A Label-Free Electrical Impedance Spectroscopy for Detection of Clusters of Extracellular Vesicles Based on Their Unique Dielectric Properties
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Nanovesicles
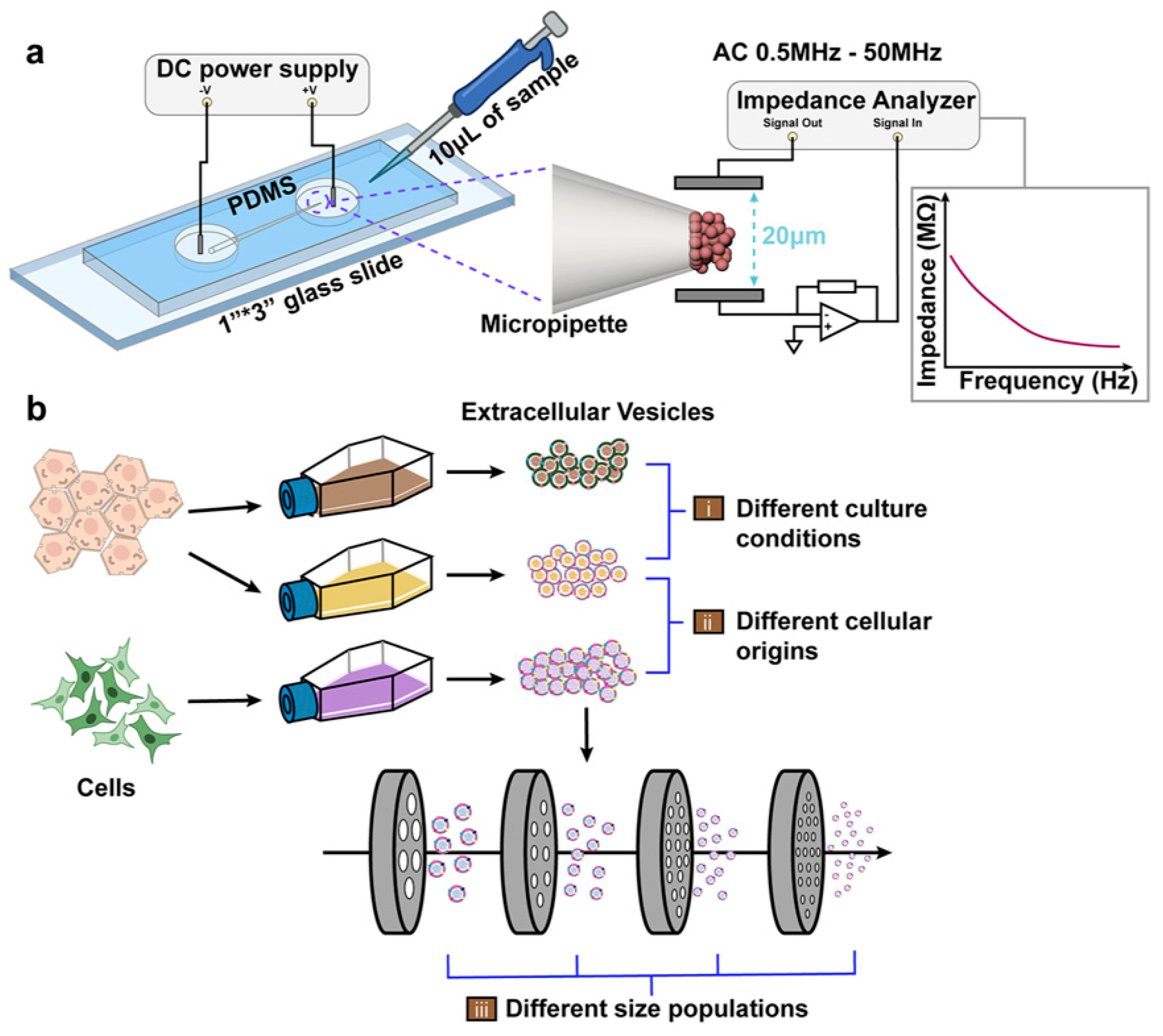
2.3. Device Assembly and Electrical Impedance Measurements
3. Results and Discussion
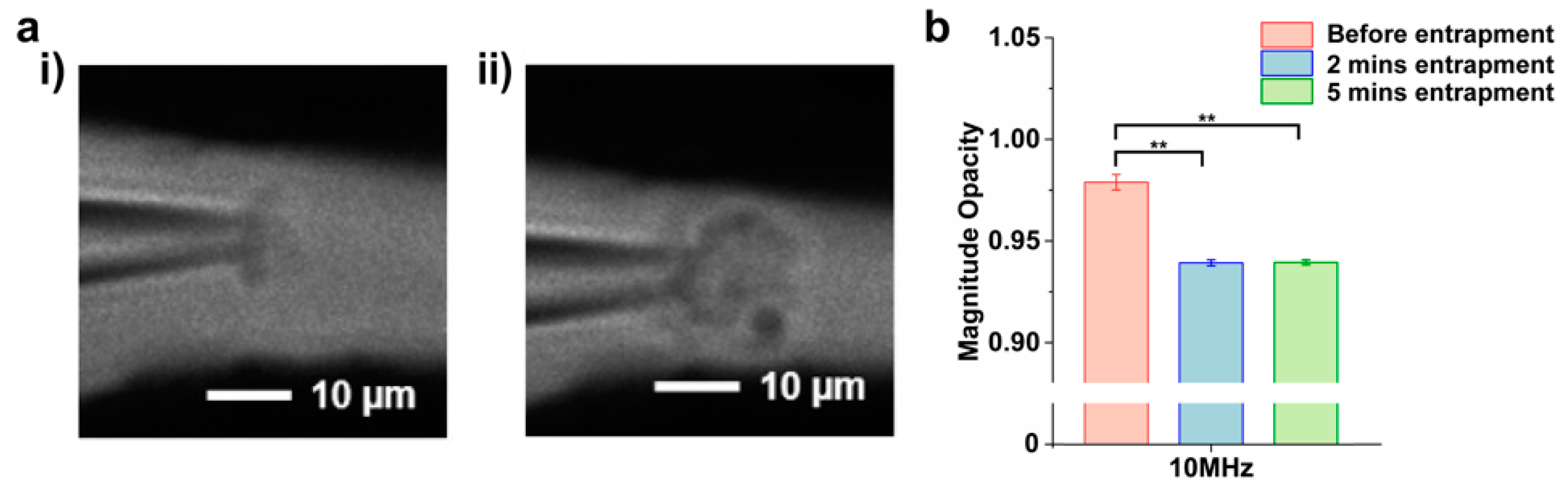
3.1. Magnitude Opacity
3.2. Detection of Nanoparticles with Known Dielectric Properties
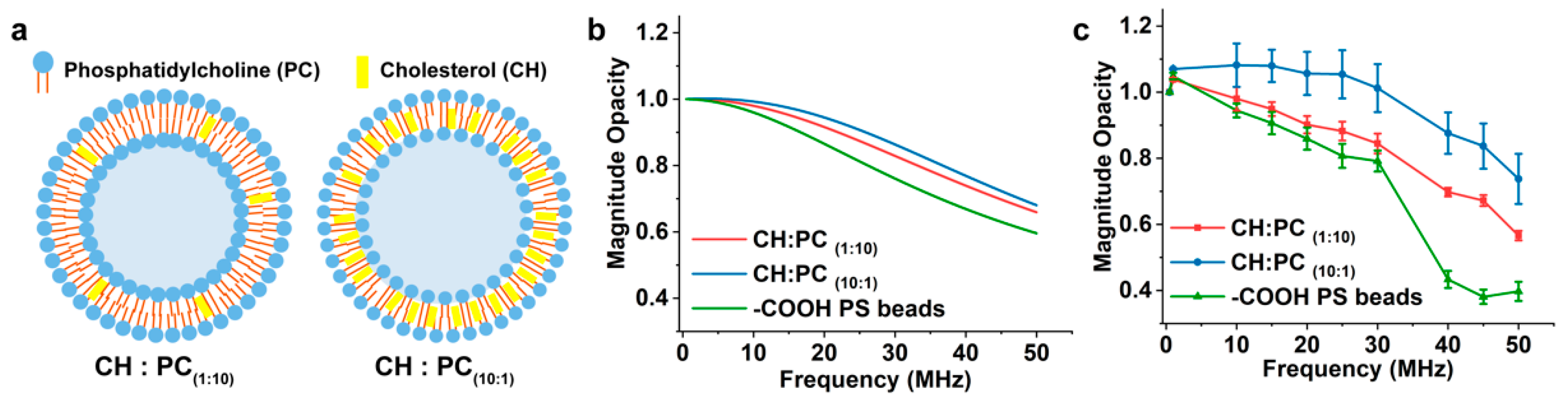
3.3. Detection of EVs with Different Membrane Compositions
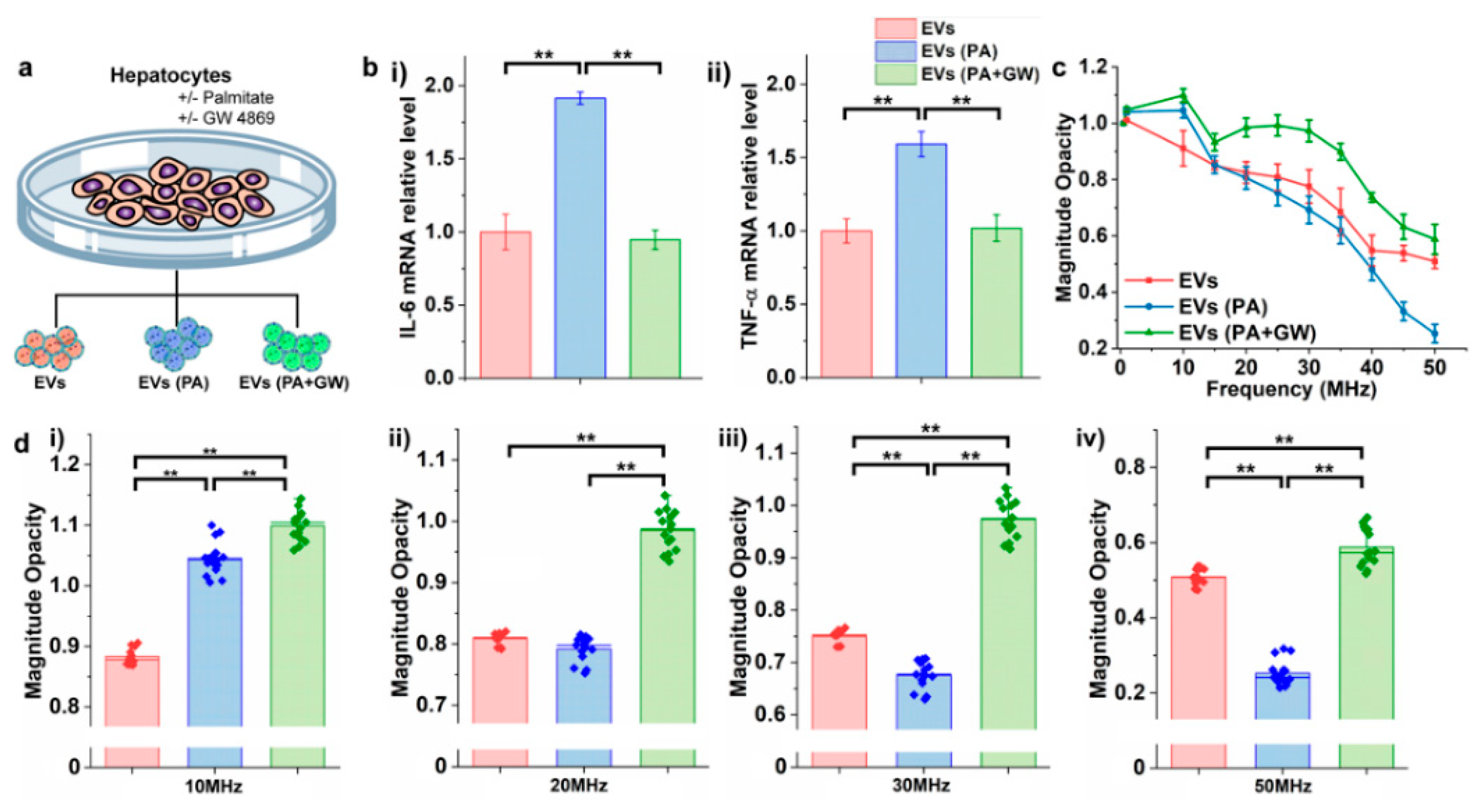
3.4. Detection of EVs Secreted from Cells Treated under Different Culture Conditions
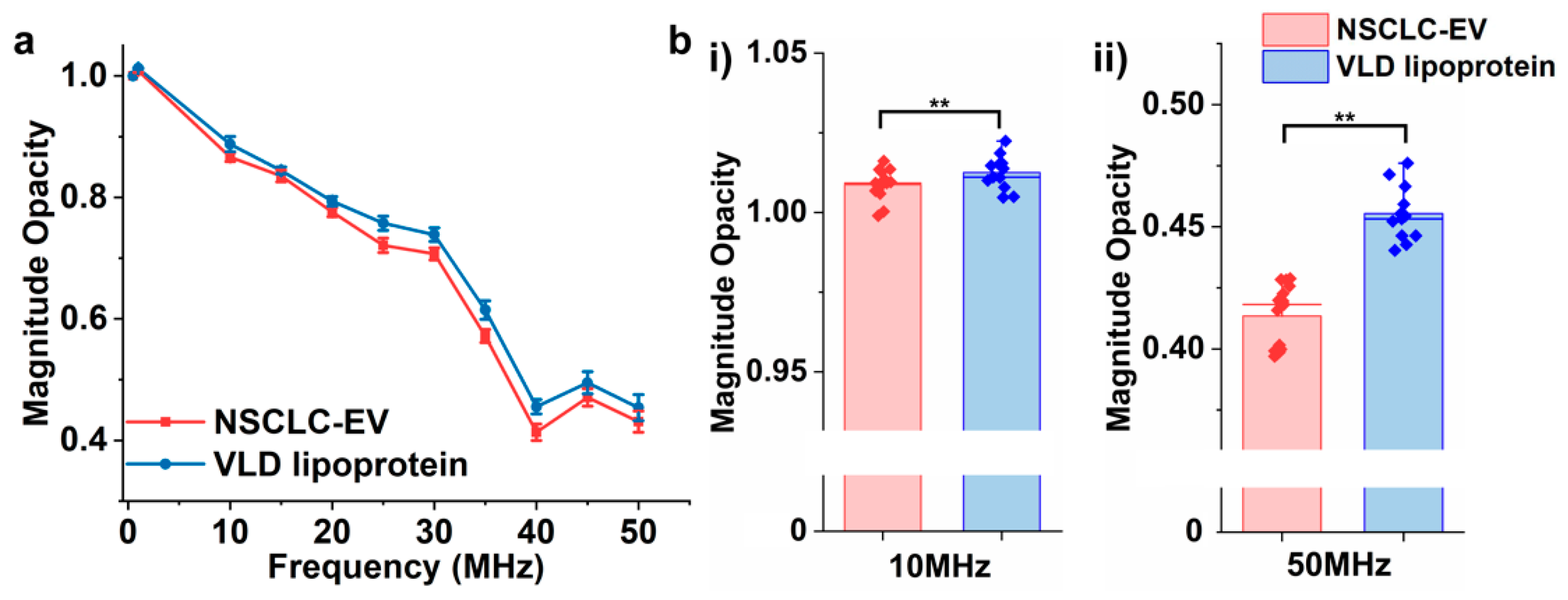
3.5. Differentiating EVs from Lipoproteins
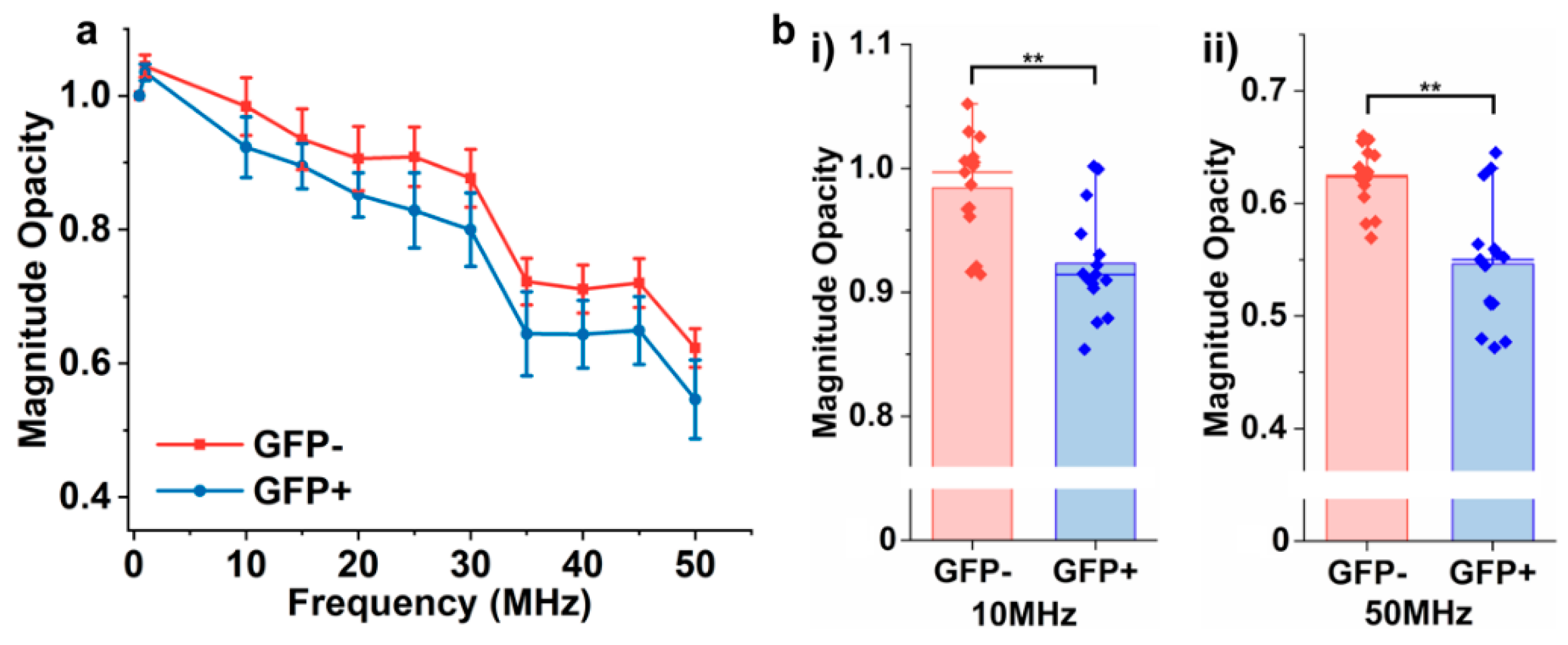
3.6. Detection of EVs Derived from Different Cellular Origins
3.7. Detection of EVs with Different Size Distributions
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Zheng, S.; Luo, Y.; Wang, B. Exosome Theranostics: Biology and Translational Medicine. Theranostics 2018, 8, 237–255. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, Functions, and Clinical Relevance of Extracellular Vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [Green Version]
- Yamashita, T.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. Biopharm. 2016, 98, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Peng, P.; Kuang, Y.; Yang, J.; Cao, D.; You, Y.; Shen, K. Characterization of exosomes derived from ovarian cancer cells and normal ovarian epithelial cells by nanoparticle tracking analysis. Tumor Biol. 2016, 37, 4213–4221. [Google Scholar] [CrossRef]
- Atay, S.; Gercel-Taylor, C.; Kesimer, M.; Taylor, D.D. Morphologic and proteomic characterization of exosomes released by cultured extravillous trophoblast cells. Exp. Cell Res. 2011, 317, 1192–1202. [Google Scholar] [CrossRef]
- Van Der Pol, E.; Van Gemert, M.J.C.; Sturk, A.; Nieuwland, R.; Van Leeuwen, T.G. Single vs. swarm detection of microparticles and exosomes by flow cytometry. J. Thromb. Haemost. 2012, 10, 919–930. [Google Scholar] [CrossRef]
- De Rond, L.; van der Pol, E.; Bloemen, P.R.; Van Den Broeck, T.; Monheim, L.; Nieuwland, R.; van Leeuwen, T.G.; Coumans, F.A.W. A Systematic Approach to Improve Scatter Sensitivity of a Flow Cytometer for Detection of Extracellular Vesicles. Cytom. A 2020, 97, 582–591. [Google Scholar] [CrossRef]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W.E. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Schade-Kampmann, G.; Huwiler, A.; Hebeisen, M.; Hessler, T.; Di Berardino, M. On-chip non-invasive and label-free cell discrimination by impedance spectroscopy. Cell Prolif. 2008, 41, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Morgan, H. Single-cell microfluidic impedance cytometry: A review. Microfluid. Nanofluid. 2010, 8, 423–443. [Google Scholar] [CrossRef]
- Feng, Y.; Huang, L.; Zhao, P.; Liang, F.; Wang, W. A Microfluidic Device Integrating Impedance Flow Cytometry and Electric Impedance Spectroscopy for High-Efficiency Single-Cell Electrical Property Measurement. Anal. Chem. 2019, 91, 15204–15212. [Google Scholar] [CrossRef] [PubMed]
- Bagnaninchi, P.O.; Drummond, N. Real-time label-free monitoring of adipose-derived stem cell differentiation with electric cell-substrate impedance sensing. Proc. Natl. Acad. Sci. USA 2011, 108, 6462–6467. [Google Scholar] [CrossRef] [Green Version]
- Suhito, I.R.; Angeline, N.; Choo, S.-S.; Woo, H.Y.; Paik, T.; Lee, T.; Kim, T.-H. Nanobiosensing Platforms for Real-Time and Non-Invasive Monitoring of Stem Cell Pluripotency and Differentiation. Sensors 2018, 18, 2755. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Kim, H.S.; Frazier, A.B.; Chen, Z.G.; Shin, D.M.; Han, A. Whole-Cell Impedance Analysis for Highly and Poorly Metastatic Cancer Cells. J. Microelectromech. Syst. 2009, 18, 808–817. [Google Scholar] [CrossRef]
- Zhang, F.; Jin, T.; Hu, Q.; He, P. Distinguishing skin cancer cells and normal cells using electrical impedance spectroscopy. J. Electroanal. Chem. 2018, 823, 531–536. [Google Scholar] [CrossRef]
- Zhou, Y.; Basu, S.; Laue, E.; Seshia, A.A. Single cell studies of mouse embryonic stem cell (mESC) differentiation by electrical impedance measurements in a microfluidic device. Biosens. Bioelectron. 2016, 81, 249–258. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Wang, Y.; Rosano, J.M.; Prabhakarpandian, B.; Garson, C.; Pant, K.; Lai, E. A microfluidic impedance flow cytometer for identification of differentiation state of stem cells. Lab Chip 2013, 13, 2300–2310. [Google Scholar] [CrossRef]
- Holmes, D.; Pettigrew, D.; Reccius, C.H.; Gwyer, J.D.; van Berkel, C.; Holloway, J.; Davies, D.E.; Morgan, H. Leukocyte analysis and differentiation using high speed microfluidic single cell impedance cytometry. Lab Chip 2009, 9, 2881–2889. [Google Scholar] [CrossRef]
- Friedrich, R.; Block, S.; Alizadehheidari, M.; Heider, S.; Fritzsche, J.; Esbjörner, E.K.; Westerlund, F.; Bally, M. A nano flow cytometer for single lipid vesicle analysis. Lab Chip 2017, 17, 830–841. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Zhu, S.; Wang, S.; Zhang, W.; Cheng, Y.; Yan, X. Multiparameter Quantification of Liposomal Nanomedicines at the Single-Particle Level by High-Sensitivity Flow Cytometry. ACS Appl. Mater. Interfaces 2017, 9, 13913–13919. [Google Scholar] [CrossRef]
- Shi, L.; Kuhnell, D.; Borra, V.J.; Langevin, S.M.; Nakamura, T.; Esfandiari, L. Rapid and label-free isolation of small extracellular vesicles from biofluids utilizing a novel insulator based dielectrophoretic device. Lab Chip 2019, 19, 3726–3734. [Google Scholar] [CrossRef]
- Shi, L.; Rana, A.; Esfandiari, L. A low voltage nanopipette dielectrophoretic device for rapid entrapment of nanoparticles and exosomes extracted from plasma of healthy donors. Sci. Rep. 2018, 8, 6751. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Vermesh, O.; Mani, V.; Ge, T.J.; Madsen, S.J.; Sabour, A.; Hsu, E.-C.; Gowrishankar, G.; Kanada, M.; Jokerst, J.V.; et al. The Exosome Total Isolation Chip. ACS Nano 2017, 11, 10712–10723. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Rana, A.; Stratton, Y.; Czyzyk-Krzeska, M.F.; Esfandiari, L. Sequence-Specific Detection of MicroRNAs Related to Clear Cell Renal Cell Carcinoma at fM Concentration by an Electroosmotically Driven Nanopore-Based Device. Anal. Chem. 2017, 89, 9201–9208. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Altman, N. Significance, p values and t-tests. Nat. Methods 2013, 10, 1041–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Biasio, A.; Ambrosone, L.; Cametti, C. The dielectric behavior of nonspherical biological cell suspensions: An analytic approach. Biophys. J. 2010, 99, 163–174. [Google Scholar] [CrossRef] [Green Version]
- Shi, L.; Esfandiari, L. An Electrokinetically-Driven Microchip for Rapid Entrapment and Detection of Nanovesicles. Micromachines 2021, 12, 11. [Google Scholar] [CrossRef]
- Hoffman, R.A.; Johnson, T.S.; Britt, W.B. Flow cytometric electronic direct current volume and radiofrequency impedance measurements of single cells and particles. Cytometry 1981, 1, 377–384. [Google Scholar] [CrossRef]
- Haandbæk, N.; Bürgel, S.C.; Heer, F.; Hierlemann, A. Characterization of subcellular morphology of single yeast cells using high frequency microfluidic impedance cytometer. Lab Chip 2014, 14, 369–377. [Google Scholar] [CrossRef]
- Norlin, A.; Pan, J.; Leygraf, C. Investigation of interfacial capacitance of Pt, Ti and TiN coated electrodes by electrochemical impedance spectroscopy. Biomol. Eng. 2002, 19, 67–71. [Google Scholar] [CrossRef]
- Naumowicz, M.; Petelska, A.D.; Figaszewski, Z.A. Capacitance and resistance of the bilayer lipid membrane formed of phosphatidylcholine and cholesterol. Cell. Mol. Biol. Lett. 2003, 8, 5–18. [Google Scholar]
- Gawad, S.; Schild, L.; Renaud, P. Micromachined impedance spectroscopy flow cytometer for cell analysis and particle sizing. Lab Chip 2001, 1, 76–82. [Google Scholar] [CrossRef]
- Yim, N.; Ryu, S.-W.; Choi, K.; Lee, K.R.; Lee, S.; Choi, H.; Kim, J.; Shaker, M.R.; Sun, W.; Park, J.-H.; et al. Exosome engineering for efficient intracellular delivery of soluble proteins using optically reversible protein–protein interaction module. Nat. Commun. 2016, 7, 12277. [Google Scholar] [CrossRef]
- Nakamura, S.; Takamura, T.; Matsuzawa-Nagata, N.; Takayama, H.; Misu, H.; Noda, H.; Nabemoto, S.; Kurita, S.; Ota, T.; Ando, H.; et al. Palmitate Induces Insulin Resistance in H4IIEC3 Hepatocytes through Reactive Oxygen Species Produced by Mitochondria. J. Biol. Chem. 2009, 284, 14809–14818. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.-S.; Kim, S.Y.; Ko, E.; Lee, J.-H.; Yi, H.-S.; Yoo, Y.J.; Je, J.; Suh, S.J.; Jung, Y.K.; Kim, J.H.; et al. Exosomes derived from palmitic acid-treated hepatocytes induce fibrotic activation of hepatic stellate cells. Sci. Rep. 2017, 7, 3710. [Google Scholar] [CrossRef]
- Essandoh, K.; Yang, L.; Wang, X.; Huang, W.; Qin, D.; Hao, J.; Wang, Y.; Zingarelli, B.; Peng, T.; Fan, G.-C. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim. Biophys. Acta Mol. Basis Dis. 2015, 1852, 2362–2371. [Google Scholar] [CrossRef] [Green Version]
- Kakazu, E.; Mauer, A.S.; Yin, M.; Malhi, H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J. Lipid Res. 2016, 57, 233–245. [Google Scholar] [CrossRef] [Green Version]
- Naumowicz, M.; Petelska, A.D.; Figaszewski, Z.A. Physicochemical analysis of phosphatidylcholine-ceramide system in bilayer lipid membranes. Acta Biochim. Pol. 2008, 55, 721–730. [Google Scholar] [CrossRef]
- McGrath, J.S.; Honrado, C.; Moore, J.H.; Adair, S.J.; Varhue, W.B.; Salahi, A.; Farmehini, V.; Goudreau, B.J.; Nagdas, S.; Blais, E.M.; et al. Electrophysiology-based stratification of pancreatic tumorigenicity by label-free single-cell impedance cytometry. Anal. Chim. Acta 2020, 1101, 90–98. [Google Scholar] [CrossRef]
- Feingold, K.R. Introduction to Lipids and Lipoproteins. In Endotext; MDText.com, Inc.: South Dartmouth, MA, USA, 2000. [Google Scholar] [PubMed]
- Sun, Y.; Saito, K.; Saito, Y. Lipid Profile Characterization and Lipoprotein Comparison of Extracellular Vesicles from Human Plasma and Serum. Metabolites 2019, 9, 259. [Google Scholar] [CrossRef] [Green Version]
- Yáñez-Mó, M.; Siljander, P.R.M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [Green Version]
- Kruger, S.; Elmageed, Z.Y.A.; Hawke, D.H.; Wörner, P.M.; Jansen, D.A.; Abdel-Mageed, A.B.; Alt, E.U.; Izadpanah, R. Molecular characterization of exosome-like vesicles from breast cancer cells. BMC Cancer 2014, 14, 44. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Hernandez, A.; Leal-Orta, E.; Ramirez-Ricardo, J.; Cortes-Reynosa, P.; Thompson-Bonilla, R.; Salazar, E.P. Linoleic acid induces secretion of extracellular vesicles from MDA-MB-231 breast cancer cells that mediate cellular processes involved with angiogenesis in HUVECs. Prostaglandins Other Lipid Mediat. 2021, 153, 106519. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Zhang, H.; Freitas, D.; Kim, H.S.; Fabijanic, K.; Li, Z.; Chen, H.; Mark, M.T.; Molina, H.; Martin, A.B.; Bojmar, L.; et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat. Cell Biol. 2018, 20, 332–343. [Google Scholar] [CrossRef]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H. Thin-Film Hydration Followed by Extrusion Method for Liposome Preparation. In Liposomes: Methods and Protocols; D’Souza, G.G.M., Ed.; Springer: New York, NY, USA, 2017; pp. 17–22. [Google Scholar]
- Zhang, Z.; Lutz, B. Cre recombinase-mediated inversion using lox66 and lox71: Method to introduce conditional point mutations into the CREB-binding protein. Nucleic Acids Res. 2002, 30, e90. [Google Scholar] [CrossRef]
- Salem, E.S.B.; Murakami, K.; Takahashi, T.; Bernhard, E.; Borra, V.; Bethi, M.; Nakamura, T. Isolation of Primary Mouse Hepatocytes for Nascent Protein Synthesis Analysis by Non-radioactive L-azidohomoalanine Labeling Method. J. Vis. Exp. 2018, 140, e58323. [Google Scholar] [CrossRef]
- Shen, B.; Wu, N.; Yang, J.M.; Gould, S.J. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J. Biol. Chem. 2011, 286, 14383–14395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwan, H.P. Electrode Polarization Impedance and Measurements in Biological Materials. Ann. N. Y. Acad. Sci. 1968, 148, 191–209. [Google Scholar] [CrossRef] [PubMed]
- Foster, K.R.; Schwan, H.P. Dielectric properties of tissues and biological materials: A critical review. Crit. Rev. Biomed. Eng. 1989, 17, 25–104. [Google Scholar] [PubMed]
- Ren, J.; He, W.; Zheng, L.; Duan, H. From structures to functions: Insights into exosomes as promising drug delivery vehicles. Biomater. Sci. 2016, 4, 910–921. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Sabuncu, A.C.; Zhuang, J.; Kolb, J.F.; Beskok, A. Microfluidic impedance spectroscopy as a tool for quantitative biology and biotechnology. Biomicrofluidics 2012, 6, 034103. [Google Scholar] [CrossRef] [Green Version]
- Raicu, V. A simple theoretical and practical approach to measuring dielectric properties with an open-ended coaxial probe. Meas. Sci. Technol. 1995, 6, 410–414. [Google Scholar] [CrossRef]
- Asami, K. Dielectric dispersion in biological cells of complex geometry simulated by the three-dimensional finite difference method. J. Phys. D Appl. Phys. 2006, 39, 492–499. [Google Scholar] [CrossRef]
- Sun, T.; Gawad, S.; Green, N.G.; Morgan, H. Dielectric spectroscopy of single cells: Time domain analysis using Maxwell’s mixture equation. J. Phys. D Appl. Phys. 2006, 40, 1–8. [Google Scholar] [CrossRef]
- Maxwell, J.C. A Treatise on Electricity and Magnetism; Clarendon Press: Oxford, UK, 1873; Volume 1. [Google Scholar]
- Sun, T.; Bernabini, C.; Morgan, H. Single-Colloidal Particle Impedance Spectroscopy: Complete Equivalent Circuit Analysis of Polyelectrolyte Microcapsules. Langmuir 2010, 26, 3821–3828. [Google Scholar] [CrossRef]
- Sun, T.; Holmes, D.; Gawad, S.; Green, N.G.; Morgan, H. High speed multi-frequency impedance analysis of single particles in a microfluidic cytometer using maximum length sequences. Lab Chip 2007, 7, 1034–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vahey, M.D.; Voldman, J. High-throughput cell and particle characterization using isodielectric separation. Anal. Chem. 2009, 81, 2446–2455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, L.; Holmes, D.; Morgan, H. The dielectrophoretic levitation and separation of latex beads in microchips. Electrophoresis 2001, 22, 3893–3901. [Google Scholar] [CrossRef]
- Zhang, G.; Zhu, R. Effect of Parasitic Capacitance on Impedance Measurement and Model Extraction. Electroanalysis 2010, 22, 351–358. [Google Scholar] [CrossRef]
- Yoon, G. Dielectric properties of glucose in bulk aqueous solutions: Influence of electrode polarization and modeling. Biosens. Bioelectron. 2011, 26, 2347–2353. [Google Scholar] [CrossRef]
- Kaatze, U. Complex permittivity of water as a function of frequency and temperature. J. Chem. Eng. Data 1989, 34, 371–374. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Murakami, K.; Borra, V.J.; Ozen, M.O.; Demirci, U.; Nakamura, T.; Esfandiari, L. A Label-Free Electrical Impedance Spectroscopy for Detection of Clusters of Extracellular Vesicles Based on Their Unique Dielectric Properties. Biosensors 2022, 12, 104. https://doi.org/10.3390/bios12020104
Zhang Y, Murakami K, Borra VJ, Ozen MO, Demirci U, Nakamura T, Esfandiari L. A Label-Free Electrical Impedance Spectroscopy for Detection of Clusters of Extracellular Vesicles Based on Their Unique Dielectric Properties. Biosensors. 2022; 12(2):104. https://doi.org/10.3390/bios12020104
Chicago/Turabian StyleZhang, Yuqian, Kazutoshi Murakami, Vishnupriya J. Borra, Mehmet Ozgun Ozen, Utkan Demirci, Takahisa Nakamura, and Leyla Esfandiari. 2022. "A Label-Free Electrical Impedance Spectroscopy for Detection of Clusters of Extracellular Vesicles Based on Their Unique Dielectric Properties" Biosensors 12, no. 2: 104. https://doi.org/10.3390/bios12020104
APA StyleZhang, Y., Murakami, K., Borra, V. J., Ozen, M. O., Demirci, U., Nakamura, T., & Esfandiari, L. (2022). A Label-Free Electrical Impedance Spectroscopy for Detection of Clusters of Extracellular Vesicles Based on Their Unique Dielectric Properties. Biosensors, 12(2), 104. https://doi.org/10.3390/bios12020104





