic Acid, Protein, and Enzyme Activity Detection. Anal. Chem. 2013, 85, 3614–3620. [Google Scholar] [CrossRef]
- Travascio, P.; Li, Y.; Sen, D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998, 5, 505–517. [Google Scholar] [CrossRef]
- Teller, C.; Shimron, S.; Willner, I. Aptamer-DNAzyme Hairpins for Amplified Biosensing. Anal. Chem. 2009, 81, 9114–9119. [Google Scholar] [CrossRef]
- Li, H.; Chang, J.; Hou, T.; Li, F. HRP-Mimicking DNAzyme-Catalyzed in Situ Generation of Polyaniline To Assist Signal Amplification for Ultrasensitive Surface Plasmon Resonance Biosensing. Anal. Chem. 2017, 89, 673–680. [Google Scholar] [CrossRef]
- Whetton, A.D.; Preston, G.W.; Abubeker, S.; Geifman, N. Proteomics and Informatics for Understanding Phases and Identifying Biomarkers in COVID-19 Disease. J. Proteome Res. 2020, 19, 4219–4232. [Google Scholar] [CrossRef]
- DeKosky, S.T.; Kochanek, P.M.; Valadka, A.B.; Clark, R.S.B.; Chou, S.H.-Y.; Au, A.K.; Horvat, C.; Jha, R.M.; Mannix, R.; Wisniewski, S.R.; et al. Blood Biomarkers for Detection of Brain Injury in COVID-19 Patients. J. Neurotrauma 2020, 38, 1–43. [Google Scholar] [CrossRef]
- Kaur, M.; Tiwari, S.; Jain, R. Protein based biomarkers for non-invasive Covid-19 detection. Sens. Bio-Sens. Res. 2020, 29, 100362. [Google Scholar] [CrossRef]
- Bartlett, J.M.S.; Stirling, D. A Short History of the Polymerase Chain Reaction. In PCR Protocols; Bartlett, J.M.S., Stirling, D., Eds.; Methods in Molecular Biology™; Humana Press: Totowa, NJ, USA, 2003; pp. 3–6. ISBN 978-1-59259-384-2. [Google Scholar] [CrossRef]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Aspects Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Hacia, J.G. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 1999, 21, 42–47. [Google Scholar] [CrossRef]
- Lequin, R.M. Enzyme Immunoassay (EIA)/Enzyme-Linked Immunosorbent Assay (ELISA). Clin. Chem. 2005, 51, 2415–2418. [Google Scholar] [CrossRef]
- Fang, X.; Liu, X.; Schuster, S.; Tan, W. Designing a Novel Molecular Beacon for Surface-Immobilized DNA Hybridization Studies. J. Am. Chem. Soc. 1999, 121, 2921–2922. [Google Scholar] [CrossRef]
- Fan, C.; Plaxco, K.W.; Heeger, A.J. Electrochemical interrogation of conformational changes as a reagentless method for the sequence-specific detection of DNA. Proc. Natl. Acad. Sci. USA 2003, 100, 9134–9137. [Google Scholar] [CrossRef]
- Rowe, A.A.; Chuh, K.N.; Lubin, A.A.; Miller, E.A.; Cook, B.; Hollis, D.; Plaxco, K.W. Electrochemical Biosensors Employing an Internal Electrode Attachment Site and Achieving Reversible, High Gain Detection of Specific Nucleic Acid Sequences. Anal. Chem. 2011, 83, 9462–9466. [Google Scholar] [CrossRef]
- Xiong, E.; Li, Z.; Zhang, X.; Zhou, J.; Yan, X.; Liu, Y.; Chen, J. Triple-Helix Molecular Switch Electrochemical Ratiometric Biosensor for Ultrasensitive Detection of Nucleic Acids. Anal. Chem. 2017, 89, 8830–8835. [Google Scholar] [CrossRef]
- Peterson, A.W.; Heaton, R.J.; Georgiadis, R.M. The effect of surface probe density on DNA hybridization. Nucleic Acids Res. 2001, 29, 5163–5168. [Google Scholar] [CrossRef]
- Hou, T.; Li, W.; Liu, X.; Li, F. Label-Free and Enzyme-Free Homogeneous Electrochemical Biosensing Strategy Based on Hybridization Chain Reaction: A Facile, Sensitive, and Highly Specific MicroRNA Assay. Anal. Chem. 2015, 87, 11368–11374. [Google Scholar] [CrossRef]
- Chen, Z.; Chengjun, S.; Zewei, L.; Kunping, L.; Xijian, Y.; Haimin, Z.; Yongxin, L.; Yixiang, D. Fiber optic biosensor for detection of genetically modified food based on catalytic hairpin assembly reaction and nanocomposites assisted signal amplification. Sens. Actuators B Chem. 2018, 254, 956–965. [Google Scholar] [CrossRef]
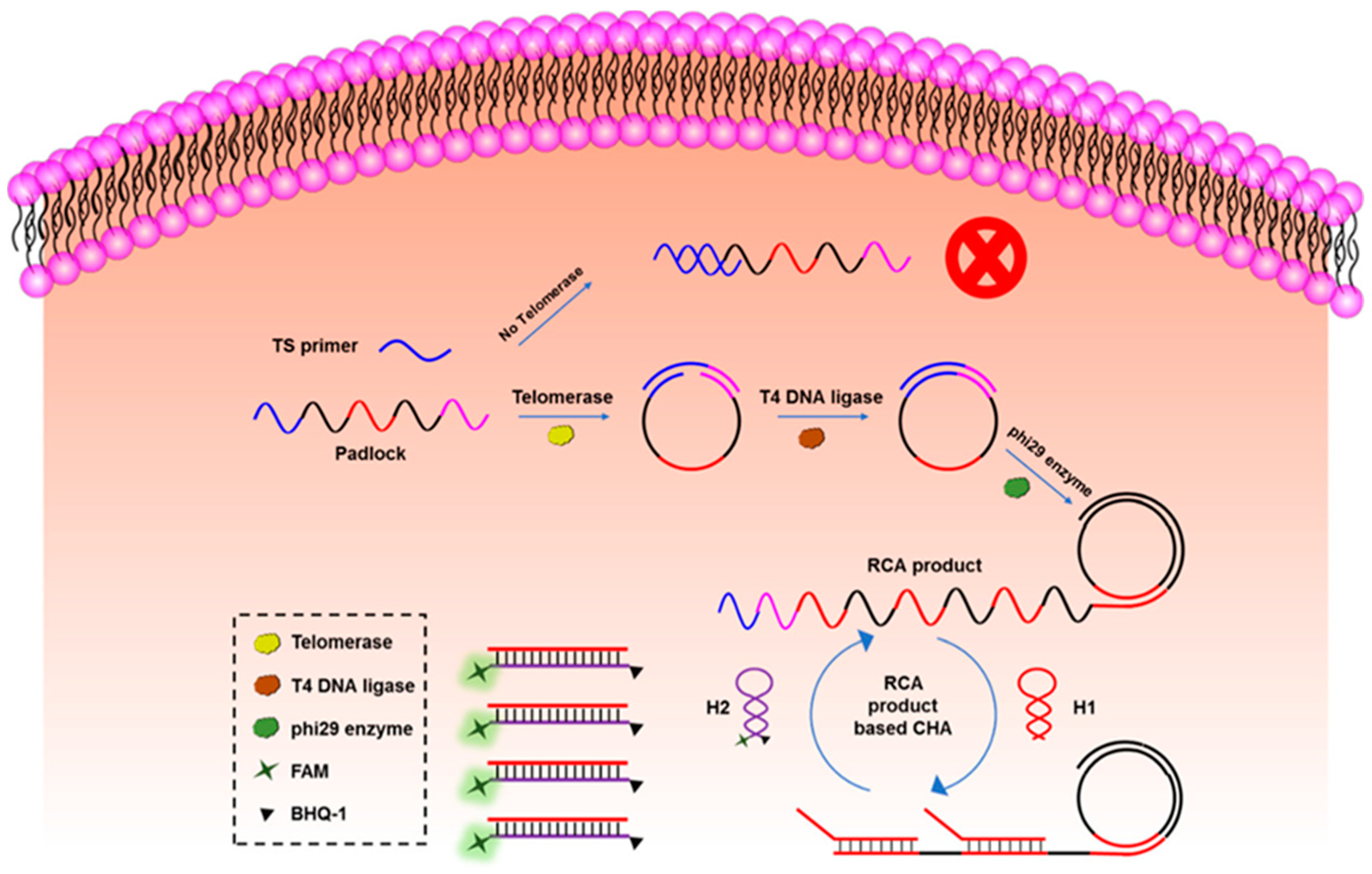
- Liu, Y.; Li, S.; Zhang, L.; Zhao, Q.; Li, N.; Wu, Y. Catalytic Hairpin Assembly-Assisted Rolling Circle Amplification for High-Sensitive Telomerase Activity Detection. ACS Omega 2020, 5, 11836–11841. [Google Scholar] [CrossRef]
- Chan, V.; Graves, D.J.; McKenzie, S.E. The biophysics of DNA hybridization with immobilized oligonucleotide probes. Biophys. J. 1995, 69, 2243–2255. [Google Scholar] [CrossRef]
- Ouldridge, T.E.; Šulc, P.; Romano, F.; Doye, J.P.K.; Louis, A.A. DNA hybridization kinetics: Zippering, internal displacement and sequence dependence. Nucleic Acids Res. 2013, 41, 8886–8895. [Google Scholar] [CrossRef]
- Liu, Y.; Kumar, S.; Taylor, R.E. Mix-and-match nanobiosensor design: Logical and spatial programming of biosensors using self-assembled DNA nanostructures. WIREs Nanomed. Nanobiotechnol. 2018, 10, e1518. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Cheng, J.; Liu, Y.; Wang, D.; Luo, T.; Dai, B.; Zhang, C.; Cui, D.; Ke, Y.; Song, J. Proximity-Induced Pattern Operations in Reconfigurable DNA Origami Domino Array. J. Am. Chem. Soc. 2020, 142, 14566–14573. [Google Scholar] [CrossRef] [PubMed]
- Kallenbach, N.R.; Ma, R.-I.; Seeman, N.C. An immobile nucleic acid junction constructed from oligonucleotides. Nature 1983, 305, 829–831. [Google Scholar] [CrossRef]
- Liu, Y.; West, S.C. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004, 5, 937–944. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Sun, W.; Seeman, N.C. Designed Two-Dimensional DNA Holliday Junction Arrays Visualized by Atomic Force Microscopy. J. Am. Chem. Soc. 1999, 121, 5437–5443. [Google Scholar] [CrossRef]
- Du, S.M.; Seeman, N.C. The construction of a trefoil knot from a DNA branched junction motif. Biopolymers 1994, 34, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Li, X.; Yang, X.; Seeman, N.C. Ligation of Triangles Built from Bulged 3-Arm DNA Branched Junctions. J. Am. Chem. Soc. 1996, 118, 6121–6130. [Google Scholar] [CrossRef]
- Lin, C.; Liu, Y.; Rinker, S.; Yan, H. DNA Tile Based Self-Assembly: Building Complex Nanoarchitectures. Chem. Phys. Chem. 2006, 7, 1641–1647. [Google Scholar] [CrossRef]
- Kim, S.; Kim, J.; Qian, P.; Shin, J.; Amin, R.; Ahn, S.J.; LaBean, T.H.; Kim, M.K.; Park, S.H. Intrinsic DNA curvature of double-crossover tiles. Nanotechnology 2011, 22, 245706. [Google Scholar] [CrossRef][Green Version]
- Sa-Ardyen, P.; Vologodskii, A.V.; Seeman, N.C. The Flexibility of DNA Double Crossover Molecules. Biophys. J. 2003, 84, 3829–3837. [Google Scholar] [CrossRef]
- Mao, C.; LaBean, T.H.; Reif, J.H.; Seeman, N.C. Logical computation using algorithmic self-assembly of DNA triple-crossover molecules. Nature 2000, 407, 493–496. [Google Scholar] [CrossRef]
- Liu, D.; Park, S.H.; Reif, J.H.; LaBean, T.H. DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc. Natl. Acad. Sci. USA 2004, 101, 717–722. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Yin, P.; Park, S.H.; Li, H.; Feng, L.; Guan, X.; Liu, D.; Reif, J.H.; LaBean, T.H. Self-assembled DNA Structures for Nanoconstruction. AIP Conf. Proc. 2004, 725, 43–52. [Google Scholar] [CrossRef]
- He, Y.; Ye, T.; Su, M.; Zhang, C.; Ribbe, A.E.; Jiang, W.; Mao, C. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature 2008, 452, 198–201. [Google Scholar] [CrossRef]
- Zhang, C.; Su, M.; He, Y.; Zhao, X.; Fang, P.; Ribbe, A.E.; Jiang, W.; Mao, C. Conformational flexibility facilitates self-assembly of complex DNA nanostructures. Proc. Natl. Acad. Sci. USA 2008, 105, 10665–10669. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.P.; Berry, R.M.; Turberfield, A.J. The Single-Step Synthesis of a DNA Tetrahedron. Chem. Commun. 2004, 1372–1373. [Google Scholar] [CrossRef]
- Goodman, R.P.; Heilemann, M.; Doose, S.; Erben, C.M.; Kapanidis, A.N.; Turberfield, A.J. Reconfigurable, Braced, Three-Dimensional DNA Nanostructures. Nat. Nanotechnol. 2008, 3, 93–96. [Google Scholar] [CrossRef] [PubMed]
- Aldaye, F.A.; Sleiman, H.F. Modular Access to Structurally Switchable 3D Discrete DNA Assemblies. J. Am. Chem. Soc. 2007, 129, 13376–13377. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Sohn, Y.S.; Nechushtai, R.; Willner, I. DNA Tetrahedra Modules as Versatile Optical Sensing Platforms for Multiplexed Analysis of miRNAs, Endonucleases, and Aptamer–Ligand Complexes. ACS Nano 2020, 14, 9021–9031. [Google Scholar] [CrossRef]
- Li, J.; Dai, J.; Jiang, S.; Xie, M.; Zhai, T.; Guo, L.; Cao, S.; Xing, S.; Qu, Z.; Zhao, Y.; et al. Encoding quantized fluorescence states with fractal DNA frameworks. Nat. Commun. 2020, 11, 2185. [Google Scholar] [CrossRef]
- Fan, Z.; Lin, Z.; Wang, Z.; Wang, J.; Xie, M.; Zhao, J.; Zhang, K.; Huang, W. Dual-Wavelength Electrochemiluminescence Ratiometric Biosensor for NF-κB p50 Detection with Dimethylthiodiaminoterephthalate Fluorophore and Self-Assembled DNA Tetrahedron Nanostructures Probe. ACS Appl. Mater. Interfaces 2020, 12, 11409–11418. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Shen, J.; Ye, D.; Dong, B.; Wang, F.; Pei, H.; Wang, J.; Shi, J.; Wang, L.; Xue, W.; et al. Programming bulk enzyme heterojunctions for biosensor development with tetrahedral DNA framework. Nat. Commun. 2020, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Dong, Y.; Liang, X. Development of a SPR aptasensor containing oriented aptamer for direct capture and detection of tetracycline in multiple honey samples. Biosens. Bioelectron. 2018, 109, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Nie, W.; Wang, Q.; Zou, L.; Zheng, Y.; Liu, X.; Yang, X.; Wang, K. Low-Fouling Surface Plasmon Resonance Sensor for Highly Sensitive Detection of MicroRNA in a Complex Matrix Based on the DNA Tetrahedron. Anal. Chem. 2018, 90, 12584–12591. [Google Scholar] [CrossRef] [PubMed]
- Diao, W.; Tang, M.; Ding, S.; Li, X.; Cheng, W.; Mo, F.; Yan, X.; Ma, H.; Yan, Y. Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices. Biosens. Bioelectron. 2018, 100, 228–234. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; He, N.; Li, Z. Recent progresses in DNA nanostructure-based biosensors for detection of tumor markers. Biosens. Bioelectron. 2018, 109, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Li, Q.; Zuo, X.; Fan, C. DNA framework-engineered electrochemical biosensors. Sci. China Life Sci. 2020, 63, 1130–1141. [Google Scholar] [CrossRef] [PubMed]
- Chidchob, P.; Sleiman, H.F. Recent advances in DNA nanotechnology. Curr. Opin. Chem. Biol. 2018, 46, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bu, N.-N.; Tang, C.-X.; He, X.-W.; Yin, X.-B. Tetrahedron-structured DNA and functional oligonucleotide for construction of an electrochemical DNA-based biosensor. Chem. Commun. 2011, 47, 7689–7691. [Google Scholar] [CrossRef]
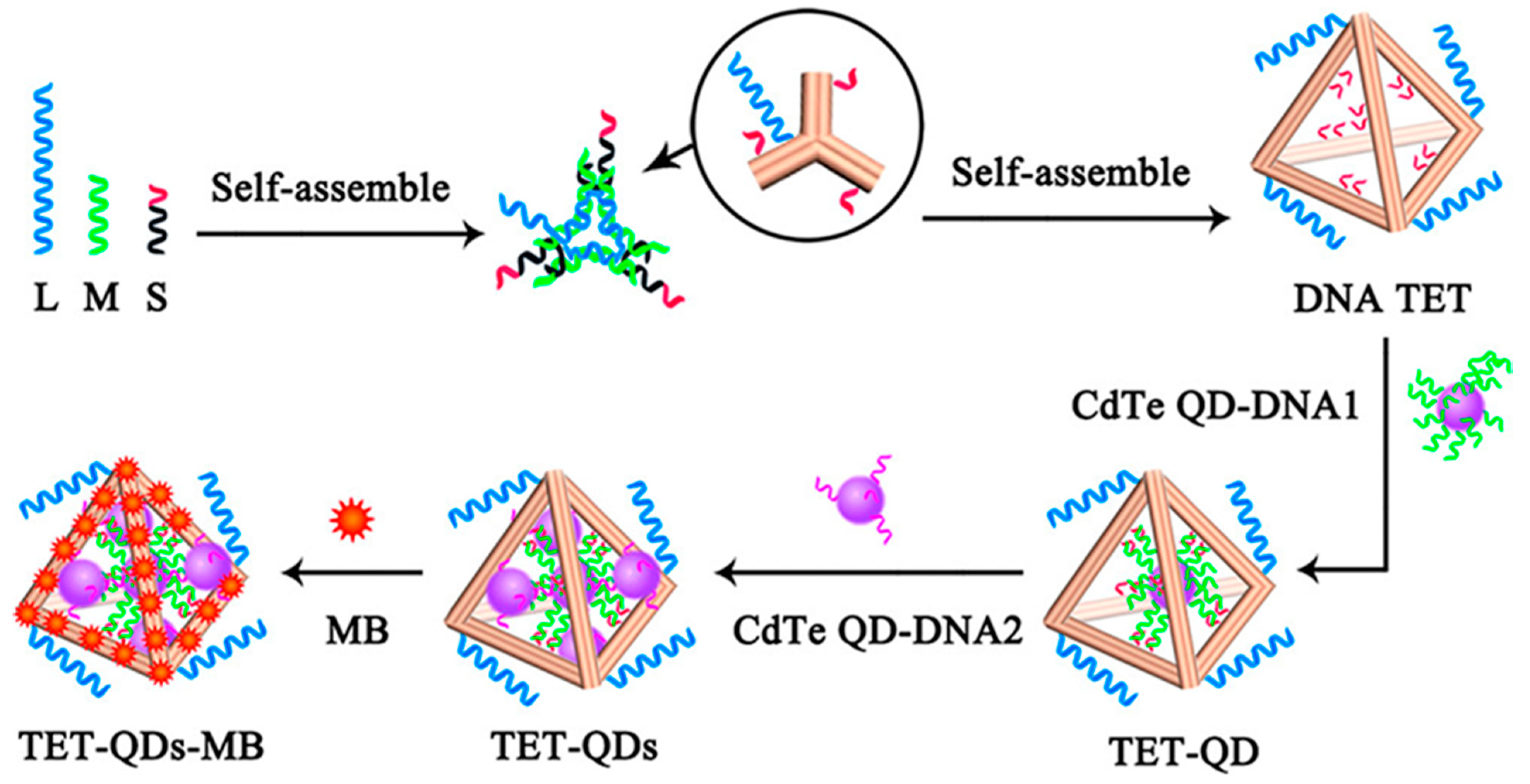
- Li, M.; Xiong, C.; Zheng, Y.; Liang, W.; Yuan, R.; Chai, Y. Ultrasensitive Photoelectrochemical Biosensor Based on DNA Tetrahedron as Nanocarrier for Efficient Immobilization of CdTe QDs-Methylene Blue as Signal Probe with Near-Zero Background Noise. Anal. Chem. 2018, 90, 8211–8216. [Google Scholar] [CrossRef]
- Su, J.; Wu, F.; Xia, H.; Wu, Y.; Liu, S. Accurate cancer cell identification and microRNA silencing induced therapy using tailored DNA tetrahedron nanostructures. Chem. Sci. 2020, 11, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Jiang, Y.; Li, S.; Zhang, Y.; Ma, X.; Wu, Z.; Wu, Z.; Qi, X. Multivalent aptamer-modified tetrahedral DNA nanocage demonstrates high selectivity and safety for anti-tumor therapy. Nanoscale 2019, 11, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Bhuckory, S.; Kays, J.C.; Dennis, A.M. In Vivo Biosensing Using Resonance Energy Transfer. Biosensors 2019, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Rothemund, P.W.K. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Daems, D.; Pfeifer, W.; Rutten, I.; Saccà, B.; Spasic, D.; Lammertyn, J. Three-dimensional DNA origami as programmable anchoring points for bioreceptors in fiber optic surface plasmon resonance biosensing. ACS Appl. Mater. Interfaces 2018, 10, 23539–23547. [Google Scholar] [CrossRef] [PubMed]
- Rutten, I.; Daems, D.; Lammertyn, J. Boosting biomolecular interactions through DNA origami nano-tailored biosensing interfaces. J. Mater. Chem. B 2020, 8, 3606–3615. [Google Scholar] [CrossRef] [PubMed]
- Kong, G.; Xiong, M.; Liu, L.; Hu, L.; Meng, H.-M.; Ke, G.; Zhang, X.-B.; Tan, W. DNA origami-based protein networks: From basic construction to emerging applications. Chem. Soc. Rev. 2021, 50, 1846–1873. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Fu, J.; Liu, M.; Jiang, S.; Andreoni, A.; Zuo, X.; Liu, Y.; Yan, H.; Fan, C. Constructing Submonolayer DNA Origami Scaffold on Gold Electrode for Wiring of Redox Enzymatic Cascade Pathways. ACS Appl. Mater. Interfaces 2019, 11, 13881–13887. [Google Scholar] [CrossRef]
- Prodan, E.; Radloff, C.; Halas, N.J.; Nordlander, P. A Hybridization Model for the Plasmon Response of Complex Nanostructures. Science 2003, 302, 419–422. [Google Scholar] [CrossRef]
- Ding, S.-Y.; Yi, J.; Li, J.-F.; Ren, B.; Wu, D.-Y.; Panneerselvam, R.; Tian, Z.-Q. Nanostructure-based plasmon-enhanced Raman spectroscopy for surface analysis of materials. Nat. Rev. Mater. 2016, 1, 16021. [Google Scholar] [CrossRef]
- Jiang, R.; Li, B.; Fang, C.; Wang, J. Metal/Semiconductor Hybrid Nanostructures for Plasmon-Enhanced Applications. Adv. Mater. 2014, 26, 5274–5309. [Google Scholar] [CrossRef] [PubMed]
- Ming, T.; Chen, H.; Jiang, R.; Li, Q.; Wang, J. Plasmon-Controlled Fluorescence: Beyond the Intensity Enhancement. J. Phys. Chem. Lett. 2012, 3, 191–202. [Google Scholar] [CrossRef]
- Wiley, B.J.; Im, S.H.; Li, Z.-Y.; McLellan, J.; Siekkinen, A.; Xia, Y. Maneuvering the Surface Plasmon Resonance of Silver Nanostructures through Shape-Controlled Synthesis. J. Phys. Chem. B 2006, 110, 15666–15675. [Google Scholar] [CrossRef] [PubMed]
- Shaukat, A.; Anaya-Plaza, E.; Julin, S.; Linko, V.; Torres, T.; de la Escosura, A.; Kostiainen, M.A. Phthalocyanine–DNA origami complexes with enhanced stability and optical properties. Chem. Commun. 2020, 56, 7341–7344. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, C.; Niu, D.; Sha, R.; Seeman, N.C.; Canary, J.W. Construction of a DNA Origami Based Molecular Electro-optical Modulator. Nano Lett. 2018, 18, 2112–2115. [Google Scholar] [CrossRef]
- Kuzyk, A.; Jungmann, R.; Acuna, G.P.; Liu, N. DNA Origami Route for Nanophotonics. ACS Photonics 2018, 5, 1151–1163. [Google Scholar] [CrossRef] [PubMed]
- Urban, M.J.; Dutta, P.K.; Wang, P.; Duan, X.; Shen, X.; Ding, B.; Ke, Y.; Liu, N. Plasmonic Toroidal Metamolecules Assembled by DNA Origami. J. Am. Chem. Soc. 2016, 138, 5495–5498. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Schreiber, R.; Zhang, H.; Govorov, A.O.; Liedl, T.; Liu, N. Reconfigurable 3D plasmonic metamolecules. Nat. Mater. 2014, 13, 862–866. [Google Scholar] [CrossRef] [PubMed]
- Hübner, K.; Pilo-Pais, M.; Selbach, F.; Liedl, T.; Tinnefeld, P.; Stefani, F.D.; Acuna, G.P. Directing Single-Molecule Emission with DNA Origami-Assembled Optical Antennas. Nano Lett. 2019, 19, 6629–6634. [Google Scholar] [CrossRef]
- Vogele, K.; List, J.; Pardatscher, G.; Holland, N.B.; Simmel, F.C.; Pirzer, T. Self-Assembled Active Plasmonic Waveguide with a Peptide-Based Thermomechanical Switch. ACS Nano 2016, 10, 11377–11384. [Google Scholar] [CrossRef] [PubMed]
- Prinz, J.; Schreiber, B.; Olejko, L.; Oertel, J.; Rackwitz, J.; Keller, A.; Bald, I. DNA Origami Substrates for Highly Sensitive Surface-Enhanced Raman Scattering. J. Phys. Chem. Lett. 2013, 4, 4140–4145. [Google Scholar] [CrossRef]
- Puchkova, A.; Vietz, C.; Pibiri, E.; Wünsch, B.; Sanz Paz, M.; Acuna, G.P.; Tinnefeld, P. DNA Origami Nanoantennas with over 5000-fold Fluorescence Enhancement and Single-Molecule Detection at 25 μM. Nano Lett. 2015, 15, 8354–8359. [Google Scholar] [CrossRef] [PubMed]
- Ijäs, H.; Nummelin, S.; Shen, B.; Kostiainen, M.A.; Linko, V. Dynamic DNA Origami Devices: From Strand-Displacement Reactions to External-Stimuli Responsive Systems. Int. J. Mol. Sci. 2018, 19, 2114. [Google Scholar] [CrossRef] [PubMed]
- Grossi, G.; Dalgaard Ebbesen Jepsen, M.; Kjems, J.; Andersen, E.S. Control of enzyme reactions by a reconfigurable DNA nanovault. Nat. Commun. 2017, 8, 992. [Google Scholar] [CrossRef] [PubMed]
- Kopperger, E.; List, J.; Madhira, S.; Rothfischer, F.; Lamb, D.C.; Simmel, F.C. A self-assembled nanoscale robotic arm controlled by electric fields. Science 2018, 359, 296–301. [Google Scholar] [CrossRef]
- Kuzyk, A.; Yang, Y.; Duan, X.; Stoll, S.; Govorov, A.O.; Sugiyama, H.; Endo, M.; Liu, N. A light-driven three-dimensional plasmonic nanosystem that translates molecular motion into reversible chiroptical function. Nat. Commun. 2016, 7, 10591. [Google Scholar] [CrossRef]
- Funck, T.; Liedl, T.; Bae, W. Dual Aptamer-Functionalized 3D Plasmonic Metamolecule for Thrombin Sensing. Appl. Sci. 2019, 9, 3006. [Google Scholar] [CrossRef]
- Lan, X.; Chen, Z.; Dai, G.; Lu, X.; Ni, W.; Wang, Q. Bifacial DNA Origami-Directed Discrete, Three-Dimensional, Anisotropic Plasmonic Nanoarchitectures with Tailored Optical Chirality. J. Am. Chem. Soc. 2013, 135, 11441–11444. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, A.R.; Pushpanathan, M.; Halvorsen, K. Evolution of DNA origami scaffolds. Mater. Lett. 2016, 170, 221–224. [Google Scholar] [CrossRef]
- Pound, E.; Ashton, J.R.; Becerril, H.A.; Woolley, A.T. Polymerase Chain Reaction Based Scaffold Preparation for the Production of Thin, Branched DNA Origami Nanostructures of Arbitrary Sizes. Nano Lett. 2009, 9, 4302–4305. [Google Scholar] [CrossRef]
- Liu, W.; Zhong, H.; Wang, R.; Seeman, N.C. Crystalline Two-Dimensional DNA-Origami Arrays. Angew. Chem. Int. Ed. 2011, 50, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Zhang, F.; Wang, F.; Peng, T.; Liu, H.; Poppleton, E.; Šulc, P.; Jiang, S.; Liu, L.; Gong, C.; et al. Meta-DNA structures. Nat. Chem. 2020, 12, 1067–1075. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Zeng, D.; Chao, J.; Jin, Y.; Zhang, Z.; Liu, H.; Li, D.; Ma, H.; Huang, Q.; Gothelf, K.V.; et al. Single-Step Rapid Assembly of DNA Origami Nanostructures for Addressable Nanoscale Bioreactors. J. Am. Chem. Soc. 2013, 135, 696–702. [Google Scholar] [CrossRef] [PubMed]

| Category | Advantages | Disadvantages | Detection Object | |
|---|---|---|---|---|
| Functional DNA strand-based biosensors | DNA aptamer | Easily accessed; easily modified; adjustable affinity; more economic; more durable lifetime | Requires multi-round selection; easily attacked by the nucleic enzyme; potential biotoxicity | IFN-γ [64,65] |
| Pb2+ [77] | ||||
| Thrombin [69,83] | ||||
| DNAzyme | High catalytic activity; small molecule detection with high sensitivity | Easily affected by temperature; needs oxidative substrate; the reaction product cannot be recycled to use | Pb2+ [102] | |
| AMP, Lyso [106] | ||||
| Bleomycin [107] | ||||
| DNA hybridization-based biosensors | DNA hairpin | Detects nucleic acids with high selectivity; easily converts the hybridization process into physical signal change | Easily damaged by temperature | DNA [117,118,119] |
| HCR | High sensitivity, especially at the biosensing interface | Easily be triggered automatically by mistake | micro-RNA [121] | |
| CHA | More stable than HCR | Not as sensitive as HCR | micro-RNA [122] | |
| DNA template-based biosensors | DNA tiles | Effectively adjust the surface density of bioprobes; suitable for in vivo biosensing | Lack of the ability to form complex and large-scale patterns | mi-RNA 141 [155] |
| DNA origami | Control the arrange of bioprobes and materials with nanoscale accuracy; programmable nanostructure | Time-consuming annealing process; expensive; difficult to design | Oligonucleotides [177] | |
| Thrombin [160,182] | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hua, Y.; Ma, J.; Li, D.; Wang, R. DNA-Based Biosensors for the Biochemical Analysis: A Review. Biosensors 2022, 12, 183. https://doi.org/10.3390/bios12030183
Hua Y, Ma J, Li D, Wang R. DNA-Based Biosensors for the Biochemical Analysis: A Review. Biosensors. 2022; 12(3):183. https://doi.org/10.3390/bios12030183
Chicago/Turabian StyleHua, Yu, Jiaming Ma, Dachao Li, and Ridong Wang. 2022. "DNA-Based Biosensors for the Biochemical Analysis: A Review" Biosensors 12, no. 3: 183. https://doi.org/10.3390/bios12030183
APA StyleHua, Y., Ma, J., Li, D., & Wang, R. (2022). DNA-Based Biosensors for the Biochemical Analysis: A Review. Biosensors, 12(3), 183. https://doi.org/10.3390/bios12030183






