A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia
Abstract
:1. Introduction
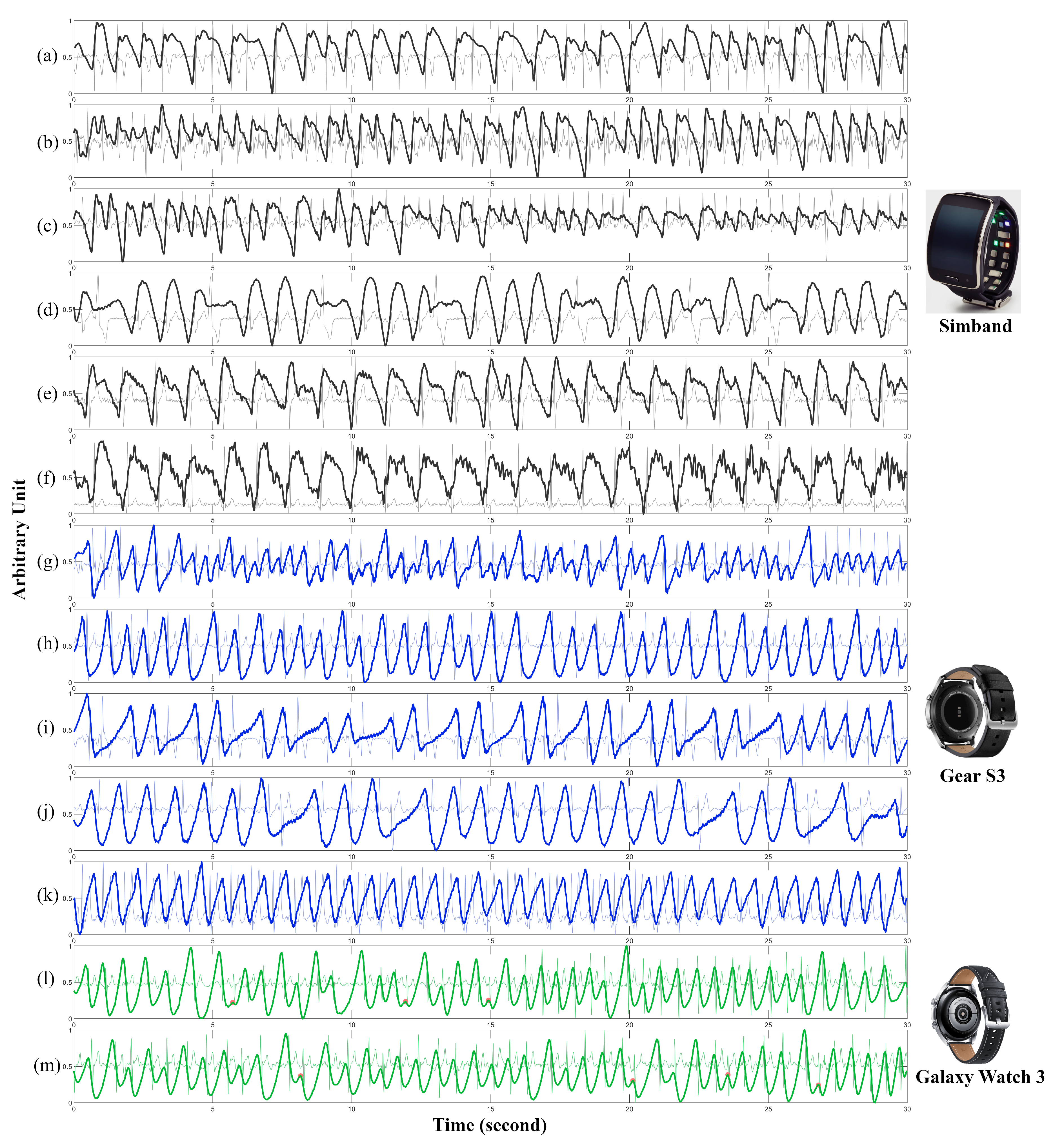
- Our proposed algorithms were tested on three different models of smartwatches: Samsung’s Simband, Gear S3, and Galaxy Watch 3. We trained our algorithms only on the PPG data recorded from Simbands, and independently tested them with large PPG datasets consisting of PPG signals from Gear S3 and Galaxy Watch 3 smartwatches. This is the main difference from our previous published conference proceedings in [14].
- Our proposed algorithms showed reliable results not only for NSR data, but also for various cardiac arrhythmias such as PAC/PVC, basal AF, and AF with RVR.
2. Materials and Methods
2.1. Dataset and Experiment Protocols
2.1.1. Peak Detection Strategy and Motivation
2.1.2. Training Dataset: UMMC Simband Dataset
2.1.3. Experiment Protocol for the Training Dataset
2.1.4. Testing Dataset: UMMC Pulsewatch Dataset (Samsung Gear S3 and Galaxy Watch 3)
2.1.5. Experiment Protocol for the Testing Dataset
2.2. Methods and Evaluation
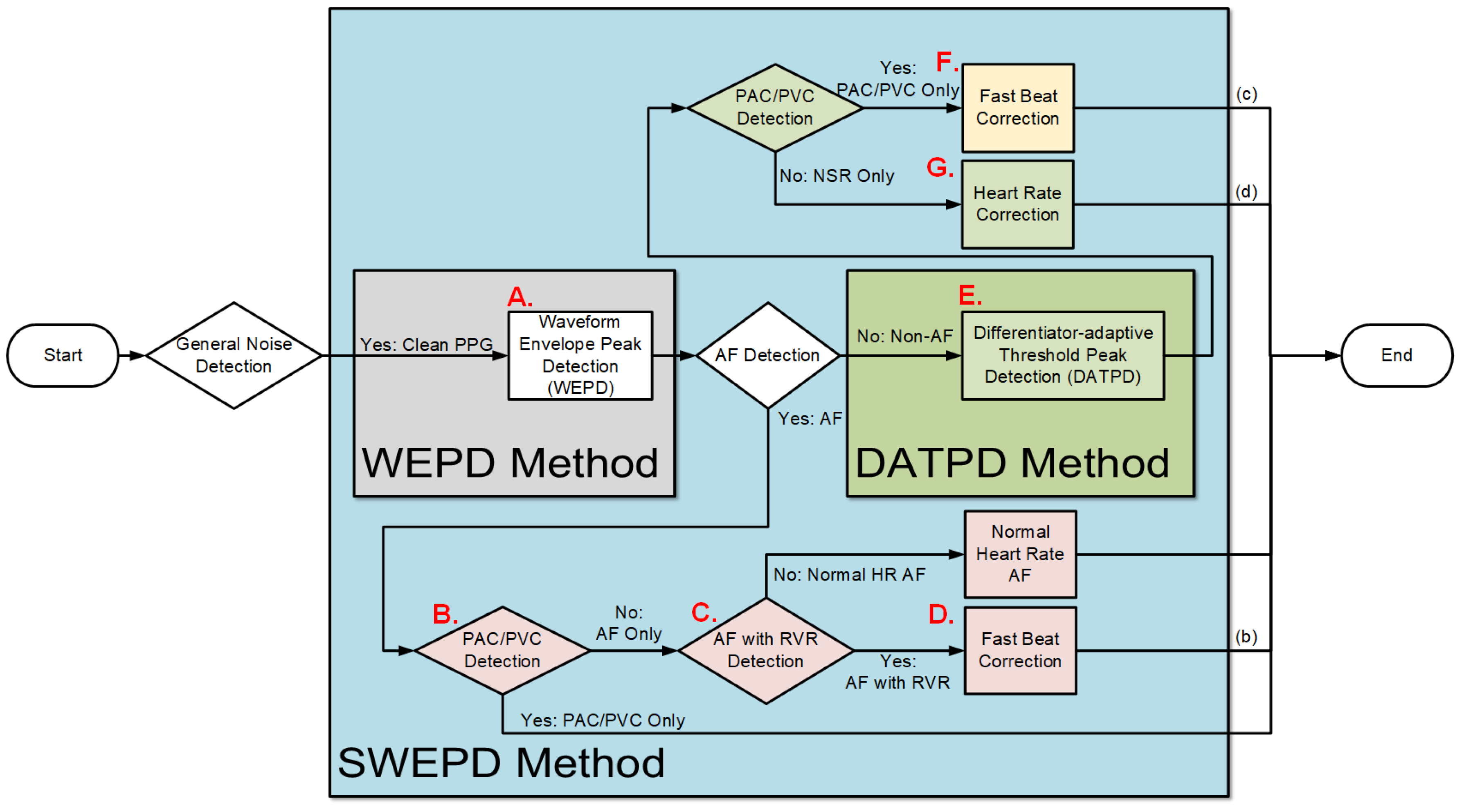
2.2.1. Overview of the Proposed (SWEPD) Method
2.2.2. Details of New WEPD Peak Detection Algorithm
- Moving Average Denoising and DifferentiationA bandpass filter with a passband of 0.5 Hz to 5 Hz is applied to each 30-s PPG data segment that has been declared to be relatively devoid of motion artifacts. We used a 5th order zero phase elliptic filter to design our bandpass filter. The design of this filter is based on the PPG pre-processing section mentioned in Chong et al. [6], which was optimized on smartphone PPG signals. The bandpass filter does not ensure smoothness of the signal, thus, a moving average filter with a proper sliding window is used to remove non-cardiac related oscillations and spikes which may have been caused by motion artifacts or poor signal quality data. This smoothing process is implemented three separate times:where with (round M to nearest integer if M is not an integer) for the first moving average process, and (round M to nearest integer if M is not an integer) for the remaining two times; denotes the raw signal after bandpass filtering; and is the sampling rate.Before the final smoothing, calculation of the first-order difference of the filtered signal is performed to accentuate small fluctuations in the peak plateau portion of the PPG waveform:
- Standardizing PPGWe subtract the mean of the signal from the above step, and then divide it by its standard deviation.This step is especially important to enable the signal’s waveform envelope calculation to remove false positive beats, which will be detailed in the subsequent section.
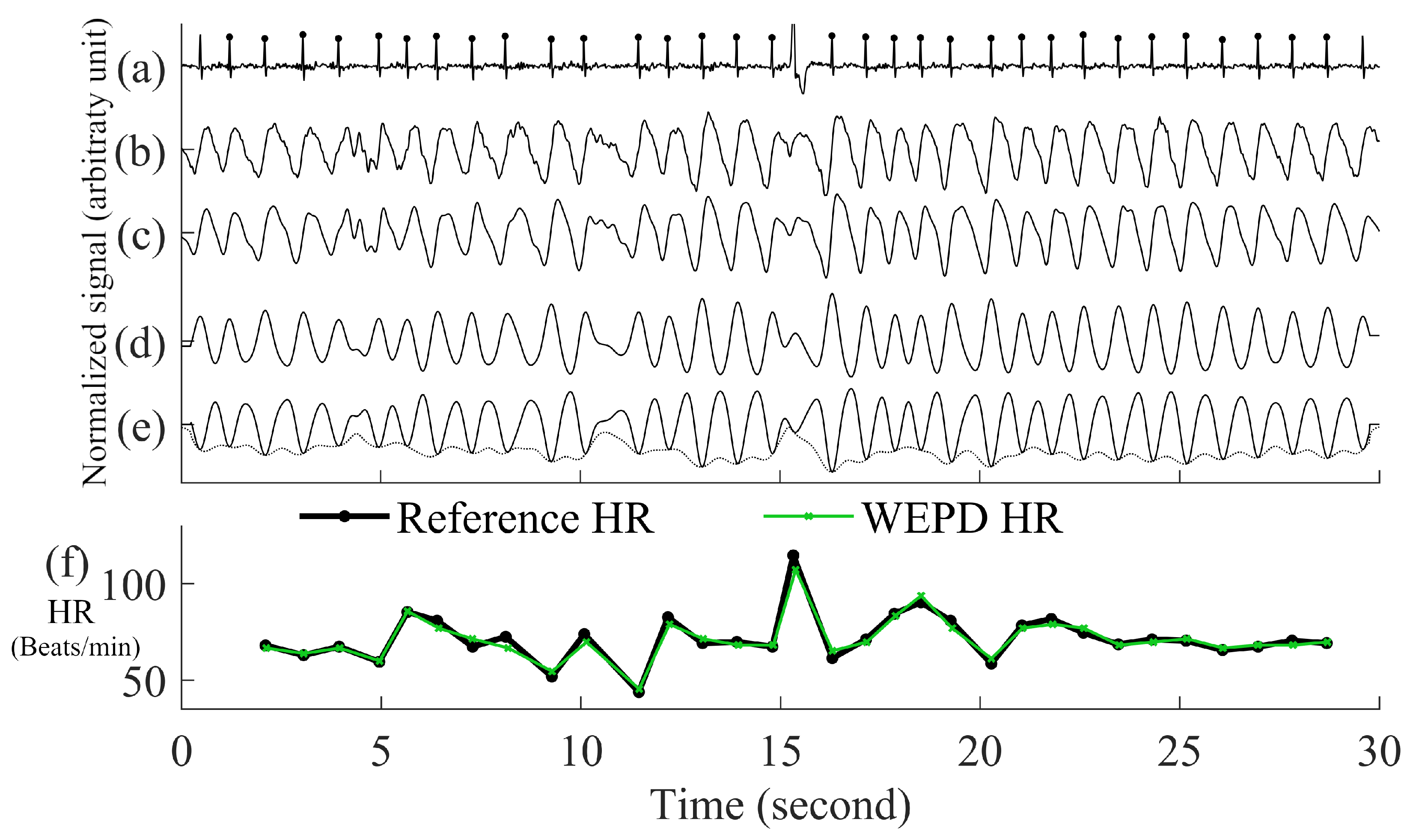
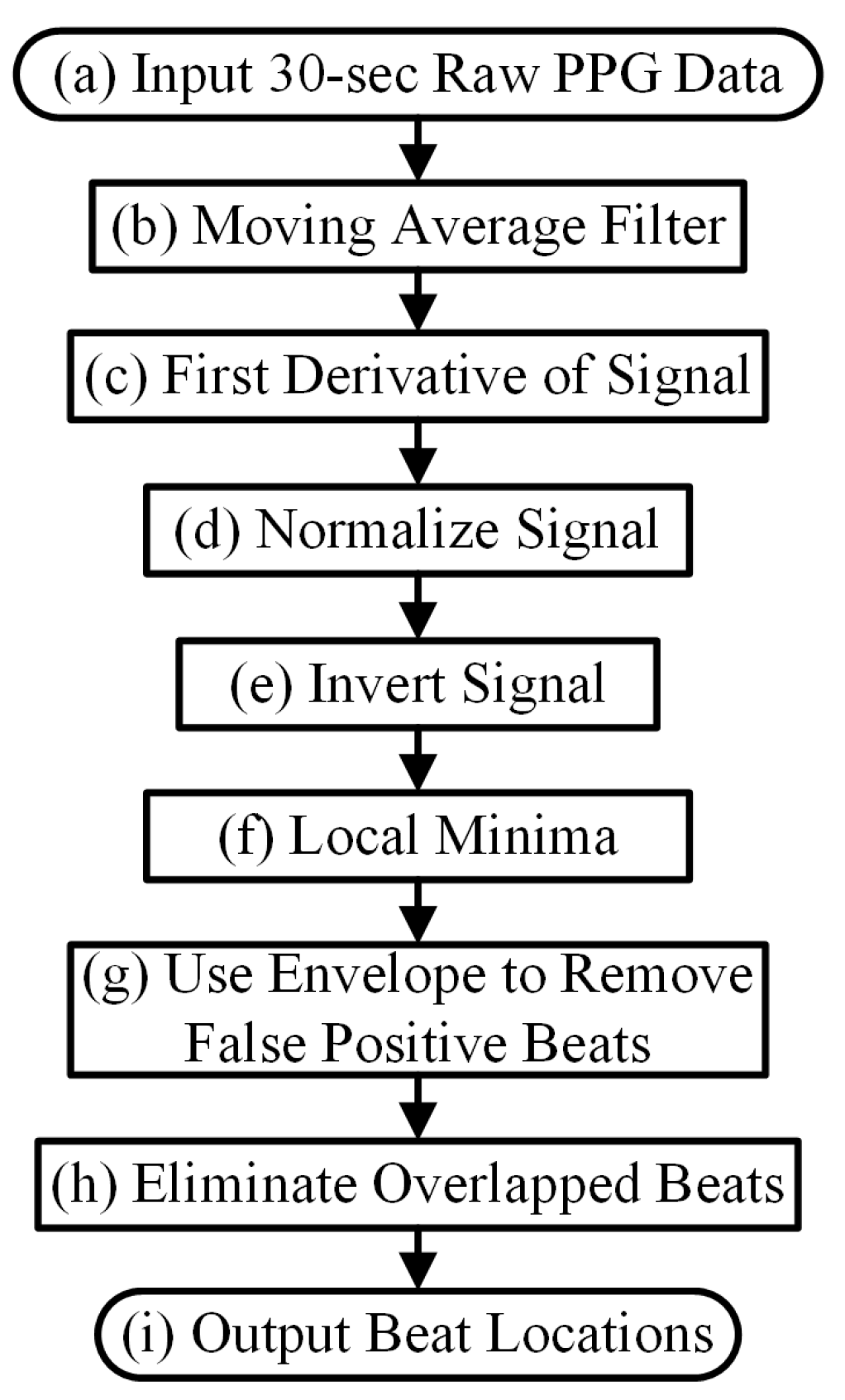
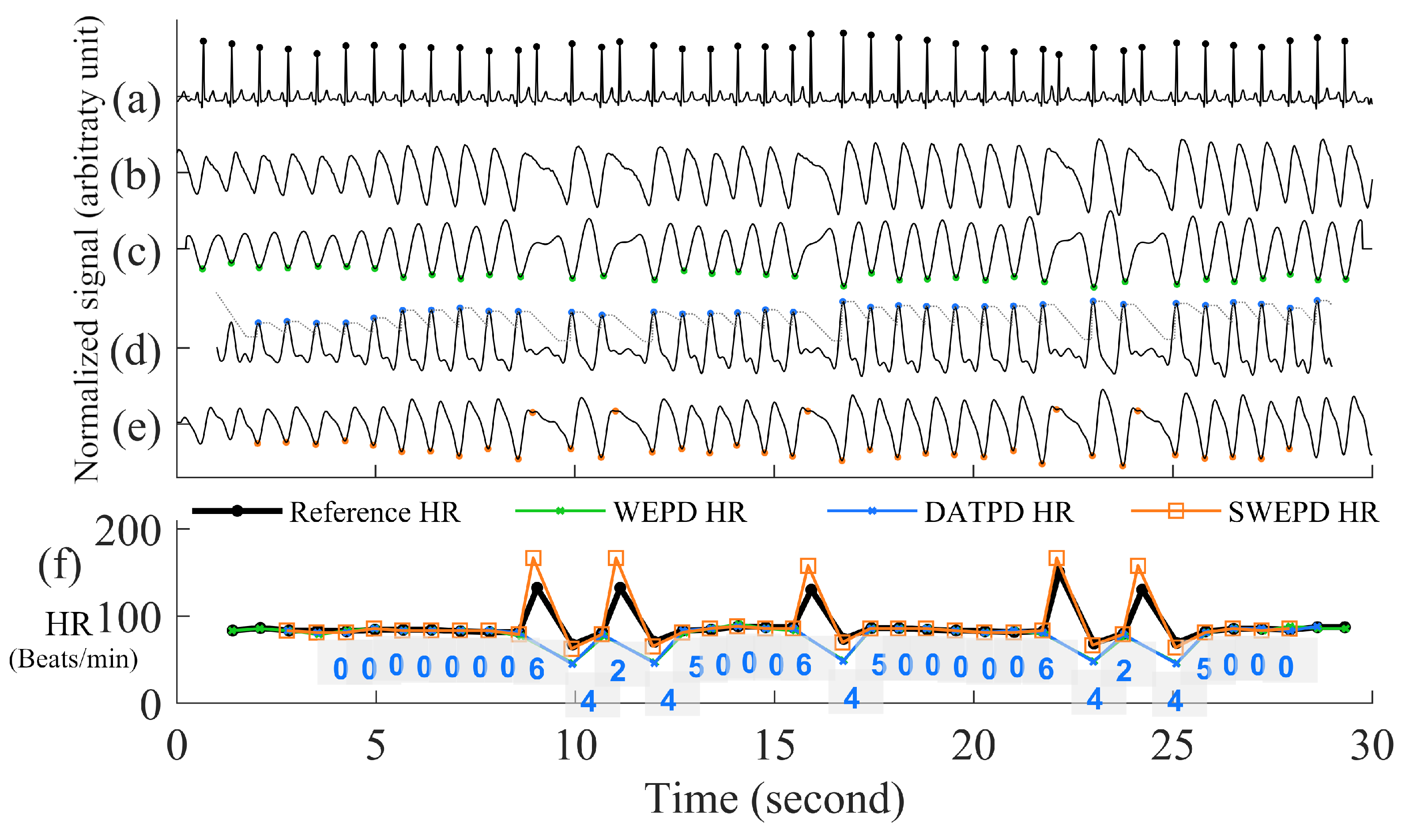
- Inverting the Signal and Performing Peak DetectionPeak detection is applied on either the PPG pulses’ top or bottom curve portion that has the sharpest peaks, to maximize the accuracy of heart rate estimation. To determine which side has the sharpest peaks (either top or bottom portion of a PPG segment), we first eliminate the side with detected number of peaks that are more than the fastest heart rate a human could have. If both top and bottom portions are relatively clean, we use the peaks from the side (top or bottom) with fewer beats. If both top and bottom portions have similar number of beats, we choose the side of the PPG has the sharper peaks by using finding the side has larger mean value of amplitude gradient around the peaks. If the top portion of the PPG signal is chosen, we flip the signal and estimate the local minima points for each beat. Example of inverting of PPG is in the panel (d) and (e) of Figure 3.Figure 3. Step by step procedures for the proposed WEPD algorithm on a basal heart rate AF signal. The top panel (a) is the raw ECG signal with the reference ECG beats; panel (b) is a raw PPG signal, panel (c) is the output of the bandpass filter; panel (d) is the output after moving average filtering and the first derivative operation; panel (e) is the output after signal standardization, inversion of the signal, and application of the envelope method for local minima detection. The bottom panel (f) is a comparison of the heart rate (unit: beats per minute, BPM) from the reference ECG signal vs. estimated heart rate from WEPD.Figure 3. Step by step procedures for the proposed WEPD algorithm on a basal heart rate AF signal. The top panel (a) is the raw ECG signal with the reference ECG beats; panel (b) is a raw PPG signal, panel (c) is the output of the bandpass filter; panel (d) is the output after moving average filtering and the first derivative operation; panel (e) is the output after signal standardization, inversion of the signal, and application of the envelope method for local minima detection. The bottom panel (f) is a comparison of the heart rate (unit: beats per minute, BPM) from the reference ECG signal vs. estimated heart rate from WEPD.
- Removing False Positive Peaks Using an EnvelopeFor high resolution PPG data from a commercial grade pulse oximeter and Simband, a dicrotic notch is prevalent in most subjects with normal sinus rhythm. Motion artifacts in the wrist can also create the appearance of a dicrotic notch. If a true or artifact dicrotic notch is not accounted for, our peak detection algorithm will detect it as a heartbeat. Consequently, this will lead to erroneously fast heart rate detection and depending on the rate of appearance of dicrotic notches, it can also lead to false detection of AF. Hence, to disregard any dicrotic notches, we estimate the envelope of the bottom portion of each beat of the PPG signal. There are two ways to calculate the envelope of the PPG signal. The first way is to interpolate the local minimas in the PPG signal with a cubic spline. This method has been used for respiratory rate detection with PPG signals [26], and it can accurately capture the general outlines of the PPG waveform. The second way is to use an N-tap Hilbert filter to obtain the upper envelope of the analytic signal; this method has been successfully used for abnormal heart sound analysis [27]. For the length of “N-tap” in the Hilbert filter, we found that it works the best on a smartwatch PPG signal with the value of 1.5 times the sampling frequency of PPG. This defined N-tap filter allows the envelope to account for dominant grooves of PPG waveforms. For each beat, the minimal amplitude that intersects between the PPG signal and two envelopes is considered as the new estimated peak.
- Eliminating Overlapping PeaksIf we obtain more than one intersection point between each beat of the PPG signal and its envelope, we retain the lowest amplitude peak and disregard the other candidate peaks. To ensure that we are not choosing two consecutive heartbeats (after going through steps (a)–(e) in Figure 3) that are too closely spaced, we stipulate that the time difference between two adjacent minima peaks has to be greater than 0.3 s, which is considered the refractory period of cardiac muscle for normal sinus rhythm [28]. A representative example of the PPG signal that has gone through the WEPD algorithm is shown in Figure 3. Note the accuracy (squared error of the mean HR: 0.21 BPM and the root mean squared error (RMSE) of beat-to-beat HR: 2.79 BPM) in the estimation of heart rates via the WEPD algorithm when compared to the reference ECG.
2.2.3. PAC/PVC Detection
2.2.4. New Peak Correction Method for AF Patients
- AF with RVR Detection for AF SubjectsNow we are in step C in Figure 1. The aim of this step is to detect AF with a rapid ventricular response, which will henceforth be called AF with RVR. Thus, we designed a supervised linear classifier to separate fast from normal heart rate AF. As this is one of the first studies to investigate AF with RVR in a smartwatch PPG, we define it based on the following criteria which all should be met:
- (a)
- A segment has to be an AF segment. In our algorithm, this segment has to be first detected as an AF segment from the AF detection algorithm [18] using the heart rates provided by the WEPD algorithm.
- (b)
- A heart rate that is greater than 140 BPM, based on our observation on the training data and [31,32,33]. For example, in [33] authors used 150 BPM as a threshold for defining faster heart rate among AF with RVR in the ICU; in [32] it was suggested that the upper limit is 130 BPM for target heart rate control among AF and heart failure patients during moderate exercise; in [31] author used 140 BPM as the minimum ventricular rate for marking patients with rapid ventricular rate in atrial fibrillation.
- (c)
- A heart rate that increases more than 40 BPM from one beat to the next. For example, ref. [34] suggested that the heart rate value cannot change more than 10 BPM from a previous value on normal sinus rhythm, and from our observation on our training data with PAC/PVC and AF with RVR subjects, we noticed that the heart rates for fast beats usually increased more than 20 BPM for the former, and more than 40 BPM for the latter.
- (d)
- The heart rate estimation from the WEPD is inaccurate for more than half the beats in each reference heart rate segment. WEPD is largely appropriate for determining irregularity of the rhythm, thus, accurate heart rate estimation requires SWEPD.
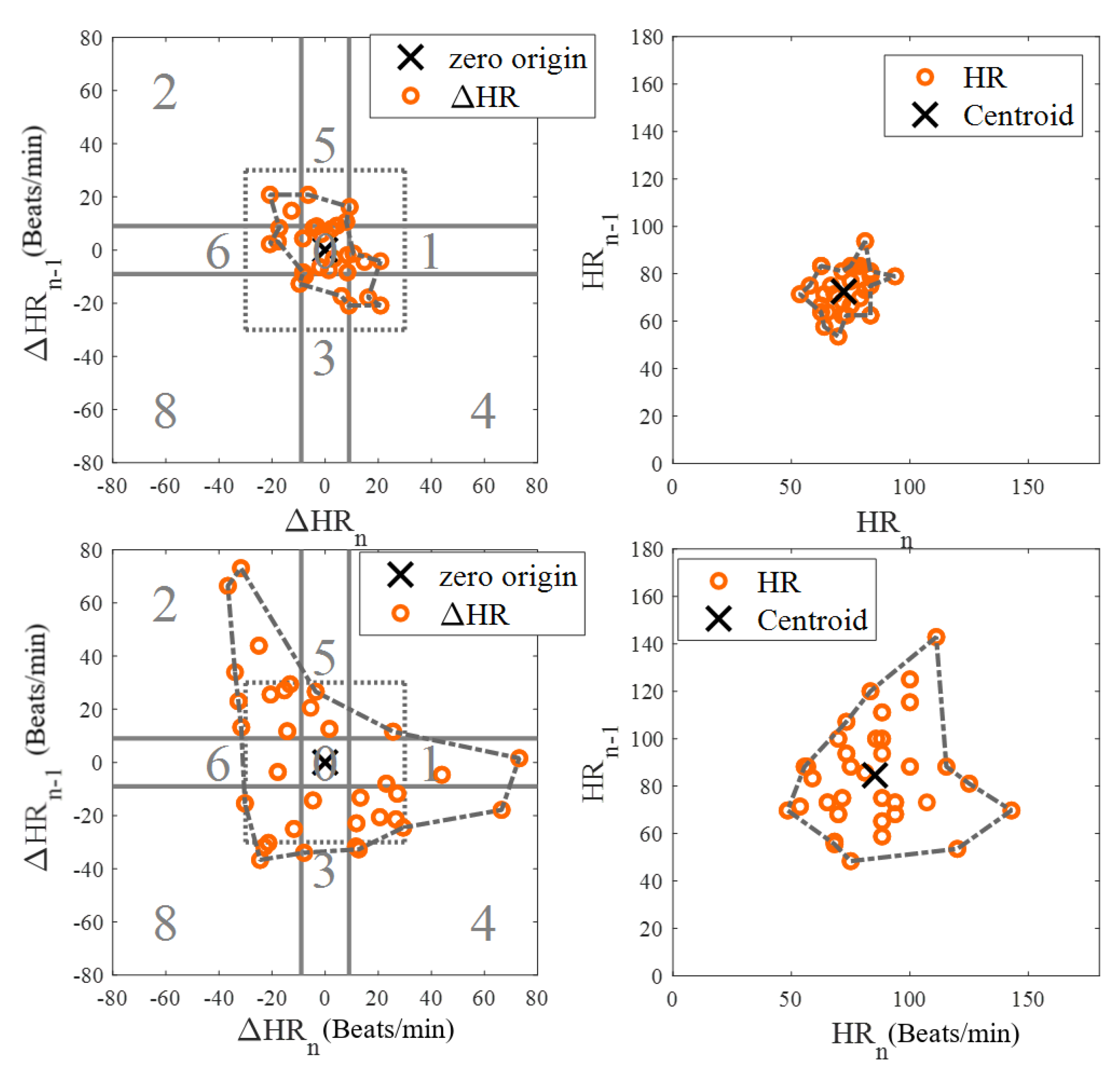
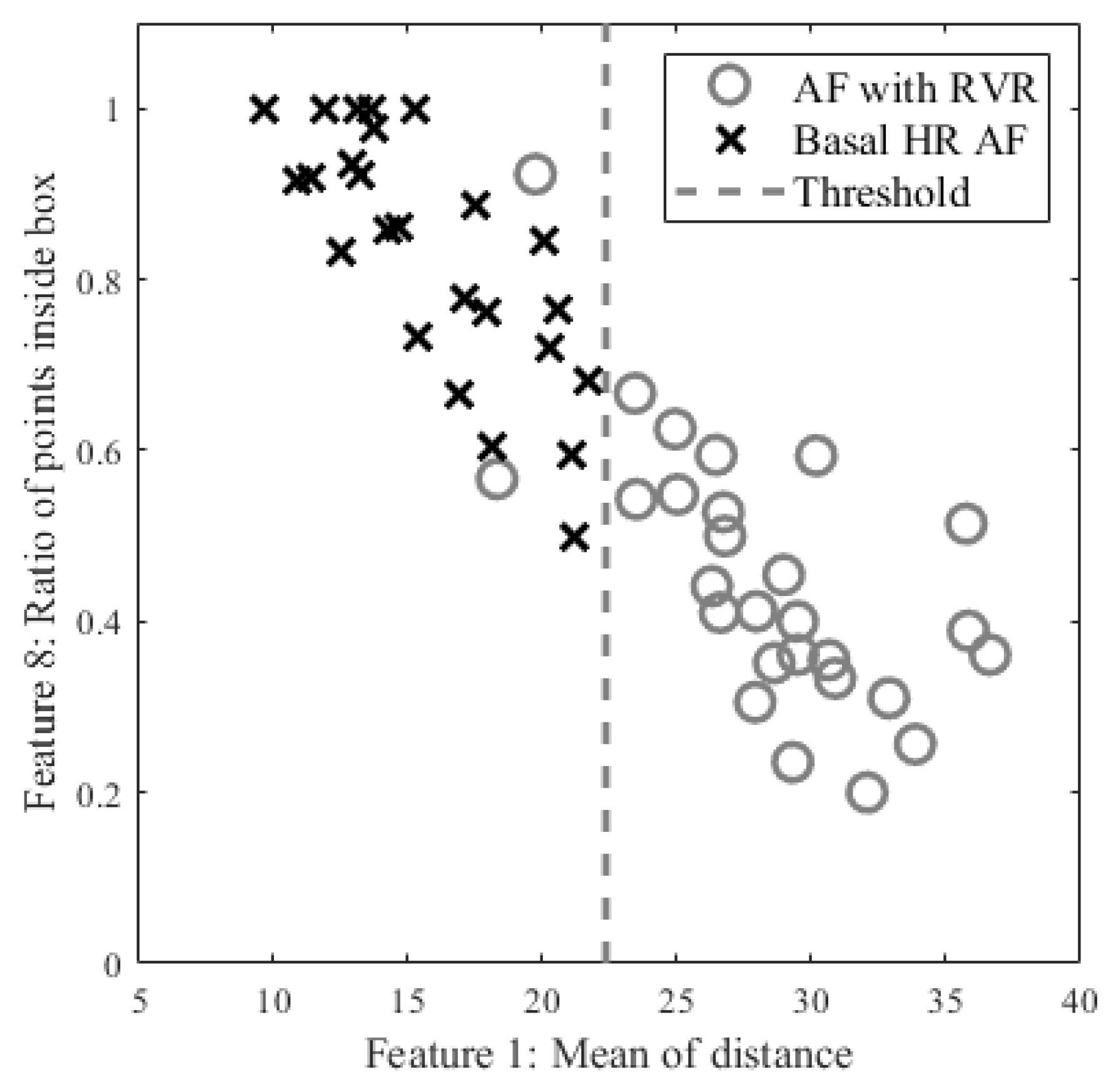
Among 10 AF subjects with AF detected in 53 segments, 25 were labelled as RVR heart rate and 28 as basal heart rate AF, based on the criteria defined above. All 53 segments were used for determining a threshold value for fast heart rate detection. The top and bottom panels of Figure 4 show Poincaré plots of basal heart rate AF and AF with RVR, respectively. As shown in Figure 4, the basal AF heart rates are more confined to the origin whereas the AF with RVR heart rates are more dispersed. To exploit this main characteristic, we selected nine candidate features to characterize the AF with RVR segments. One feature is the number of beats exceeding 140 BPM, five other features are derived from the Poincaré plot of the heart rate difference (HRn and HRn+1), and the remaining three features are derived from the Poincaré plot of heart rates (not difference of heart rates). The left panels of Figure 4 show Poincaré plots of the heart rate difference. The heart rate range for both X and Y axes are set to [−80, 80] BPM. Four of the five features derived from these plots are based on calculation of the distance of each point to the origin, from which we determine: the mean value of all distances, variance of all distances, boundary of the covered area of all points, and a ratio. This last is the ratio of points that have a distance to other points that is lower than 5 BPM to the number of combinations to calculate the distance between all of the points in the Poincaré plot. The fifth feature is the ratio of points that fall inside a confined square box (the dotted line square boxes in the left panels of Figure 4) to the entire number of points in the Poincaré plot; points outside of this dotted-line box indicate rapidly changing heart rates. To find the optimized limits of the confined box, we tested various heart rates ranging from [−10, 10] BPM to [−45, 45] BPM by increasing the X- and Y-axes by 5 BPM. The box’s confined limit of [−15, 15] BPM showed the best results to separate between the fast and normal heart rates of AF segments. The right panels of Figure 4 show the Poincaré plots without the difference of heart rates, which were derived from the WEPD approach. The X-axis represents the n+1 heart rate and Y-axis represents the current heart rate. The heart rate boundary for both axes was set to [0, 180] BPM. The centroid of each point in the Poincaré plot is calculated by averaging coordinates of all points. The distance between the centroid and all points was then calculated. Three features were calculated from this distance: the mean of distance, variance of distances, and the area in the boundary of all points as shown in the right panel of Figure 4. Of the nine features described above, we found that the mean value of all distances from the Poincaré plot of heart rate difference was the best in separating the basal from the RVR heart rate AF. Figure 5 shows the threshold value to separate RVR and basal heart rate, which was found to be 22.4 BPM.Figure 4. Poincaré plots for basal (top panels) and RVR (bottom panels) heart rate AF detection. The left panels are the Poincaré plot of heart rate difference, and the right panels are the Poincaré plot of heart rate. In the left panels, the entire Poincaré plot is separated into 9 regions, which are also used to locate erroneous peaks for peak correction during PAC/PVC detection.Figure 4. Poincaré plots for basal (top panels) and RVR (bottom panels) heart rate AF detection. The left panels are the Poincaré plot of heart rate difference, and the right panels are the Poincaré plot of heart rate. In the left panels, the entire Poincaré plot is separated into 9 regions, which are also used to locate erroneous peaks for peak correction during PAC/PVC detection.Figure 5. Scatter plot of candidate features for heart rate estimation among individuals with AF stratified by high vs. basal ventricular response. The X axis is the best feature value of mean of distance calculated from the Poincaré plot of heart rate difference; the Y axis is the second-best feature value of ratio of points inside a confined box from a Poincaré plot of heart rate.Figure 5. Scatter plot of candidate features for heart rate estimation among individuals with AF stratified by high vs. basal ventricular response. The X axis is the best feature value of mean of distance calculated from the Poincaré plot of heart rate difference; the Y axis is the second-best feature value of ratio of points inside a confined box from a Poincaré plot of heart rate. - Peak Correction for AF with RVR SegmentsIn the last section, we described how the AF with RVR segments can be discriminated from the basal HR AF data. Given this information, the next task is to search for additional peaks in the PPG signal to address the AF with RVR data segments. In the left panels of Figure 4, we section the entire Poincaré plot into nine regions, which represent all possible permutations of basal and RVR heart rate changes within the three consecutive heartbeats [6]. We set the size of region zero to [−9, 9] BPM (this represents the limits of the normal heart rates), and the other region boundaries can be inferred from Figure 4. Region two represents a true scenario where a fast heartbeat is surrounded by two slow heartbeats. This is a typical scenario for a RVR heart rate AF. This section is to describe how we look for fast heart rates in step D in Figure 1 based on the progression of the flow chart. As detailed in the preceding section, when a heartbeat is located in the other 8 regions (other than the second region), it is incorrectly detected as a basal heartbeat. Thus, the next task is to re-examine a filtered PPG signal between two detected beats to look for additional local minima. The last row of Figure 6 illustrates the above-described concept. The heart rates estimated from the WEPD method are shown in gray circles. For each of these beats, we also label the regions they are associated with. The dark circle lines are the reference heart rates. As shown, the heartbeats labelled region two coincide with the reference heart rates, as expected. However, heartbeats labelled with other regions (1,3,4,5,7,8) deviate significantly from the reference heartbeats, as expected. Hence, these beats are all candidates for having been incorrectly detected as normal heart rates that need to be corrected if they are actually fast heart rates.To account for heart rates mis-detected as normal, we modify the WEPD method’s procedures. The first step involves increasing the upper band of the bandpass filter from [0.5, 5] Hz to [0.5, 10] Hz. This procedure will lead to additional peaks in the PPG signal, if they exist. Additionally, we assume that the fastest heart rate of an AF subject should be less than 200 BPM (based on data collected from AF subjects in this study), and that the refractory period of cardiac muscle for AF should be faster than the normal refractory period of 0.3 s [28]. Given these assumptions, any additional peaks detected via the increased bandpass filter will not be considered as candidates if they are less than 0.3 s apart from the original peak detected via the first pass of the WEPD algorithm prior to AF detection, as noted in Figure 1. If there is a peak that is greater than 0.3 s apart, a local minima (assuming the signal is already inverted to look for the minima points) detection is applied. If there is no local minima point, the middle time point of the candidate interval is selected as a new fast heartbeat. If there are several local minima points that are greater than 0.3 s apart from the original peak, we use the first local minima location as the new fast heartbeat. The aforementioned additional fast beat locations are combined with the peaks detected from the first pass of the WEPD algorithm’s results. Rows (c–d) in Figure 6 show the additional peaks detected using the second pass of the WEPD method via the modified bandpass filter ([0.5, 10] Hz), and the resultant heart rates, respectively. As shown in row (e) of Figure 6, the modified heart rates labelled as the gray crossed line match the reference heart rates significantly better than the heart rates derived from the first pass of the WEPD algorithm, shown as the gray circled line.
2.2.5. Details of New Peak Detection for PAC/PVC Detection
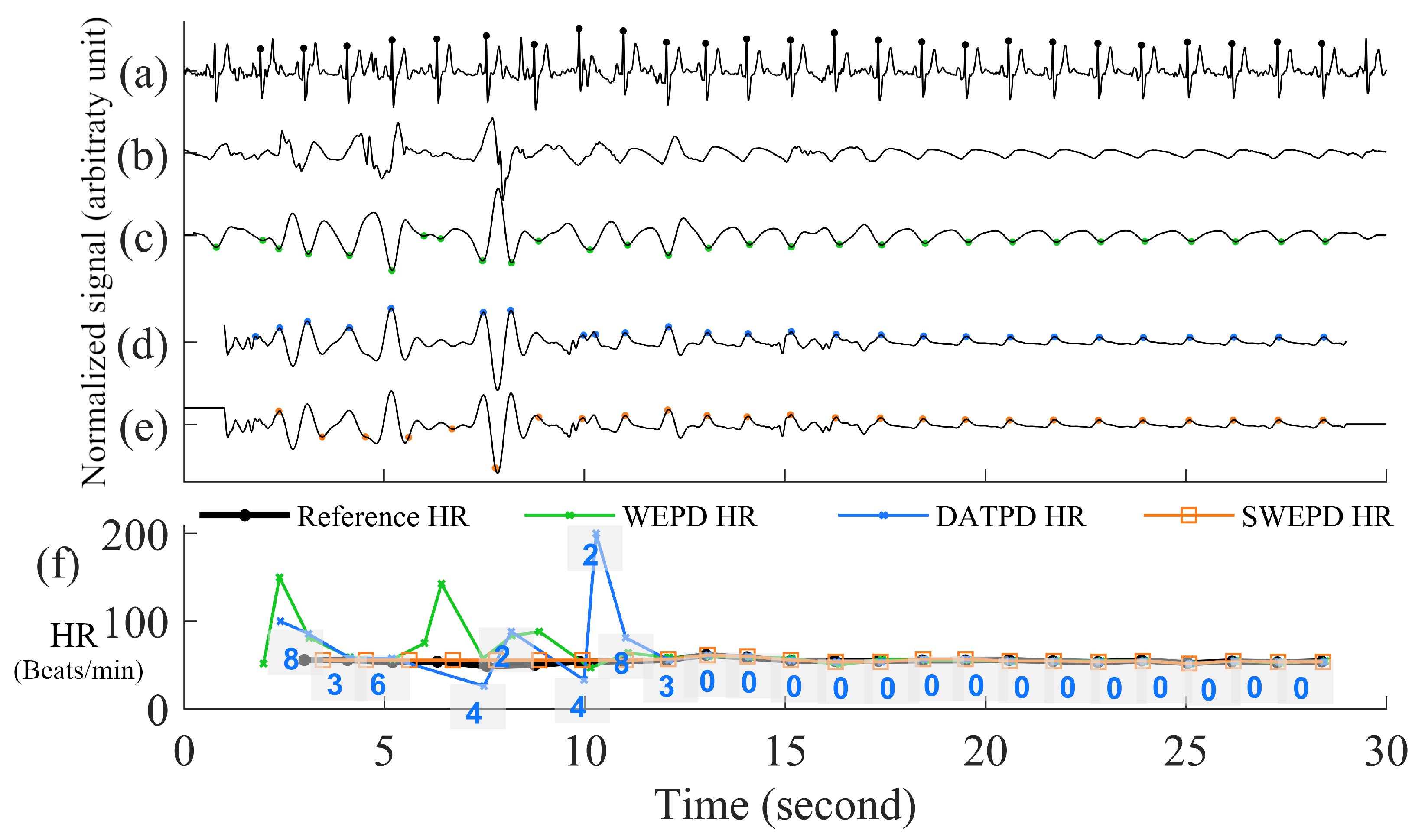
- The DATPD AlgorithmNow we are in the step E in Figure 1. This section describes an approach that better reflects heart rate patterns of NSR including PAC/PVC using a similar approach previously described [5]. The motivation for the approaches for better peak detection even with PAC/PVC or fast NSR beats is shown in Figure 7. The top row (panel (a)) shows an example of PVC beats. The panel (b) shows the peak detection estimated from the WEPD method, while the WEPD does well in detecting most of the PVCs, it incorrectly assigned two beats for the 4th PVC beat instead of a single beat. To mitigate this issue, even for PAC/PVC and the fast heart rate NSR beats, we describe in the next section various procedures to overcome low heart rate estimation in the event of fast heart rates.
- Reconstruction of Clean Signal Using VFCDMLázaro et al. used a FIR low-pass-differentiator filter with the pass band (0–7.8 Hz) [5] to filter the PPG signal. However, this linear filter was specifically designed for fingertip PPG signals. The motion artifact characteristics of a smartwatch PPG are often observed in the frequency band from 3.5 Hz to 7.8 Hz [35,36]. Moreover, the differentiator filter will accentuate the rapid changes in the PPG signal caused by motion artifacts, resulting in erroneous high amplitude peaks in the first derivative of the filtered signal. Consequently, an adaptive threshold method to detect peaks becomes more difficult. To overcome these issues, we illustrate our preferred approach.The first step involves the use of a time-frequency technique named Variable Frequency Complex Demodulation Method (VFCDM) [37]. The VFCDM technique has been described in detail and tested with different physiological signals [38]. Only the first two components are within a realistic heart rate range (e.g., 0.5–4 Hz). The higher components are beyond the heart rate frequency range, thus, are considered to represent higher frequency motion artifact dynamics. Thus, typically, a signal reconstruction is based on only the first two components. A representative signal reconstruction based on the first two components is shown in panel (d) of Figure 6.
- Adaptive Thresholding Peak Detection
2.2.6. New Peak Correction for Non-AF Windows
- Peak Correction for PAC/PVC SubjectsNow we are in the step F in Figure 1. The next step involves PAC/PVC detection so that their heartbeats are correctly accounted for. In the event of a mis-detected PAC/PVC beat, the next subsequent beat (let n = 1) will be located in the 4th region of the Poincaré plot of the heart rate difference. This is because for a fast PAC/PVC heartbeat the difference of the heart rate at n = 1 to the prior normal sinus heart rate at n = 0 will be a relatively large but negative value. In addition, the difference between the heart rate at n = 1 to that at the next normal sinus beat (n = 2) will be a large but positive value. Thus, the beat at n = 1 will be located in region 4. This is shown in panel (f) of Figure 7 where the location of the region is provided for each PPG beat. Note in this example that all beats immediately after PAC events are located in the 4th region. When the 4th region along with a HR < 60 BPM are detected, this indicates a missed PAC/PVC beat. We then rectify this problem by using the same approach as detailed in the section Peak Correction for Fast HR AF Segments. Specifically, we used the bandpass [0.5, 10] Hz filtered PPG data to look for the first minimum point in the time location where PAC/PVC occurred. This correction is shown in panel (f) of Figure 7 by the light gray line with “x” marks. With these correction procedures, the PPG-derived heart rates match well with the ECG-derived heart rate, as shown in panel (f) of Figure 7.
- Heart Rate Correction for NSR SubjectsFor those segments that are detected as NSR after PAC/PVC detection procedures, as shown in step G in the flowchart of Figure 1, we examine if there exist highly deviated heartbeats when they are compared to either the previous or future heartbeats. An example of this is provided in panel (e) of Figure 8. In the Poincaré plot, NSR heartbeats are mostly confined within the zeroth region as there is not much difference in heartbeats from one beat to the next. If there is any heartbeat outside the zeroth region, it is most likely due to a brief episode of motion artifact or bad signal-to-noise ratio, thus, these contaminated or erroneous heartbeats need to be corrected. To correct for the erroneous heartbeats, we take the average heart rate from two neighboring heartbeats of the mis-detected beats, that are located in the zeroth region. If there is no good heartbeat either before or after the noisy heartbeats, we use the mean heart rate of all beats that reside in the zeroth region. In the case where there is a large portion of incorrect beats (e.g., in Figure 8, starting from 2 s to 12 s), we calculate how many beats need to be inserted in the affected time segment. For example, the number of beats that need to be inserted is equal to mean heart rate (computed as beats/s) multiplied by the affected time segment.
2.2.7. Evaluation Methods
| Evaluation Method | Subject Type | SWEPD 1 | WEPD 2 | DATPD 3 | Method 1 4 | Method 2 5 | Method 3 6 | Method 4 7 | Method 5 8 | Method 6 9 | Method 7 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Average beat-to-beat RMSE (BPM/segment) (IBI in millisecond) | NSR | 2.21 (29.64) | 5.65 * (61.98 *) | 8.77 * (88.62 *) | 12.10 * (264.60 *) | 11.55 * (226.54 *) | 12.23 * (118.68 *) | 12.70 * (102.66 *) | 7.61 * (124.89 *) | 12.54 * (113.30 *) | 8.32 * (79.54 *) |
| PAC/PVC | 12.53 (97.85) | 10.48 * (135.28 *) | 21.02 * (300.94 *) | 27.34 * (526.13 *) | 28.30 * (530.20 *) | 15.19 (266.84 *) | 14.20 (269.08 *) | 16.34 (337.61 *) | 16.65 (285.17 *) | 15.48 (269.24) | |
| Basal AF | 7.69 (101.58) | 7.69 (101.58) | 22.11 * (276.09 *) | 22.67 * (397.77 *) | 23.91 * (426.28 *) | 22.55 * (208.66 *) | 23.97 * (214.24 *) | 14.31 * (245.26 *) | 24.04 * (217.47 *) | 12.34 (168.78) | |
| AF with RVR | 35.49 (164.28) | 34.31 * (289.99 *) | 50.52 * (480.23 *) | 53.95 * (547.95 *) | 61.52 * (713.14 *) | 40.25 * (295.21 *) | 41.18 * (296.12 *) | 47.27 * (440.27 *) | 43.96 * (335.49 *) | 43.33 * (327.31 *) | |
| RMSE of the mean HR (BPM/segment) (IBI in millisecond) | NSR | 0.46 (5.78) | 2.27 * (23.62 *) | 4.32 * (33.15 *) | 10.77 * (243.04 *) | 12.00 * (260.22 *) | 29.53 * (139.97 *) | 27.28 * (139.91 *) | 3.92 * (42.26 *) | 23.00 * (133.40 *) | 9.92 * (67.37 *) |
| PAC/PVC | 3.06 (23.30) | 8.63 * (94.90 *) | 10.33 * (142.96 *) | 20.17 * (332.85 *) | 26.31 * (400.47 *) | 7.77 * (91.67 *) | 7.77 * (90.33 *) | 10.37 * (205.62) | 8.43 * (109.92 *) | 8.51 * (92.24 *) | |
| Basal AF | 3.81 (44.18) | 3.81 (44.18) | 10.95 (97.57 *) | 11.25 * (197.11 *) | 14.81 * (242.83 *) | 40.03 * (178.34 *) | 33.83 * (179.33 *) | 5.82 (86.07) | 34.42 * (187.13 *) | 9.90 (73.45) | |
| AF with RVR | 16.90 (87.20) | 31.38 * (231.25 *) | 41.71 * (381.64 *) | 42.75 * (397.93 *) | 51.63 * (532.93 *) | 21.11 (162.94 *) | 21.63 (163.91 *) | 33.30 * (269.30 *) | 25.09 * (193.96 *) | 26.77 * (197.89 *) | |
| Extra Beats (Beats) | NSR | 50 | 85 * | 116 * | 205 * | 63 * | 677 * | 507 * | 172 * | 529 * | 211 * |
| PAC/PVC | 4 | 7 | 22 | 7 | 3 | 24 * | 16 | 4 | 24 * | 13 | |
| Basal AF | 9 | 9 | 55 * | 20 | 7 | 136 * | 134 * | 10 | 162 * | 30 * | |
| AF with RVR | 114 | 12 * | 14 * | 6 * | 1 * | 35 * | 44 * | 8 * | 23 * | 22 * | |
| Undetected Beats (Beats) | NSR | 18 | 30 * | 68 * | 697 * | 637 * | 195 * | 54 * | 168 * | 70 * | 61 * |
| PAC/PVC | 19 | 63 * | 126 * | 222 * | 267 * | 65 * | 63 * | 92 * | 74 * | 64 * | |
| Basal AF | 17 | 17 | 94 * | 128 * | 166 * | 11 | 15 | 50 * | 17 | 19 | |
| AF with RVR | 33 | 367 * | 442 * | 523 * | 617 * | 297 * | 310 * | 420 * | 342 * | 341 * |
| Evaluation Method | Subject Type | SWEPD 1 | WEPD 2 | DATPD 3 | Method 1 4 | Method 2 5 | Method 3 6 | Method 4 7 | Method 5 8 | Method 6 9 | Method 7 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Average beat-to-beat RMSE (BPM/segment) (IBI in millisecond) | NSR | 3.69 (41.16) | 5.56 (64.13 *) | 6.56 (94.08 *) | 10.72 * (242.75 *) | 10.94 * (240.94 *) | 5.96 (66.73 *) | 7 * (82.2 *) | 21.18 * (1027.26 *) | 44.3 * (605.89 *) | 7.07 * (78.37 *) |
| PAC/PVC | 8.2 (95.21) | 8.2 (106.29) | 17.25 (241.06) | 17.36 * (341.21 *) | 15.64 * (303.75 *) | 8.35 (105.38) | 10.11 (122.93) | 27.61 * (1869.61 *) | 45.11 * (702.63 *) | 9.04 (107.01) | |
| Basal AF | 12.38 (98.17) | 16.31 (148.87 *) | 29.4 * (303.03 *) | 40.63 * (571.8 *) | 32.83 * (503.88 *) | 14.74 (125.94) | 15.56 (134.85) | 30.27 * (665.65 *) | 49.31 * (960.74 *) | 14.06 (123.15) | |
| AF with RVR | 23.4 (152.5) | 28.45 * (212.72 *) | 41.48 * (368.73 *) | 53.86 * (640.06 *) | 45.43 * (502.85 *) | 26.71 (189.85 *) | 25.37 (180.24 *) | 42.11 * (501.88 *) | 51.25 * (793.82 *) | 23.06 (161.05) | |
| RMSE of the mean HR (BPM/segment) (IBI in millisecond) | NSR | 4.12 (25.54) | 6 * (62.2 *) | 5.22 * (53.63 *) | 12.58 * (335.74 *) | 12.24 * (295.5 *) | 7.45 * (67.99 *) | 8.56 * (70.39 *) | 10.43 * (788.84 *) | 58.6 * (629.45 *) | 8.58 * (67.08 *) |
| PAC/PVC | 7.1 (115.75) | 5.08 * (105.46 *) | 6.95 * (130.79 *) | 14 * (302.67 *) | 12.02 * (286.72 *) | 8.02 (110.21) | 9.03 * (113.28 *) | 15.4 * (1432.1 *) | 52.56 * (693.07 *) | 7.12 (104.77) | |
| Basal AF | 7.19 (50.76) | 7.79 * (78.36 *) | 13.82 * (143.3 *) | 30.24 * (448.42 *) | 22.4 * (515.48 *) | 8.12 * (64.65 *) | 7.52 * (65.07 *) | 16.69 * (462.89 *) | 35.16 * (836.51 *) | 8.15 (95.21) | |
| AF with RVR | 9.48 (58.79) | 12.06 * (92.88 *) | 27.88 * (241.39 *) | 43.81 * (499.38 *) | 33.84 * (356.92 *) | 13.23 (95.11 *) | 12.06 (93.82) | 27.47 * (297.5 *) | 23.15 * (316.2 *) | 8.06 (70.83) | |
| Extra Beats (Beats) | NSR | 27 | 234 * | 163 * | 110 * | 155 * | 345 * | 390 * | 947 * | 9620 * | 477 * |
| PAC/PVC | 192 | 93 * | 43 * | 27 * | 64 * | 109 * | 136 | 177 | 2936 * | 150 | |
| Basal AF | 604 | 427 | 2492 * | 120 * | 254 * | 465 * | 786 * | 363 * | 5478 * | 842 * | |
| AF with RVR | 43 | 33 | 99 * | 1 * | 25 | 32 | 94 * | 15 * | 432 * | 105 * | |
| Undetected Beats (Beats) | NSR | 132 | 387 * | 498 * | 2306 * | 2118 * | 402 * | 478 * | 5361 * | 3006 * | 444 * |
| PAC/PVC | 197 | 315 * | 506 * | 1290 * | 1057 * | 314 * | 370 * | 3074 * | 1572 * | 320 * | |
| Basal AF | 978 | 1763 * | 6467 * | 10103 * | 6802 * | 1482 * | 1692 * | 6512 * | 9296 * | 1521 * | |
| AF with RVR | 236 | 354 * | 864 * | 1213 * | 929 * | 322 | 371 * | 820 * | 946 * | 321 * |
3. Results
3.1. Results of AF with RVR Detection
3.2. Results of Proposed Peak Detection Method
3.3. Results of Proposed Peak Detection Method on Testing Dataset
4. Discussion
4.1. AF with RVR Detection Accuracy
4.2. Peak Detection Accuracy for the WEPD and SWEPD Approaches
4.3. Limitation of Small Sample Size
5. Conclusions
6. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PPG | photoplethysmogram |
| NSR | normal sinus rhythm |
| PAC | premature atrial contraction |
| PVC | premature ventricle contraction |
| AF | atrial fibrillation |
| RVR | rapid ventricular response |
| HR | heart rate |
| SNR | signal-to-noise ratio |
| WEPD | waveform envelope peak detection |
| DATPD | differentiator-adaptive threshold peak detection |
| SWEPD | the overall sequence using WEPD |
| UMMC | University of Massachusetts Medical Center |
| BPM | beats per minute |
| VFCDM | variable frequency complex demodulation method |
| RMSE | root mean squared error |
| IBI | interbeat interval |
Appendix A
| Index | UID | NSR | PAC/PVC | Basal AF | AF with RVR | Watch Model |
|---|---|---|---|---|---|---|
| 1 | 002 | 102 | 7 | 0 | 0 | Gear S3 |
| 2 | 005 | 107 | 194 | 0 | 0 | Gear S3 |
| 3 | 017 | 0 | 0 | 167 | 3 | Gear S3 |
| 4 | 029 | 31 | 3 | 0 | 0 | Gear S3 |
| 5 | 038 | 241 | 117 | 0 | 0 | Gear S3 |
| 6 | 045 | 232 | 4 | 0 | 0 | Gear S3 |
| 7 | 054 | 98 | 25 | 0 | 0 | Gear S3 |
| 8 | 301 | 0 | 0 | 17 | 0 | Gear S3 |
| 9 | 302 | 0 | 0 | 2 | 0 | Gear S3 |
| 10 | 305 | 0 | 0 | 10 | 0 | Gear S3 |
| 11 | 306 | 0 | 0 | 1 | 1 | Gear S3 |
| 12 | 307 | 0 | 0 | 39 | 0 | Gear S3 |
| 13 | 310 | 0 | 0 | 6 | 0 | Gear S3 |
| 14 | 311 | 0 | 0 | 19 | 0 | Gear S3 |
| 15 | 312 | 0 | 0 | 3 | 0 | Gear S3 |
| 16 | 318 | 0 | 0 | 0 | 8 | Gear S3 |
| 17 | 319 | 0 | 0 | 13 | 0 | Gear S3 |
| 18 | 320 | 0 | 0 | 8 | 0 | Gear S3 |
| 19 | 321 | 0 | 0 | 11 | 0 | Gear S3 |
| 20 | 322 | 0 | 0 | 7 | 0 | Gear S3 |
| 21 | 324 | 0 | 0 | 6 | 0 | Gear S3 |
| 22 | 325 | 13 | 0 | 0 | 0 | Gear S3 |
| 23 | 327 | 1 | 6 | 0 | 0 | Gear S3 |
| 24 | 329 | 0 | 0 | 10 | 5 | Gear S3 |
| 25 | 400 | 0 | 0 | 550 | 50 | Galaxy Watch 3 |
| Total | 824 | 352 | 869 | 67 |
References
- Elgendi, M.; Norton, I.; Brearley, M.; Abbott, D.; Schuurmans, D. Systolic peak detection in acceleration photoplethysmograms measured from emergency responders in tropical conditions. PLoS ONE 2013, 8, e76585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tison, G.H.; Sanchez, J.M.; Ballinger, B.; Singh, A.; Olgin, J.E.; Pletcher, M.J.; Vittinghoff, E.; Lee, E.S.; Fan, S.M.; Gladstone, R.A.; et al. Passive Detection of Atrial Fibrillation Using a Commercially Available Smartwatch. JAMA Cardiol. 2018, 3, 409–416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bumgarner, J.M.; Lambert, C.T.; Hussein, A.A.; Cantillon, D.J.; Baranowski, B.; Wolski, K.; Lindsay, B.D.; Wazni, O.M.; Tarakji, K.G. Smartwatch Algorithm for Automated Detection of Atrial Fibrillation. J. Am. Coll. Cardiol. 2018, 71, 2381–2388. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.S.; Lee, C.; Lee, M. Adaptive threshold method for the peak detection of photoplethysmographic waveform. Comput. Biol. Med. 2009, 39, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Lázaro, J.; Gil, E.; Vergara, J.M.; Laguna, P. Pulse Rate Variability Analysis for Discrimination of Sleep-Apnea-Related Decreases in the Amplitude Fluctuations of Pulse Photoplethysmographic Signal in Children. IEEE J. Biomed. Health Inform. 2014, 18, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.W.; Esa, N.; McManus, D.D.; Chon, K.H. Arrhythmia discrimination using a smart phone. IEEE J. Biomed. Health Inform. 2015, 19, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Harju, J.; Tarniceriu, A.; Parak, J.; Vehkaoja, A.; Yli-Hankala, A.; Korhonen, I. Monitoring of heart rate and inter-beat intervals with wrist plethysmography in patients with atrial fibrillation. Physiol. Meas. 2018, 39, 065007. [Google Scholar] [CrossRef]
- Lu, S.; Zhao, H.; Ju, K.; Shin, K.; Lee, M.; Shelley, K.; Chon, K.H. Can photoplethysmography variability serve as an alternative approach to obtain heart rate variability information? J. Clin. Monit. Comput. 2008, 22, 23–29. [Google Scholar] [CrossRef]
- Renevey, P.; Solà, J.; Theurillat, P.; Bertschi, M.; Krauss, J.; Andries, D.; Sartori, C. Validation of a wrist monitor for accurate estimation of RR intervals during sleep. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 5493–5496. [Google Scholar] [CrossRef]
- January, C.T.; Wann, L.S.; Alpert, J.S.; Calkins, H.; Cigarroa, J.E.; Cleveland, J.C.; Conti, J.B.; Ellinor, P.T.; Ezekowitz, M.D.; Field, M.E.; et al. 2014 AHA/ACC/HRS guideline for the management of patients with atrial fibrillation: Executive summary: A report of the American College of Cardiology/American Heart Association Task Force on practice guidelines and the Heart Rhythm Society. Circulation 2014, 130, 2071–2104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firoozabadi, R.; Helfenbein, E.D.; Babaeizadeh, S. Efficient noise-tolerant estimation of heart rate variability using single-channel photoplethysmography. J. Electrocardiol. 2017, 50, 841–846. [Google Scholar] [CrossRef]
- Firoozabadi, R.; Babaeizadeh, S. Artifact-Tolerant Pulse Rate Variability Measurement. U.S. Patent US20210113104A1, 22 April 2021. [Google Scholar]
- Kwon, S.; Hong, J.; Choi, E.K.; Lee, E.; Hostallero, D.E.; Kang, W.J.; Lee, B.; Jeong, E.R.; Koo, B.K.; Oh, S.; et al. Deep Learning Approaches to Detect Atrial Fibrillation Using Photoplethysmographic Signals: Algorithms Development Study. JMIR mHealth uHealth 2019, 7, e12770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, D.; Bashar, S.K.; Lazaro, J.; Ding, E.; Whitcomb, C.; McManus, D.D.; Chon, K.H. Smartwatch PPG Peak Detection Method for Sinus Rhythm and Cardiac Arrhythmia. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 4310–4313. [Google Scholar] [CrossRef]
- Biswas, D.; Everson, L.; Liu, M.; Panwar, M.; Verhoef, B.E.; Patki, S.; Kim, C.H.; Acharyya, A.; Van Hoof, C.; Konijnenburg, M.; et al. CorNET: Deep Learning Framework for PPG-Based Heart Rate Estimation and Biometric Identification in Ambulant Environment. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Ko, H.; Lee, H.; Lee, J. Deep Learning for Heart Rate Estimation From Reflectance Photoplethysmography with Acceleration Power Spectrum and Acceleration Intensity. IEEE Access 2020, 8, 63390–63402. [Google Scholar] [CrossRef]
- Reiss, A.; Indlekofer, I.; Schmidt, P.; Van Laerhoven, K. Deep PPG: Large-Scale Heart Rate Estimation with Convolutional Neural Networks. Sensors 2019, 19, 3079. [Google Scholar] [CrossRef] [Green Version]
- Bashar, S.K.; Han, D.; Hajeb-Mohammadalipour, S.; Ding, E.; Whitcomb, C.; McManus, D.D.; Chon, K.H. Atrial Fibrillation Detection from Wrist Photoplethysmography Signals Using Smartwatches. Sci. Rep. 2019, 9, 15054. [Google Scholar] [CrossRef]
- Ding, E.Y.; Han, D.; Whitcomb, C.; Bashar, S.K.; Adaramola, O.; Soni, A.; Saczynski, J.; Fitzgibbons, T.P.; Moonis, M.; Lubitz, S.A.; et al. Accuracy and Usability of a Novel Algorithm for Detection of Irregular Pulse Using a Smartwatch Among Older Adults: Observational Study. JMIR Cardio 2019, 3, e13850. [Google Scholar] [CrossRef]
- Simsense: 2nd Generation-Simband. Available online: https://www.simband.io/documentation/sensor-module-documentation/simsense/simsense-2nd-gen.html#ppg (accessed on 30 June 2021).
- Abboud, S.; Sadeh, D. The Use of Cross-Correlation Function for the Alignment of ECG Waveforms and Rejection of Extrasystoles. Comput. Biomed. Res. 1984, 17, 258–266. [Google Scholar] [CrossRef]
- Hossain, M.B.; Bashar, S.K.; Walkey, A.J.; McManus, D.D.; Chon, K.H. An Accurate QRS Complex and P Wave Detection in ECG Signals Using Complete Ensemble Empirical Mode Decomposition with Adaptive Noise Approach. IEEE Access 2019, 7, 128869–128880. [Google Scholar] [CrossRef]
- Dickson, E.L.; Ding, E.Y.; Saczynski, J.S.; Han, D.; Moonis, M.; Fitzgibbons, T.P.; Barton, B.; Chon, K.; McManus, D.D. Smartwatch Monitoring for Atrial Fibrillation After Stroke –The Pulsewatch Study: Protocol for a Multi-Phase Randomized Controlled Trial. Cardiovasc. Digit. Health J. 2021, 2, 231–241. [Google Scholar] [CrossRef]
- Kotzen, K.; Charlton, P.H.; Landesberg, A.; Behar, J. Benchmarking Photoplethysmography Peak Detection Algorithms Using the Electrocardiogram Signal as a Reference. In Proceedings of the Computing in Cardiology 2021, Brno, Czech Republic, 13–15 September 2021. [Google Scholar]
- Han, D.; Bashar, S.K.; Mohagheghian, F.; Ding, E.; Whitcomb, C.; McManus, D.D.; Chon, K.H. Premature Atrial and Ventricular Contraction Detection Using Photoplethysmographic Data from a Smartwatch. Sensors 2020, 20, 5683. [Google Scholar] [CrossRef]
- Kuwalek, P.; Burlaga, B.; Jesko, W.; Konieczka, P. Research on Methods for Detecting Respiratory Rate from Photoplethysmographic Signal. Biomed. Signal Process. Control 2021, 66, 102483. [Google Scholar] [CrossRef]
- Milani, M.; Abas, P.E.; De Silva, L.C. Identification of Normal and Abnormal Heart Sounds by Prominent Peak Analysis. In Proceedings of the 2019 International Symposium on Signal Processing Systems, Beijing, China, 20–22 September 2019; Association for Computing Machinery: New York, NY, USA, 2019; pp. 31–35. [Google Scholar] [CrossRef]
- Strobel, R.E.; Fisher, J.D.; Katz, G.; Kim, S.G.; Mercando, A.D. Time dependence of ventricular refractory periods: Implications for electrophysiologic protocols. J. Am. Coll. Cardiol. 1990, 15, 402–411. [Google Scholar] [CrossRef]
- Lee, J.; Reyes, B.A.; McManus, D.D.; Maitas, O.; Mathias, O.; Chon, K.H. Atrial fibrillation detection using an iPhone 4S. IEEE Trans.-Bio-Med. Eng. 2013, 60, 203–206. [Google Scholar] [CrossRef] [PubMed]
- McManus, D.D.; Lee, J.; Maitas, O.; Esa, N.; Pidikiti, R.; Carlucci, A.; Harrington, J.; Mick, E.; Chon, K.H. A novel application for the detection of an irregular pulse using an iPhone 4S in patients with atrial fibrillation. Heart Rhythm 2013, 10, 315–319. [Google Scholar] [CrossRef] [Green Version]
- Rawles, J.M. What Is Meant by a “Controlled” Ventricular Rate in Atrial Fibrillation? Heart 1990, 63, 157–161. [Google Scholar] [CrossRef] [Green Version]
- Van Gelder, I.C.; Rienstra, M.; Crijns, H.J.G.M.; Olshansky, B. Rate Control in Atrial Fibrillation. Lancet 2016, 388, 818–828. [Google Scholar] [CrossRef]
- Bosch, N.A.; Cimini, J.; Walkey, A.J. Atrial Fibrillation in the ICU. Chest 2018, 154, 1424–1434. [Google Scholar] [CrossRef]
- Salehizadeh, S.M.A.; Dao, D.; Bolkhovsky, J.; Cho, C.; Mendelson, Y.; Chon, K.H. A Novel Time-Varying Spectral Filtering Algorithm for Reconstruction of Motion Artifact Corrupted Heart Rate Signals During Intense Physical Activities Using a Wearable Photoplethysmogram Sensor. Sensors 2015, 16, 10. [Google Scholar] [CrossRef] [Green Version]
- Yu, W.C.; Chen, S.A.; Lee, S.H.; Tai, C.T.; Feng, A.N.; Kuo, B.I.T.; Ding, Y.A.; Chang, M.S. Tachycardia-Induced Change of Atrial Refractory Period in Humans. Circulation 1998, 97, 2331–2337. [Google Scholar] [CrossRef] [Green Version]
- Dao, D.; Salehizadeh, S.M.A.; Noh, Y.; Chong, J.W.; Cho, C.H.; McManus, D.; Darling, C.E.; Mendelson, Y.; Chon, K.H. A Robust Motion Artifact Detection Algorithm for Accurate Detection of Heart Rates From Photoplethysmographic Signals Using Time–Frequency Spectral Features. IEEE J. Biomed. Health Inform. 2017, 21, 1242–1253. [Google Scholar] [CrossRef]
- Chon, K.H.; Dash, S.; Ju, K. Estimation of respiratory rate from photoplethysmogram data using time-frequency spectral estimation. IEEE Trans.-Bio-Med. Eng. 2009, 56, 2054–2063. [Google Scholar] [CrossRef] [PubMed]
- Posada-Quintero, H.F.; Florian, J.P.; Orjuela-Cañón, A.D.; Chon, K.H. Highly sensitive index of sympathetic activity based on time-frequency spectral analysis of electrodermal activity. Am. J.-Physiol.-Regul. Integr. Comp. Physiol. 2016, 311, R582–R591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidaurre, C.; Sander, T.H.; Schlögl, A. BioSig: The free and open source software library for biomedical signal processing. Comput. Intell. Neurosci. 2011, 2011, 935364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, B.L.; Liu, X.; McDuff, D. Beat-To-Beat Cardiac Pulse Rate Measurement From Video. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 2739–2742. [Google Scholar]
- Hai, J.; Chan, P.H.; Tse, H.F.; Siu, C.W. Slow Heart Rate Predicts New Occurrence of Atrial Fibrillation. Hear. Lung Circ. 2015, 24, 1087–1093. [Google Scholar] [CrossRef]
- Li, B.N.; Dong, M.C.; Vai, M.I. On an automatic delineator for arterial blood pressure waveforms. Biomed. Signal Process. Control 2010, 5, 76–81. [Google Scholar] [CrossRef]
- Zong, W.; Heldt, T.; Moody, G.; Mark, R. An open-source algorithm to detect onset of arterial blood pressure pulses. In Computers in Cardiology; IEEE: Thessaloniki, Greece, 2003; Volume 1, pp. 259–262. ISSN 0276-6547. [Google Scholar] [CrossRef] [Green Version]
- Argüello Prada, E.J.; Serna Maldonado, R.D. A Novel and Low-Complexity Peak Detection Algorithm for Heart Rate Estimation from Low-Amplitude Photoplethysmographic (PPG) Signals. J. Med. Eng. Technol. 2018, 42, 569–577. [Google Scholar] [CrossRef]
- Parak, J.; Tarniceriu, A.; Renevey, P.; Bertschi, M.; Delgado-Gonzalo, R.; Korhonen, I. Evaluation of the beat-to-beat detection accuracy of PulseOn wearable optical heart rate monitor. In Proceedings of the 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Milan, Italy, 25–29 August 2015; Volume 2015, pp. 8099–8102. [Google Scholar] [CrossRef]
- Camm, A.J.; Malik, M.; Bigger, J.T.; Breithardt, G.; Cerutti, S.; Cohen, R.J.; Coumel, P.; Fallen, E.L.; Kennedy, H.L.; Kleiger, R.E.; et al. Heart rate variability: Standards of measurement, physiological interpretation and clinical use. Task Force of the European Society of Cardiology and the North American Society of Pacing and Electrophysiology. Circulation 1996, 93, 1043–1065. [Google Scholar]
- Peak Detection Using MATLAB (Non-Derivative Local Extremum, Maximum, Minimum). Available online: http://billauer.co.il/peakdet.html (accessed on 30 June 2021).

| Subject Type | ||||
|---|---|---|---|---|
| NSR | PAC/PVC | Basal AF | AF with RVR | |
| Subjects | 8 | 7 | 16 | 5 |
| 30-s segments | 824 | 352 | 869 | 67 |
| Reference ECG Beats | 22,597 | 9011 | 30,117 | 2909 |
| Subject Type | ||||
|---|---|---|---|---|
| NSR | PAC/PVC | Basal AF | AF with RVR | |
| Subjects | 23 | 5 | 5 | 4 |
| 30-s segments | 190 | 28 | 25 | 28 |
| Reference ECG Beats | 5740 | 897 | 818 | 1334 |
| Actual Class (30-s Segments) | |||
|---|---|---|---|
| Basal AF | AF with RVR | ||
| Predicted Class | Basal AF | 25 | 2 |
| AF with RVR | 0 | 26 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, D.; Bashar, S.K.; Lázaro, J.; Mohagheghian, F.; Peitzsch, A.; Nishita, N.; Ding, E.; Dickson, E.L.; DiMezza, D.; Scott, J.; et al. A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia. Biosensors 2022, 12, 82. https://doi.org/10.3390/bios12020082
Han D, Bashar SK, Lázaro J, Mohagheghian F, Peitzsch A, Nishita N, Ding E, Dickson EL, DiMezza D, Scott J, et al. A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia. Biosensors. 2022; 12(2):82. https://doi.org/10.3390/bios12020082
Chicago/Turabian StyleHan, Dong, Syed Khairul Bashar, Jesús Lázaro, Fahimeh Mohagheghian, Andrew Peitzsch, Nishat Nishita, Eric Ding, Emily L. Dickson, Danielle DiMezza, Jessica Scott, and et al. 2022. "A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia" Biosensors 12, no. 2: 82. https://doi.org/10.3390/bios12020082
APA StyleHan, D., Bashar, S. K., Lázaro, J., Mohagheghian, F., Peitzsch, A., Nishita, N., Ding, E., Dickson, E. L., DiMezza, D., Scott, J., Whitcomb, C., Fitzgibbons, T. P., McManus, D. D., & Chon, K. H. (2022). A Real-Time PPG Peak Detection Method for Accurate Determination of Heart Rate during Sinus Rhythm and Cardiac Arrhythmia. Biosensors, 12(2), 82. https://doi.org/10.3390/bios12020082