Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. ELISA Assays
2.3. LOC Fabrication and Functionalization
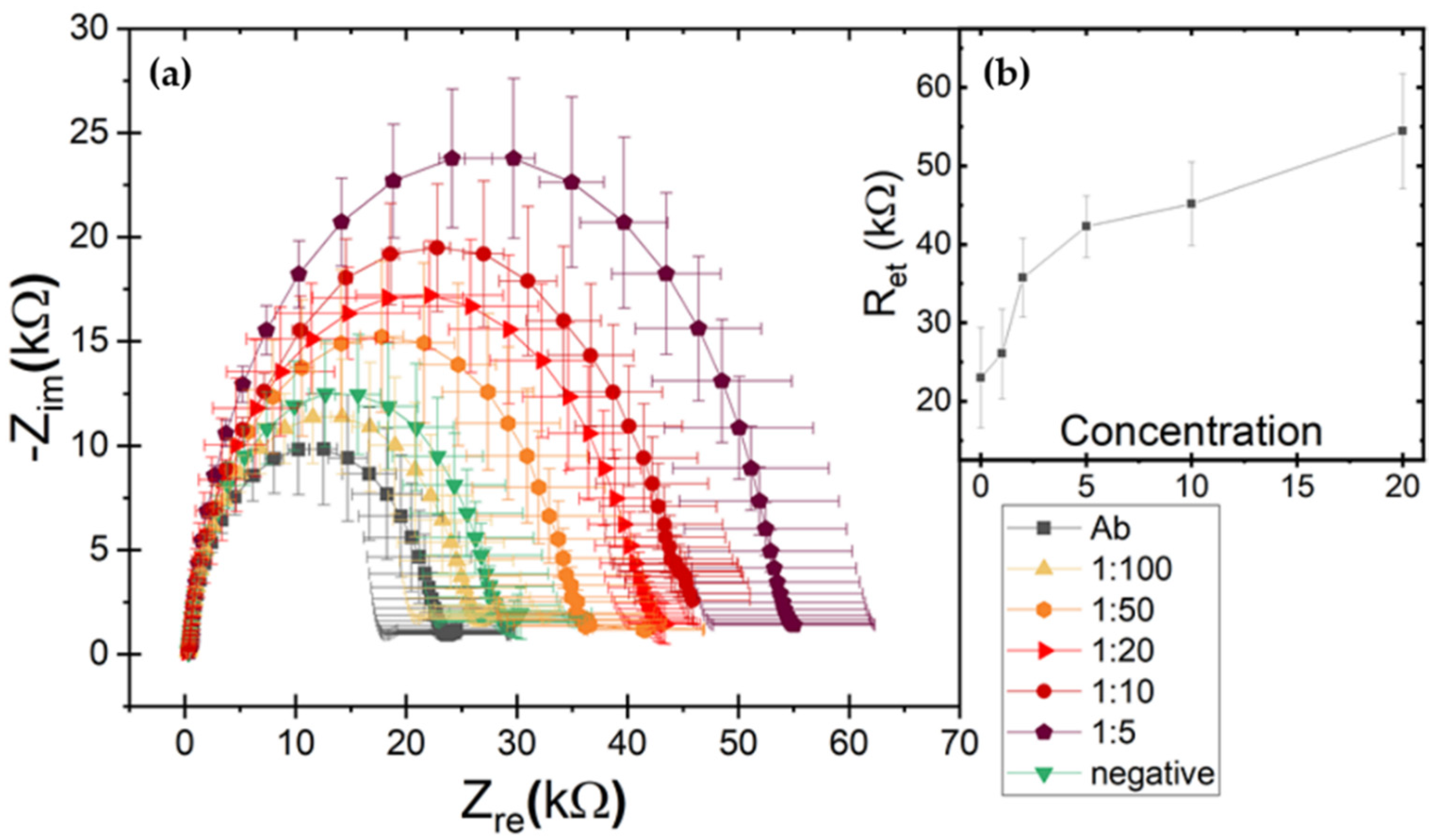
2.4. LOC Analysis of Samples
2.5. Statistical Analysis
3. Results
3.1. ELISA Assay
3.2. LOC Assay
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Martelli, G.P. Grapevine leafroll. J. Plant Pathol. 2014, 96, S51–S70. [Google Scholar] [CrossRef]
- Martelli, G.P. Infectious degeneration (Grapevine fanleaf virus). J. Plant Pathol. 2014, 96, S11–S27. [Google Scholar] [CrossRef]
- Almeida, R.P.P.; Daane, K.M.; Bell, V.A.; Eblaisdell, G.K.; Cooper, M.L.; Eherrbach, E.; Epietersen, G. Ecology and management of grapevine leafroll disease. Front. Microbiol. 2013, 4, 94. [Google Scholar] [CrossRef] [Green Version]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freeborough, M.-J.; Burger, J. Leaf Roll: Economic Implications. Wynboer. 2006. Available online: http://www.wynboer.co.za/recentarticles/200812-leafroll.php3 (accessed on 16 August 2013).
- Nimmo-Bell. The Economic Effects and Financial Impact of GLRaV3. Available online: http://www.nzwine.com/the-economic-effects-and-financialimpact/;jsessionid=D7464D6CE6F261FF32EEB266A042DB53 (accessed on 25 July 2016).
- Martelli, G.P.; Savino, V. Fanleaf degeneration. In Compendium of Grape Diseases; Pearson, R., Goheen, A.C., Eds.; APS Press: Saint Paul, MN, USA, 1990; pp. 48–49. [Google Scholar]
- Panno, S.; Caruso, A.; Bertacca, S.; Pisciotta, A.; Lorenzo, R.; Marchione, S.; Matić, S.; Davino, S. Genetic Structure and Molecular Variability of Grapevine Fanleaf Virus in Sicily. Agriculture 2021, 11, 496. [Google Scholar] [CrossRef]
- Andret-Link, P.; Hoffmann, C.; Valat, L.; Ritzenthaler, C.; Demangeat, G.; Vigne, E.; Laval, V.; Pfeiffer, P.; Stussi-Garaud, C.; Fuchs, M. Grapevine fanleaf virus: Still a major threat to the grapevine industry. J. Plant Pathol. 2004, 86, 183–195. [Google Scholar]
- Tsai, C.-W.; Rowhani, A.; Golino, D.A.; Daane, K.M.; Almeida, R.P.P. Mealybug Transmission of Grapevine Leafroll Viruses: An Analysis of Virus-Vector Specificity. Phytopathology 2010, 100, 830–834. [Google Scholar] [CrossRef] [Green Version]
- Le Maguet, J.; Beuve, M.; Herrbach, E.; Lemaire, O. Transmission of Six Ampeloviruses and Two Vitiviruses to Grapevine by Phenacoccus aceris. Phytopathology 2012, 102, 717–723. [Google Scholar] [CrossRef] [Green Version]
- Wine Production—Oiv First Estimates. 2020. Available online: http://www.oiv.int/public/medias/7541/en-oiv-2020-world-wine-production-first-estimates.pdf (accessed on 23 December 2021).
- EUR-Lex. Council Directive 68/193/EEC of 9 April 1968 on the Marketing of Material for the Vegetative Propagation of the Vine. Available online: http://data.europa.eu/eli/dir/1968/193/oj (accessed on 23 December 2021).
- EUR-Lex. Council Directive 2002/11/EC of 14 February 2002 Amending Directive 68/193/EEC on the Marketing of Material for the Vegetative Propagation of the Vine and Repealing Directive 74/649/EEC. Available online: http://data.europa.eu/eli/dir/2002/11/oj (accessed on 7 October 2020).
- EUR-Lex. Commission Directive 2005/43/EC of 23 June 2005 Amending the Annexes to Council Directive 68/193/EEC on the Marketing of Material for the Vegetative Propagation of the Vine. Available online: http://data.europa.eu/eli/dir/2005/43/oj (accessed on 7 October 2020).
- Rowhani, A.; Uyemoto, J.K.; Golino, D.A.; Martelli, G.P. Pathogen Testing and Certification of Vitis and Prunus Species. Annu. Rev. Phytopathol. 2005, 43, 261–278. [Google Scholar] [CrossRef] [Green Version]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine Leafroll: A Complex Viral Disease Affecting a High-Value Fruit Crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef] [Green Version]
- Julich, S.; Riedel, M.; Kielpinski, M.; Urban, M.; Kretschmer, R.; Wagner, S.; Fritzsche, W.; Henkel, T.; Möller, R.; Werres, S. Development of a lab-on-a-chip device for diagnosis of plant pathogens. Biosens. Bioelectron. 2011, 26, 4070–4075. [Google Scholar] [CrossRef]
- Nezhad, A.S. Future of portable devices for plant pathogen diagnosis. Lab Chip 2014, 14, 2887–2904. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Luvisi, A.; Primiceri, E.; Sabella, E.; De Bellis, L.; Maruccio, G. Development of a lab-on-a-chip method for rapid assay of Xylella fastidiosa subsp. pauca strain CoDiRO. Sci. Rep. 2018, 8, 7376. [Google Scholar] [CrossRef]
- Rizzato, S.; Leo, A.; Monteduro, A.G.; Chiriacò, M.S.; Primiceri, E.; Sirsi, F.; Milone, A.; Maruccio, G. Advances in the Development of Innovative Sensor Platforms for Field Analysis. Micromachines 2020, 11, 491. [Google Scholar] [CrossRef]
- Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in Plant Disease Detection and Monitoring: From Traditional Assays to In-Field Diagnostics. Sensors 2021, 21, 2129. [Google Scholar] [CrossRef]
- Khater, M.; de la Escosura-Muñiz, A.; Quesada-González, D.; Merkoçi, A. Electrochemical detection of plant virus using gold nanoparticle-modified electrodes. Anal. Chim. Acta 2019, 1046, 123–131. [Google Scholar] [CrossRef] [Green Version]
- Jarocka, U.; Wąsowicz, M.; Radecka, H.; Malinowski, T.; Michalczuk, L.; Radecki, J. Impedimetric Immunosensor for Detection of Plum Pox Virus in Plant Extracts. Electroanalysis 2011, 23, 2197–2204. [Google Scholar] [CrossRef]
- Jarocka, U.; Radecka, H.; Malinowski, T.; Michalczuk, L.; Radecki, J. Detection of Prunus Necrotic Ringspot Virus in Plant Extracts with Impedimetric Immunosensor based on Glassy Carbon Electrode. Electroanalysis 2013, 25, 433–438. [Google Scholar] [CrossRef]
- Byzova, N.A.; Vinogradova, S.V.; Porotikova, E.V.; Terekhova, U.D.; Zherdev, A.V.; Dzantiev, B.B. Lateral Flow Immunoassay for Rapid Detection of Grapevine Leafroll-Associated Virus. Biosensors 2018, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Tereshchenko, A.; Fedorenko, V.; Smyntyna, V.; Konup, I.; Konup, A.; Eriksson, M.; Yakimova, R.; Ramanavicius, A.; Balme, S.; Bechelany, M. ZnO films formed by atomic layer deposition as an optical biosensor platform for the detection of Grapevine virus A-type proteins. Biosens. Bioelectron. 2017, 92, 763–769. [Google Scholar] [CrossRef]
- Tereshchenko, A.; Yazdi, G.R.; Konup, I.; Smyntyna, V.; Khranovskyy, V.; Yakimova, R.; Ramanavicius, A. Application of ZnO Nanorods Based Whispering Gallery Mode Resonator in Optical Immunosensors. Colloids Surf. B Biointerfaces 2020, 191, 110999. [Google Scholar] [CrossRef]
- Vashpanov, Y.; Son, J.Y.; Kwack, K.D. Mesoporous Silicon with Modified Surface for Plant Viruses and Their Protein Particle Sensing. Sensors 2008, 8, 6225–6234. [Google Scholar] [CrossRef]
- Fang, X.; Chen, H.; Yu, S.; Jiang, X.; Kong, J. Predicting Viruses Accurately by a Multiplex Microfluidic Loop-Mediated Isothermal Amplification Chip. Anal. Chem. 2011, 83, 690–695. [Google Scholar] [CrossRef]
- Piccinno, E.; Monteduro, A.G.; Dituri, F.; Rizzato, S.; Giannelli, G.; Maruccio, G. Validation of a Lab-on-Chip Assay for Measuring Sorafenib Effectiveness on HCC Cell Proliferation. Int. J. Mol. Sci. 2021, 22, 13090. [Google Scholar] [CrossRef]
- Tourlousse, D.M.; Ahmad, F.; Stedtfeld, R.D.; Seyrig, G.; Tiedje, J.M.; Hashsham, S.A. A polymer microfluidic chip for quantitative detection of multiple water- and foodborne pathogens using real-time fluorogenic loop-mediated isothermal amplification. Biomed. Microdevices 2012, 14, 769–778. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; de Feo, F.; Primiceri, E.; Monteduro, A.G.; de Benedetto, G.E.; Pennetta, A.; Rinaldi, R.; Maruccio, G. Portable gliadin-immunochip for contamination control on the food production chain. Talanta 2015, 142, 57–63. [Google Scholar] [CrossRef]
- Primiceri, E.; Chiriacò, M.S.; de Feo, F.; Santovito, E.; Fusco, V.; Maruccio, G. A multipurpose biochip for food pathogen detection. Anal. Methods 2016, 8, 3055–3060. [Google Scholar] [CrossRef]
- Katz, E.; Willner, I. Probing Biomolecular Interactions at Conductive and Semiconductive Surfaces by Impedance Spectroscopy: Routes to Impedimetric Immunosensors, DNA-Sensors, and Enzyme Biosensors. Electroanalysis 2003, 15, 913–947. [Google Scholar] [CrossRef]
- Chiriacò, M.S.; Primiceri, E.; Monteduro, A.G.; Bove, A.; Leporatti, S.; Capello, M.; Ferri-Borgogno, S.; Rinaldi, R.; Novelli, F.; Maruccio, G. Towards pancreatic cancer diagnosis using EIS biochips. Lab Chip 2013, 13, 730–734. [Google Scholar] [CrossRef] [Green Version]
- Chiriacò, M.S.; Primiceri, E.; De Feo, F.; Montanaro, A.; Monteduro, A.G.; Tinelli, A.; Megha, M.; Carati, D.; Maruccio, G. Simultaneous detection of multiple lower genital tract pathogens by an impedimetric immunochip. Biosens. Bioelectron. 2016, 79, 9–14. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Carstens, E.B.; Lefkowitz, E.J. (Eds.) Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses; Elsevier-Academic Press: Amsterdam, The Netherlands, 2011; pp. 987–1001. [Google Scholar]




| Sample Dilution | GLRaV-3 | GFLV | ||
|---|---|---|---|---|
| R | Result | R | Result | |
| 1:3 | 5.42 | + | 3.49 | + |
| 1:5 | 4.61 | + | 1.83 | − |
| 1:10 | 2.55 | − | 1.12 | − |
| 1:20 | 1.49 | − | 0.86 | − |
| 1:50 | 0.92 | − | 0.71 | − |
| 1:100 | 0.89 | − | 0.69 | − |
| Negative control | 1.00 | − | 1.00 | − |
| Bibliographic Reference | Target | LOD | Sensing Element | Transduction | Fluidics |
|---|---|---|---|---|---|
| Our biochip | GLRaV-3- or GFLV-infected woody tissues | 1:50 for GFLV; 1:100 for GLRaV-3 | Ab immobilized on gold microelectrodes | Electrochemical impedance | Yes |
| Byzova et al., 2018 | GLRaV-3-, GLRaV-1-, GVA- and GFLV-infected leaf tissues | Sensitivity of 100% vs. ELISA; sensitivity of 93% vs. PCR; | Immobilized antibody–antigen–GNP-labeled antibody | Visual detection | No |
| Tereshchenko et al., 2017 | GVA-infected woody tissues | Unstable operation point | Immobilized GVA-antibodies | Optical/photoluminescence | No |
| Tereshchenko et al., 2020 | GVA-infected woody tissues | Detection without dose-response curve | Silicon/ZnO-NRs/anti-GVA immune-sensing | Optical/Whispering Gallery Mode Resonators | No |
| Vashpanov et al., 2008 | Purified ToRSV and GFLV | Detection without dose-response curve | Absorption on mesoporous silicon | Changes in drain-source current | No |
| Sample Dilution | GLRaV-3 | GFLV | ||
|---|---|---|---|---|
| ELISA | LOC | ELISA | LOC | |
| 1:3 | + | n.p. | + | n.p. |
| 1:5 | + | n.p. | − | + |
| 1:10 | − | + | − | + |
| 1:20 | − | + | − | + |
| 1:50 | − | + | − | + |
| 1:100 | − | + | − | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Buja, I.; Sabella, E.; Monteduro, A.G.; Rizzato, S.; Bellis, L.D.; Elicio, V.; Formica, L.; Luvisi, A.; Maruccio, G. Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine. Biosensors 2022, 12, 147. https://doi.org/10.3390/bios12030147
Buja I, Sabella E, Monteduro AG, Rizzato S, Bellis LD, Elicio V, Formica L, Luvisi A, Maruccio G. Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine. Biosensors. 2022; 12(3):147. https://doi.org/10.3390/bios12030147
Chicago/Turabian StyleBuja, Ilaria, Erika Sabella, Anna Grazia Monteduro, Silvia Rizzato, Luigi De Bellis, Vito Elicio, Lilia Formica, Andrea Luvisi, and Giuseppe Maruccio. 2022. "Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine" Biosensors 12, no. 3: 147. https://doi.org/10.3390/bios12030147
APA StyleBuja, I., Sabella, E., Monteduro, A. G., Rizzato, S., Bellis, L. D., Elicio, V., Formica, L., Luvisi, A., & Maruccio, G. (2022). Detection of Ampelovirus and Nepovirus by Lab-on-a-Chip: A Promising Alternative to ELISA Test for Large Scale Health Screening of Grapevine. Biosensors, 12(3), 147. https://doi.org/10.3390/bios12030147












