Evaluation of Hydroxycarboxylic Acid Receptor 1 (HCAR1) as a Building Block for Genetically Encoded Extracellular Lactate Biosensors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Engineering of Constructs
2.2. Cell Culture and Transfection
2.3. Widefield Fluorescence Imaging
2.4. Confocal Imaging
2.5. Two-Photon Imaging
2.6. Data Analysis
2.7. Chemicals and Drugs
3. Results and Discussion
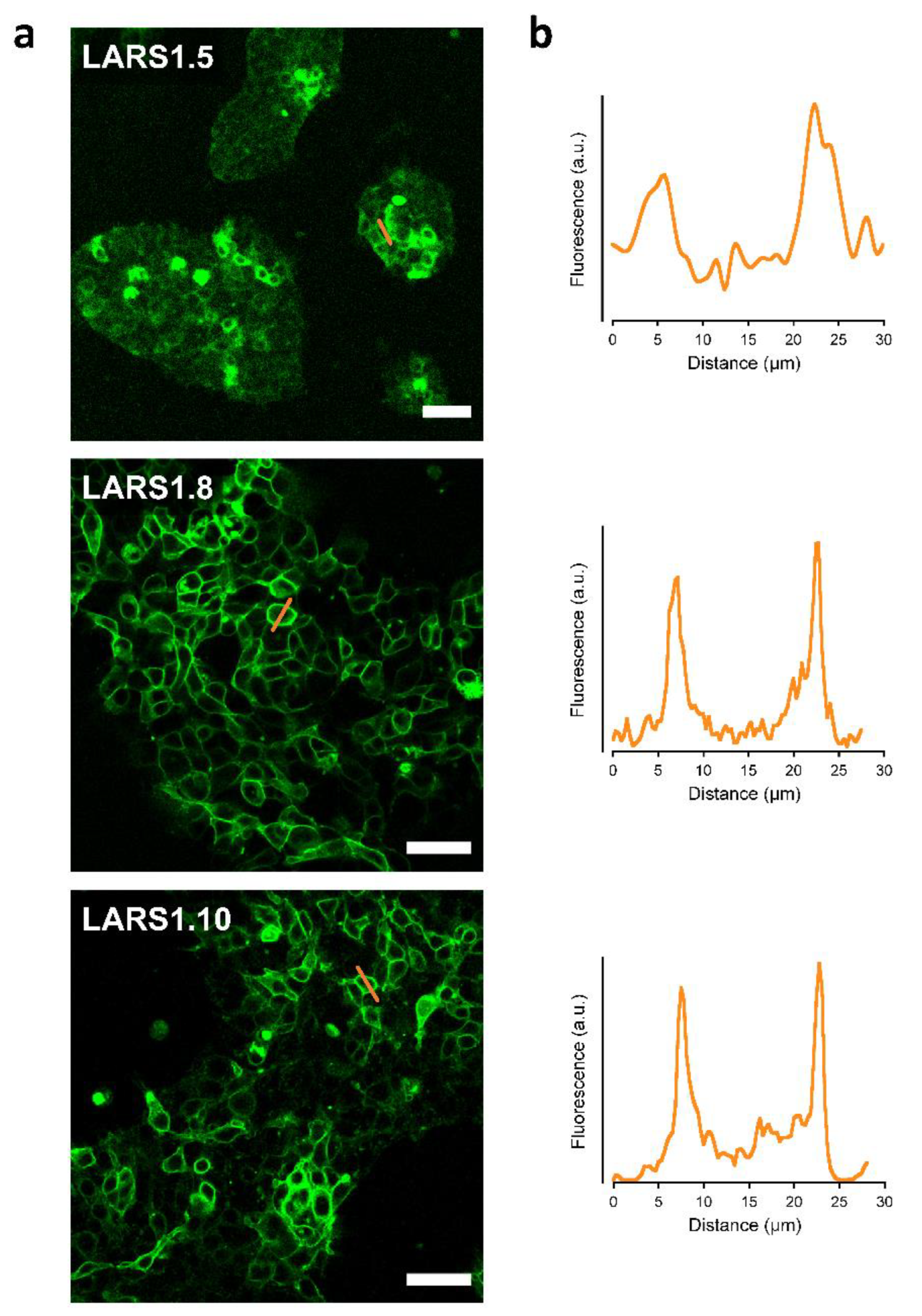
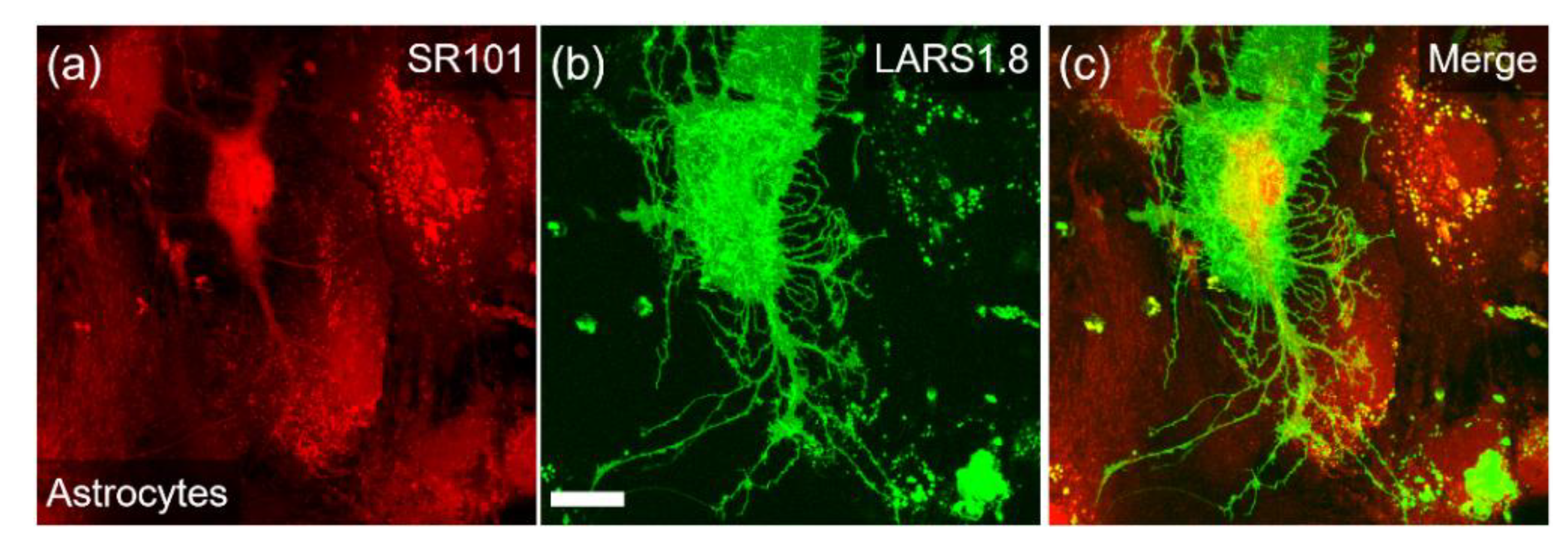
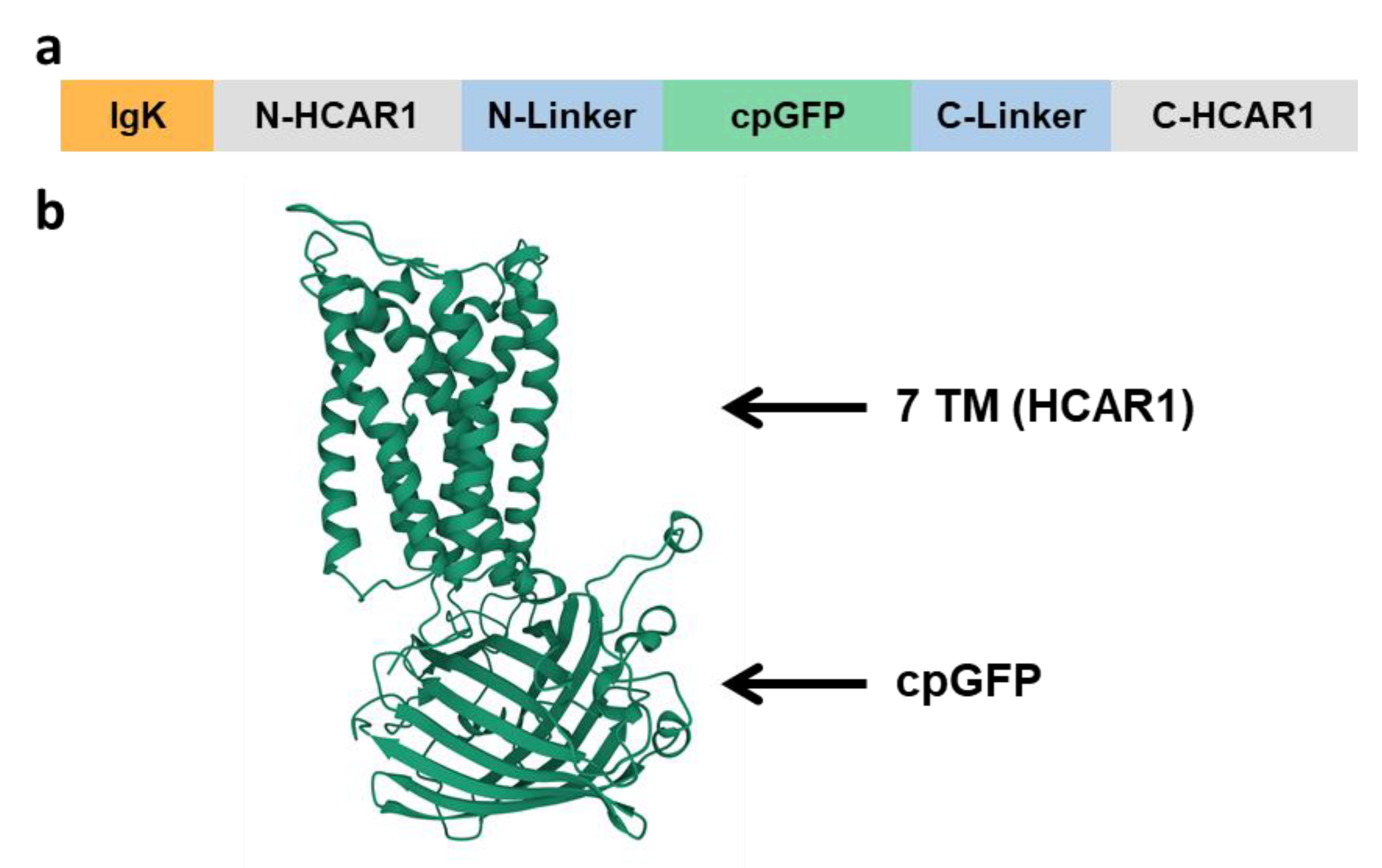
3.1. Design and Characterization of HCAR1-Based Lactate Sensors
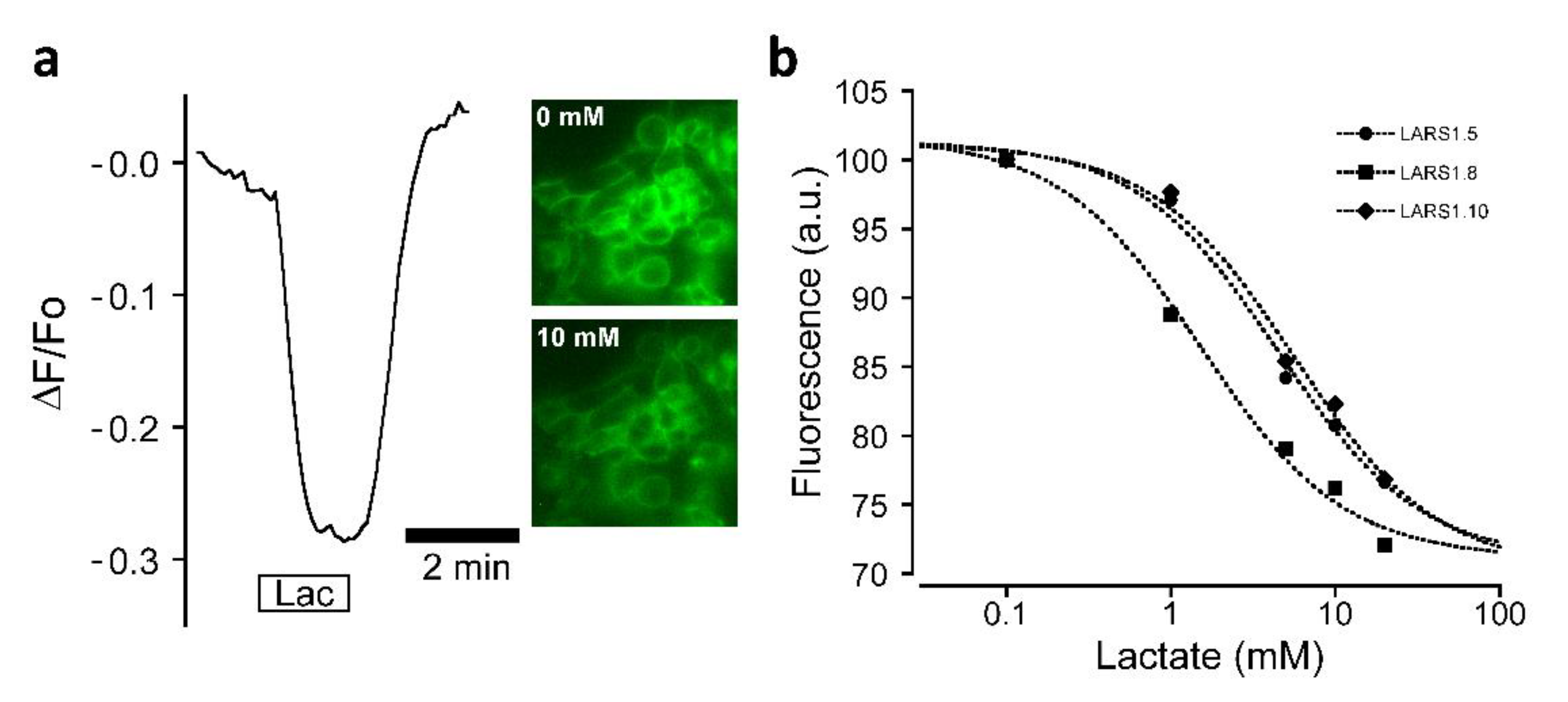
3.2. Biosensor Sensitivity to Lactate
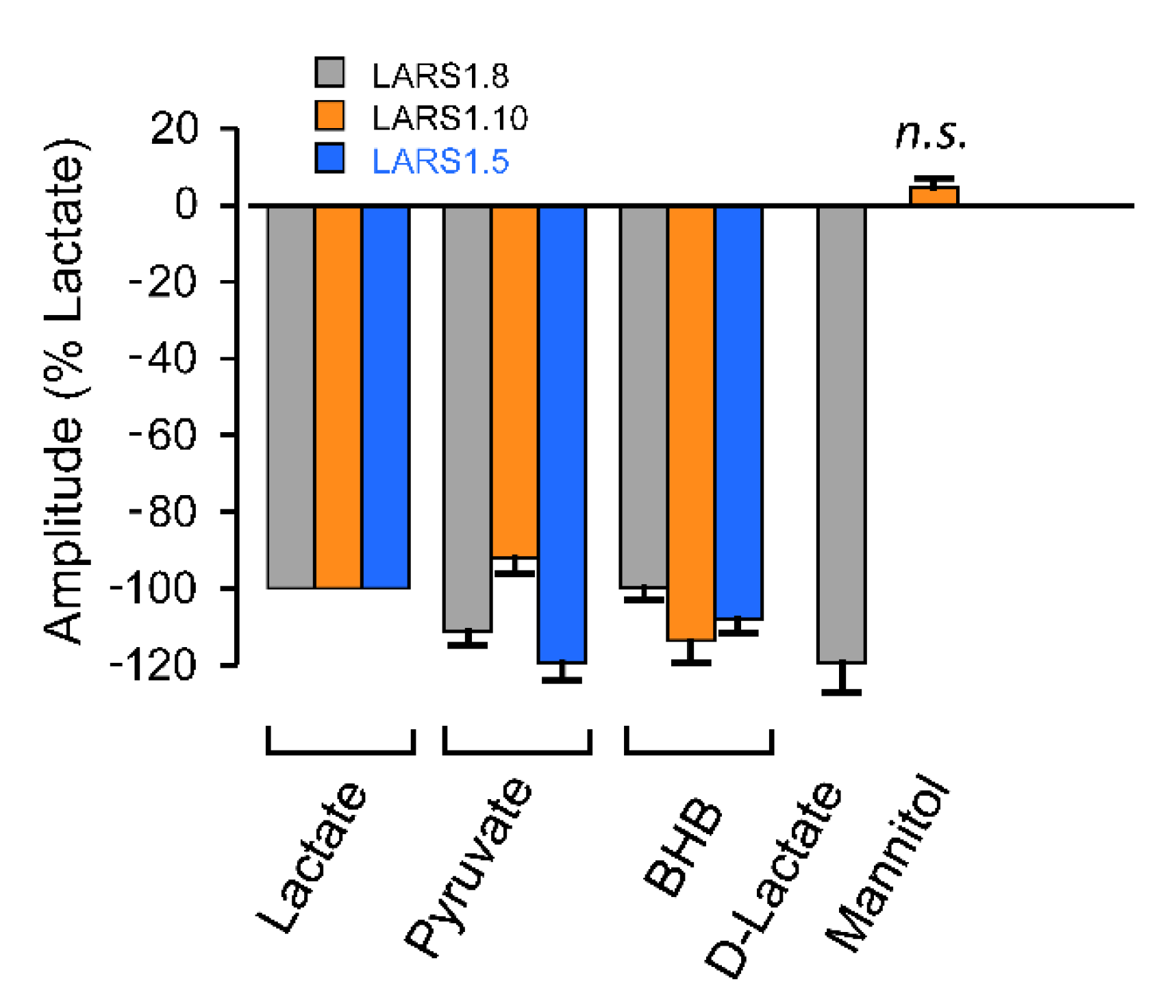
3.3. Biosensor Selectivity
4. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Brooks, G.A. Lactate as a fulcrum of metabolism. Redox Biol. 2020, 35, 101454. [Google Scholar] [CrossRef] [PubMed]
- Gladden, L.B. 200th anniversary of lactate research in muscle. Exerc. Sport Sci. Rev. 2008, 36, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Barros, L.F. Metabolic signaling by lactate in the brain. Trends Neurosci. 2013, 36, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Pellerin, L.; Magistretti, P.J. Glutamate uptake into astrocytes stimulates aerobic glycolysis: A mechanism coupling neuronal activity to glucose utilization. Proc. Natl. Acad. Sci. USA 1994, 91, 10625–10629. [Google Scholar] [CrossRef] [Green Version]
- Abi-Saab, W.M.; Maggs, D.G.; Jones, T.; Jacob, R.; Srihari, V.; Thompson, J.; Kerr, D.; Leone, P.; Krystal, J.H.; Spencer, D.D.; et al. Striking differences in glucose and lactate levels between brain extracellular fluid and plasma in conscious human subjects: Effects of hyperglycemia and hypoglycemia. J. Cereb. Blood Flow Metab. 2002, 22, 271–279. [Google Scholar] [CrossRef] [Green Version]
- Dienel, G.A.; Ball, K.K.; Cruz, N.F. A glycogen phosphorylase inhibitor selectively enhances local rates of glucose utilization in brain during sensory stimulation of conscious rats: Implications for glycogen turnover. J. Neurochem. 2007, 102, 466–478. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Wilson, G.S. A temporary local energy pool coupled to neuronal activity: Fluctuations of extracellular lactate levels in rat brain monitored with rapid-response enzyme-based sensor. J. Neurochem. 1997, 69, 1484–1490. [Google Scholar] [CrossRef]
- Offermanns, S. Hydroxy-Carboxylic Acid Receptor Actions in Metabolism. Trends Endocrinol. Metab. 2017, 28, 227–236. [Google Scholar] [CrossRef]
- Blad, C.C.; Tang, C.; Offermanns, S. G protein-coupled receptors for energy metabolites as new therapeutic targets. Nat. Rev. Drug Discov. 2012, 11, 603–619. [Google Scholar] [CrossRef]
- Husted, A.S.; Trauelsen, M.; Rudenko, O.; Hjorth, S.A.; Schwartz, T.W. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017, 25, 777–796. [Google Scholar] [CrossRef] [Green Version]
- Cai, T.Q.; Ren, N.; Jin, L.; Cheng, K.; Kash, S.; Chen, R.; Wright, S.D.; Taggart, A.K.; Waters, M.G. Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem. Biophys. Res. Commun. 2008, 377, 987–991. [Google Scholar] [CrossRef]
- Liu, C.; Wu, J.; Zhu, J.; Kuei, C.; Yu, J.; Shelton, J.; Sutton, S.W.; Li, X.; Yun, S.J.; Mirzadegan, T.; et al. Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 2009, 284, 2811–2822. [Google Scholar] [CrossRef] [Green Version]
- Bozzo, L.; Puyal, J.; Chatton, J.-Y. Lactate modulates the activity of primary cortical neurons through a receptor-mediated pathway. PLoS ONE 2013, 8, e71721. [Google Scholar] [CrossRef]
- De Castro Abrantes, H.; Briquet, M.; Schmuziger, C.; Restivo, L.; Puyal, J.; Rosenberg, N.; Rocher, A.B.; Offermanns, S.; Chatton, J.Y. The lactate receptor HCAR1 modulates neuronal network activity through the activation of Galpha and Gbeta subunits. J. Neurosci. 2019, 39, 4422–4433. [Google Scholar] [CrossRef] [Green Version]
- Briquet, M.; Rocher, A.B.; Alessandri, M.; Rosenberg, N.; de Castro Abrantes, H.; Wellbourne-Wood, J.; Schmuziger, C.; Ginet, V.; Puyal, J.; Pralong, E.; et al. Activation of lactate receptor HCAR1 down-modulates neuronal activity in rodent and human brain tissue. J. Cereb. Blood Flow Metab. 2022; in press. [Google Scholar]
- Sotelo-Hitschfeld, T.; Niemeyer, M.I.; Machler, P.; Ruminot, I.; Lerchundi, R.; Wyss, M.T.; Stobart, J.; Fernandez-Moncada, I.; Valdebenito, R.; Garrido-Gerter, P.; et al. Channel-mediated lactate release by K(+)-stimulated astrocytes. J. Neurosci. 2015, 35, 4168–4178. [Google Scholar] [CrossRef] [Green Version]
- Nasu, Y.; Shen, Y.; Kramer, L.; Campbell, R.E. Structure- and mechanism-guided design of single fluorescent protein-based biosensors. Nat. Chem. Biol. 2021, 17, 509–518. [Google Scholar] [CrossRef]
- San Martin, A.; Ceballo, S.; Ruminot, I.; Lerchundi, R.; Frommer, W.B.; Barros, L.F. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS ONE 2013, 8, e57712. [Google Scholar] [CrossRef]
- Nasu, Y.; Murphy-Royal, C.; Wen, Y.; Haidey, J.N.; Molina, R.S.; Aggarwal, A.; Zhang, S.; Kamijo, Y.; Paquet, M.E.; Podgorski, K.; et al. A genetically encoded fluorescent biosensor for extracellular L-lactate. Nat. Commun. 2021, 12, 7058. [Google Scholar] [CrossRef]
- Patriarchi, T.; Cho, J.R.; Merten, K.; Howe, M.W.; Marley, A.; Xiong, W.H.; Folk, R.W.; Broussard, G.J.; Liang, R.; Jang, M.J.; et al. Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 2018, 360, eaat4422. [Google Scholar] [CrossRef] [Green Version]
- Wan, J.; Peng, W.; Li, X.; Qian, T.; Song, K.; Zeng, J.; Deng, F.; Hao, S.; Feng, J.; Zhang, P.; et al. A genetically encoded sensor for measuring serotonin dynamics. Nat. Neurosci. 2021, 24, 746–752. [Google Scholar] [CrossRef] [PubMed]
- Cvicek, V.; Goddard, W.A., 3rd; Abrol, R. Structure-Based Sequence Alignment of the Transmembrane Domains of All Human GPCRs: Phylogenetic, Structural and Functional Implications. PLoS Comput. Biol. 2016, 12, e1004805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McGuffin, L.J.; Adiyaman, R.; Maghrabi, A.H.A.; Shuid, A.N.; Brackenridge, D.A.; Nealon, J.O.; Philomina, L.S. IntFOLD: An integrated web resource for high performance protein structure and function prediction. Nucleic Acids Res. 2019, 47, W408–W413. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.reading.ac.uk/bioinf/IntFOLD (accessed on 9 February 2022).
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodova, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koca, J.; Rose, A.S. Mol* Viewer: Modern web app for 3D visualization and analysis of large biomolecular structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef]
- 3D View. Available online: https://www.rcsb.org/3d-view (accessed on 9 February 2022).
- Sorg, O.; Magistretti, P.J. Vasoactive intestinal peptide and noradrenaline exert long-term control on glycogen levels in astrocytes: Blockade by protein synthesis inhibition. J. Neurosci. 1992, 12, 4923–4931. [Google Scholar] [CrossRef] [Green Version]
- Barnett, L.M.; Hughes, T.E.; Drobizhev, M. Deciphering the molecular mechanism responsible for GCaMP6m’s Ca2+-dependent change in fluorescence. PLoS ONE 2017, 12, e0170934. [Google Scholar] [CrossRef]

| Construct Name | Membrane Targeting Sequence | Linkers | Description | Results/Observations |
|---|---|---|---|---|
| LARS1.1 | HA secretory sequence | Based on dLight 1.1 or 1.2 1 N-linker: LSSLI C-linker: NHDQL | Use mouse HCAR1 gene; replace entire IC3 loop | No measurable fluorescence |
| LARS 1.2 | HA secretory sequence | Based on dLight 1.1 or 1.2 1 N-linker: LSSLI C-linker: NHDQL | Use human HCAR1 gene; replace entire IC3 loop | Some fluorescence, intracellular localization, lysosomes or ER |
| LARS 1.3 | HA secretory sequence | Based on dLight 1.1 or 1.2 1 N-linker: LSSLI C-linker: NHDQL | Using mouse HCAR1 gene; replace part of IC3 loop | Weak fluorescence, intracellular localization, lysosomes or ER |
| LARS 1.5 * | HA secretory sequence | Optimized linkers for B2AR and MT2R 1 N-linker: QLQKIDLSSLI C-linker: NHDQDIKQLQ | Use mouse HCAR1 gene; replace part of IC3 loop | Fluorescence partly intracellular and plasma membrane in several cells |
| LARS 1.7 | HA secretory sequence | Based on dLight 1.1 or 1.2 1 N-linker: LSSLI C-linker: NHDQL | Use human HCAR1 gene; replace entire IC2 loop | Weak and sparse fluorescence, intracellular localization |
| LARS 1.8 * | IgK secretory sequence | Optimized linkers for B2A and MT2 receptors 1 N-linker: QLQKIDLSSLI C-linker: NHDQDIKQLQ | Use mouse HCAR1 gene; replace part of IC3 loop; use IgK secretory sequence and mutated cpGFP of GRAB5-HT 2 | Robust plasma membrane fluorescence |
| LARS 1.10 * | IgK secretory sequence | Based on GRAB5HT 2 N-linker: MFLNG C-linker: GFATA | Use mouse HCAR1 gene; replace part of IC3 loop; use IgK secretory sequence; use GRAB5-HT linkers and mutated cpGFP 2 | Robust fluorescence with mixed intracellular and membrane localization |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wellbourne-Wood, J.; Briquet, M.; Alessandri, M.; Binda, F.; Touya, M.; Chatton, J.-Y. Evaluation of Hydroxycarboxylic Acid Receptor 1 (HCAR1) as a Building Block for Genetically Encoded Extracellular Lactate Biosensors. Biosensors 2022, 12, 143. https://doi.org/10.3390/bios12030143
Wellbourne-Wood J, Briquet M, Alessandri M, Binda F, Touya M, Chatton J-Y. Evaluation of Hydroxycarboxylic Acid Receptor 1 (HCAR1) as a Building Block for Genetically Encoded Extracellular Lactate Biosensors. Biosensors. 2022; 12(3):143. https://doi.org/10.3390/bios12030143
Chicago/Turabian StyleWellbourne-Wood, Joel, Marc Briquet, Maxime Alessandri, Francesca Binda, Maylis Touya, and Jean-Yves Chatton. 2022. "Evaluation of Hydroxycarboxylic Acid Receptor 1 (HCAR1) as a Building Block for Genetically Encoded Extracellular Lactate Biosensors" Biosensors 12, no. 3: 143. https://doi.org/10.3390/bios12030143
APA StyleWellbourne-Wood, J., Briquet, M., Alessandri, M., Binda, F., Touya, M., & Chatton, J.-Y. (2022). Evaluation of Hydroxycarboxylic Acid Receptor 1 (HCAR1) as a Building Block for Genetically Encoded Extracellular Lactate Biosensors. Biosensors, 12(3), 143. https://doi.org/10.3390/bios12030143





