State-of-the-Art Fluorescent Probes: Duplex-Specific Nuclease-Based Strategies for Early Disease Diagnostics
Abstract
1. Introduction
2. Diagnostic and Prognostic of miRNA Biomarkers
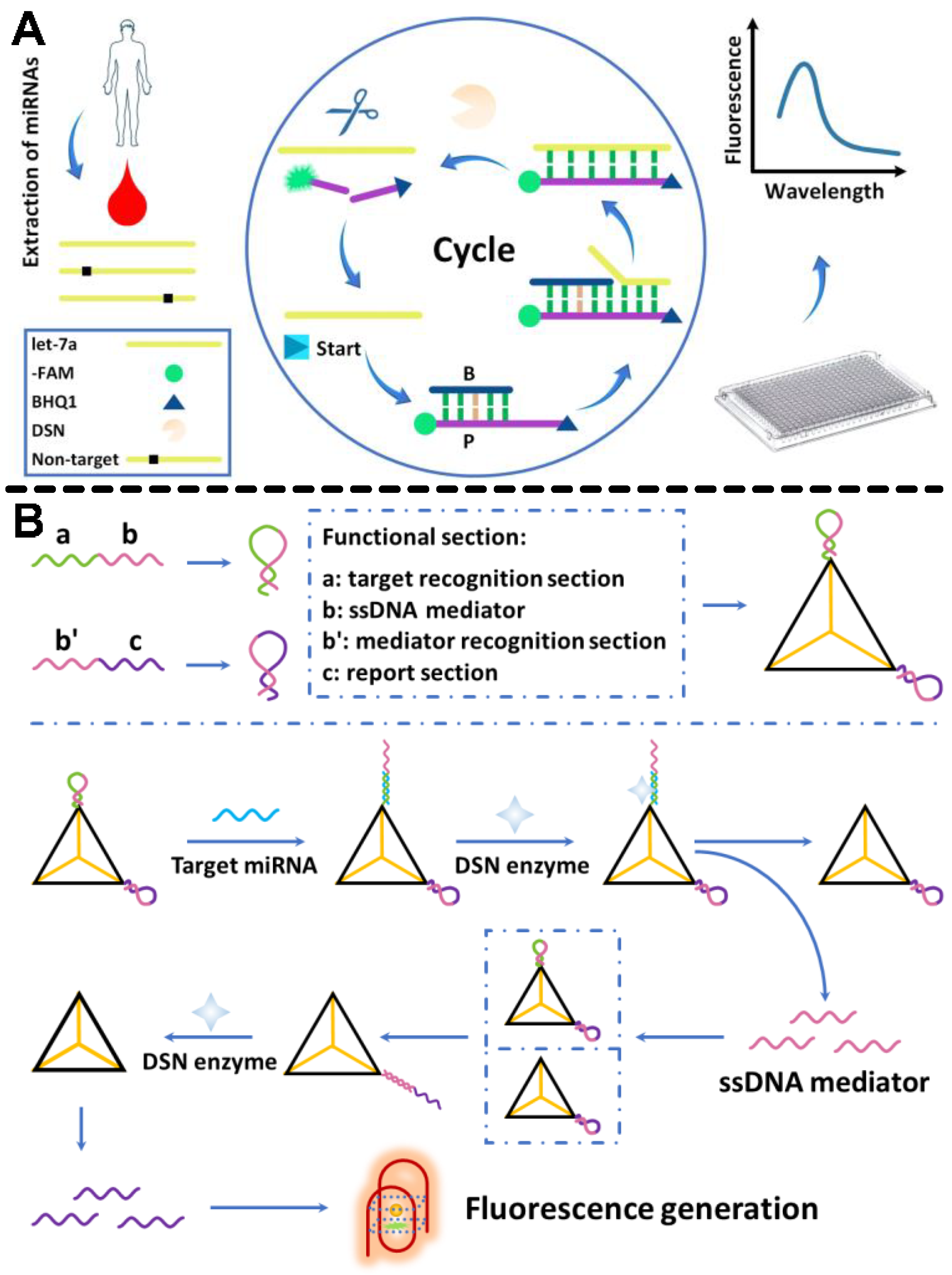
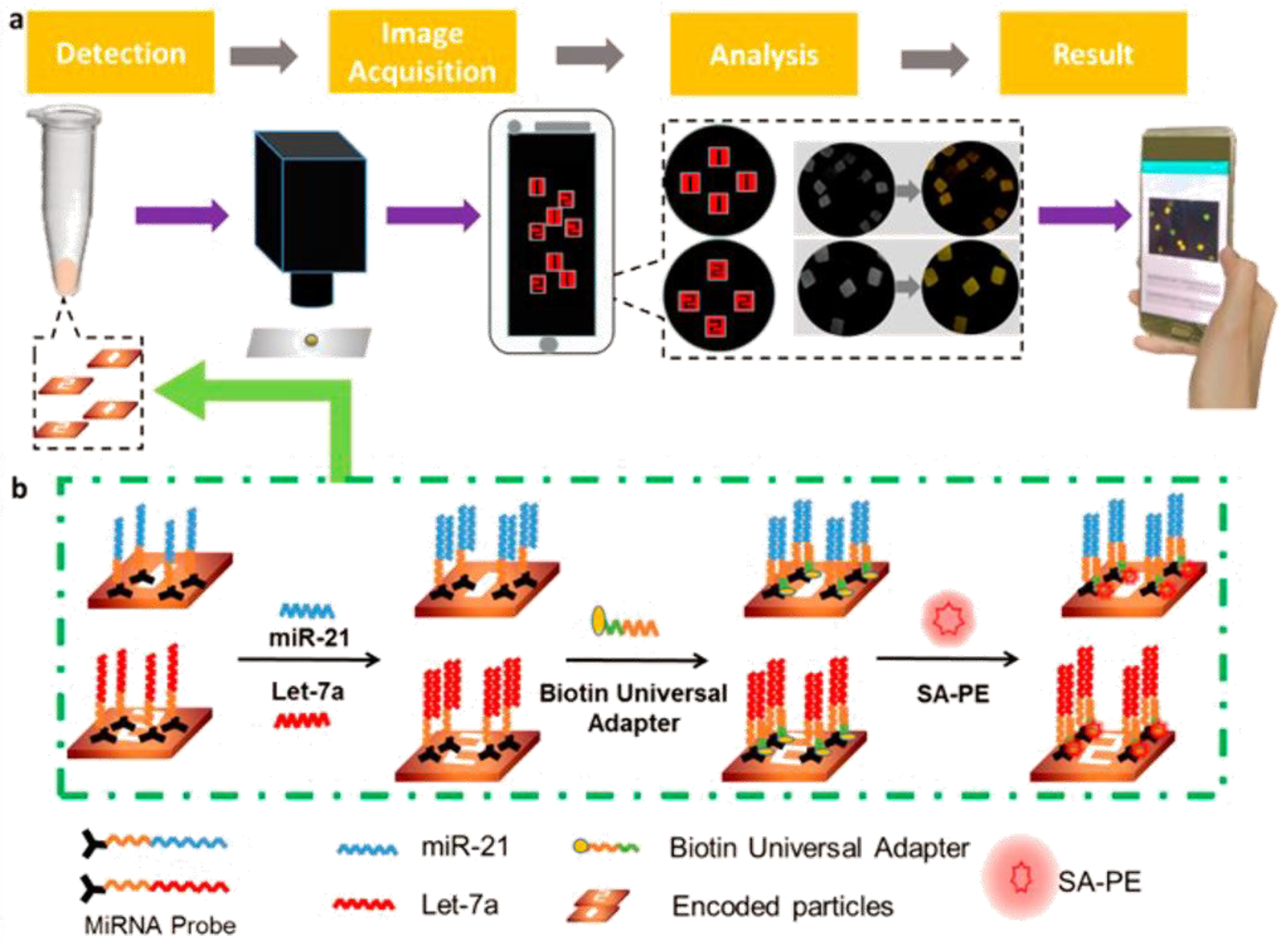
2.1. DSN Platforms for miRNA Cancer Biomarkers’ Detection
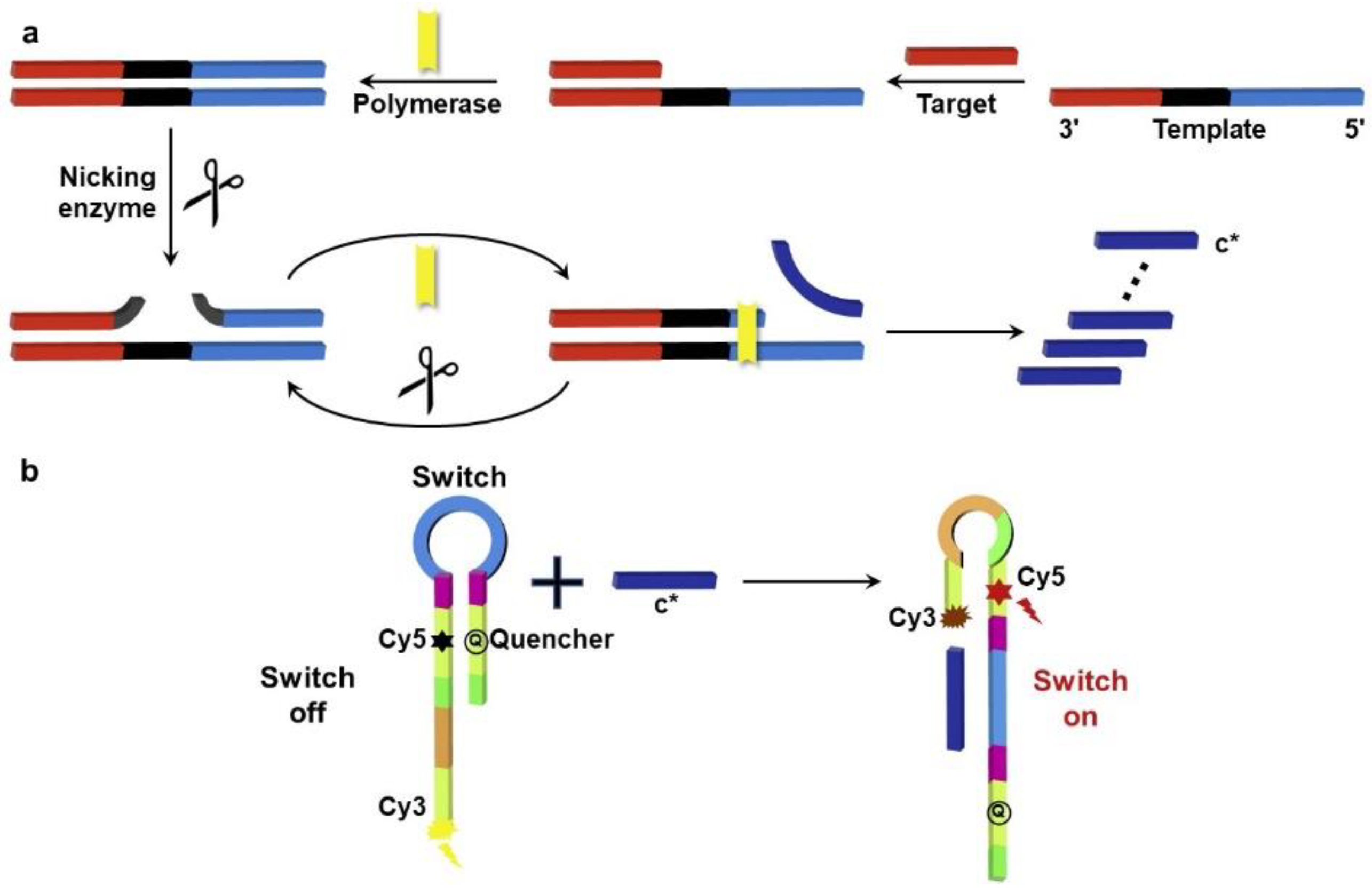
2.2. DSN Platforms for miRNA Cardiac Biomarkers’ Detection
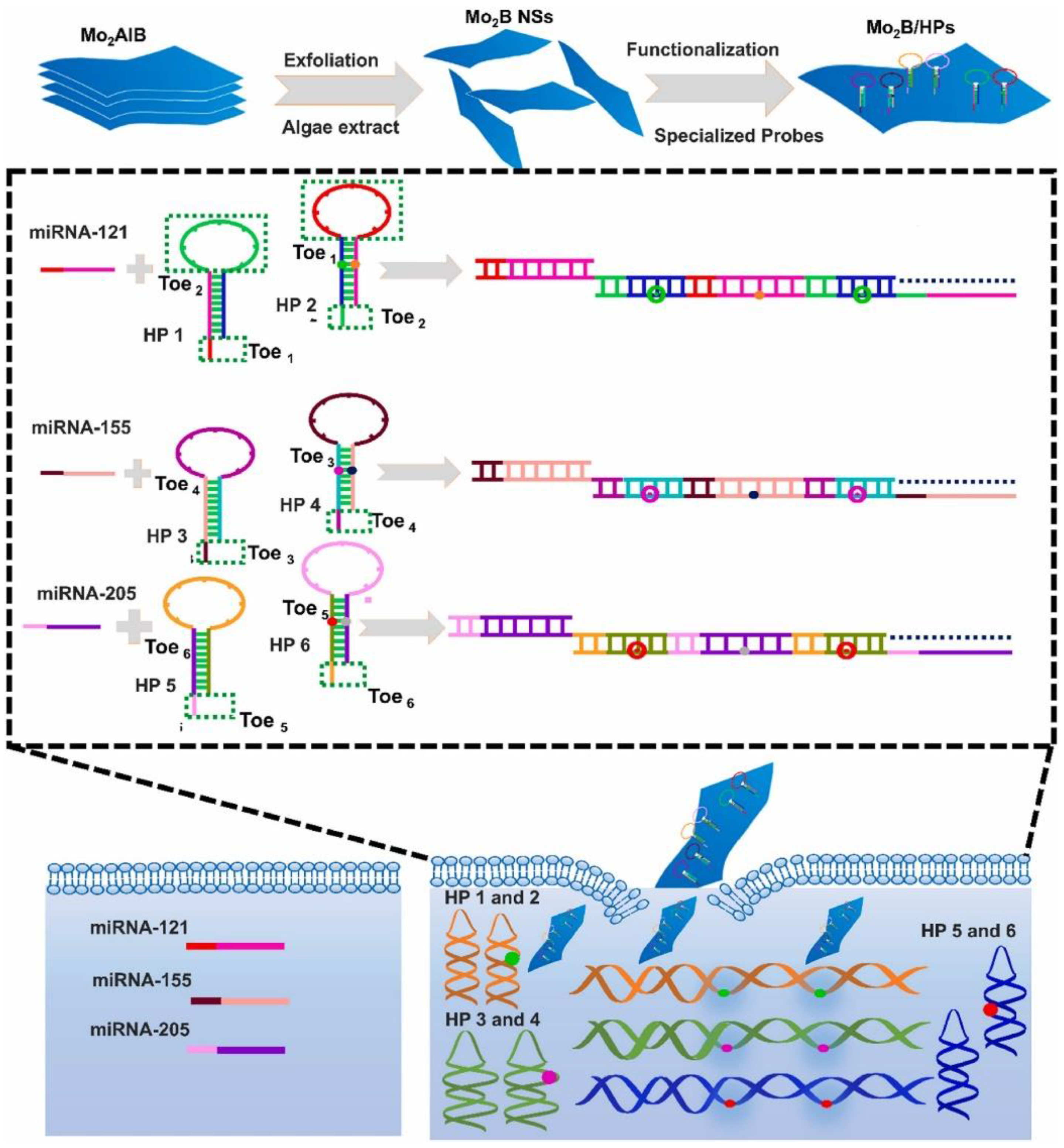
2.3. DSN Strategies for miRNA Neural Biomarker Detection
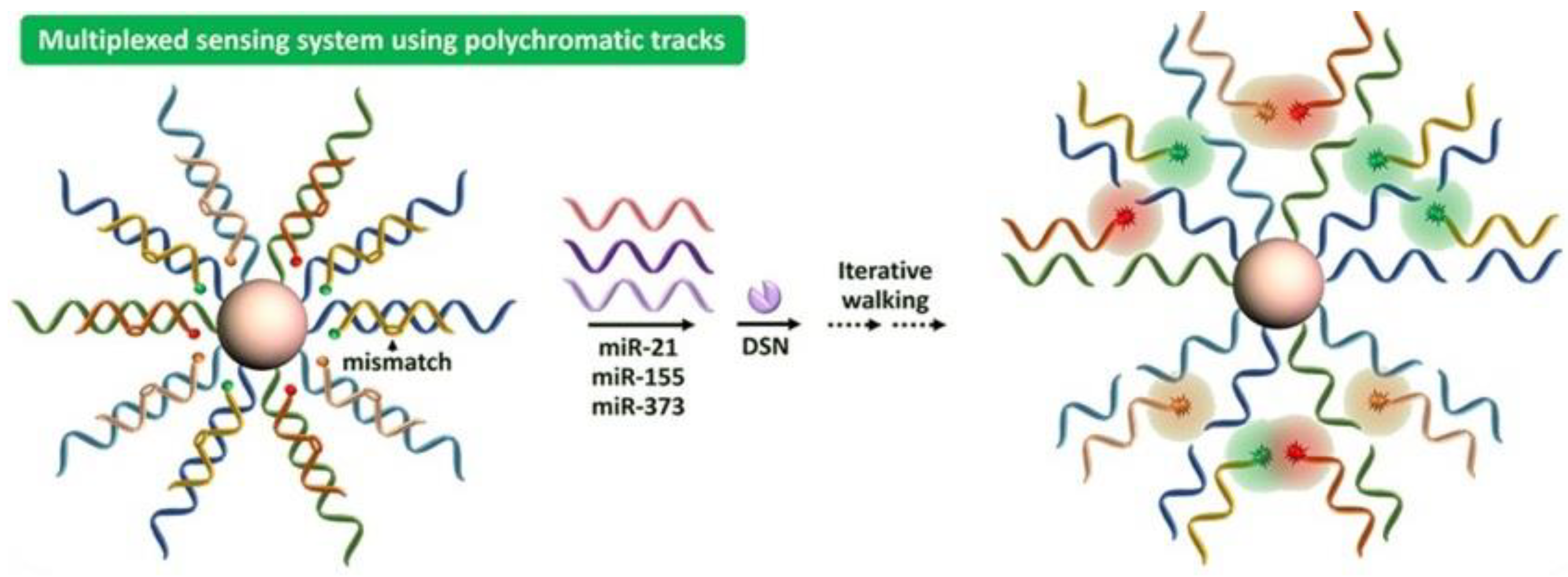
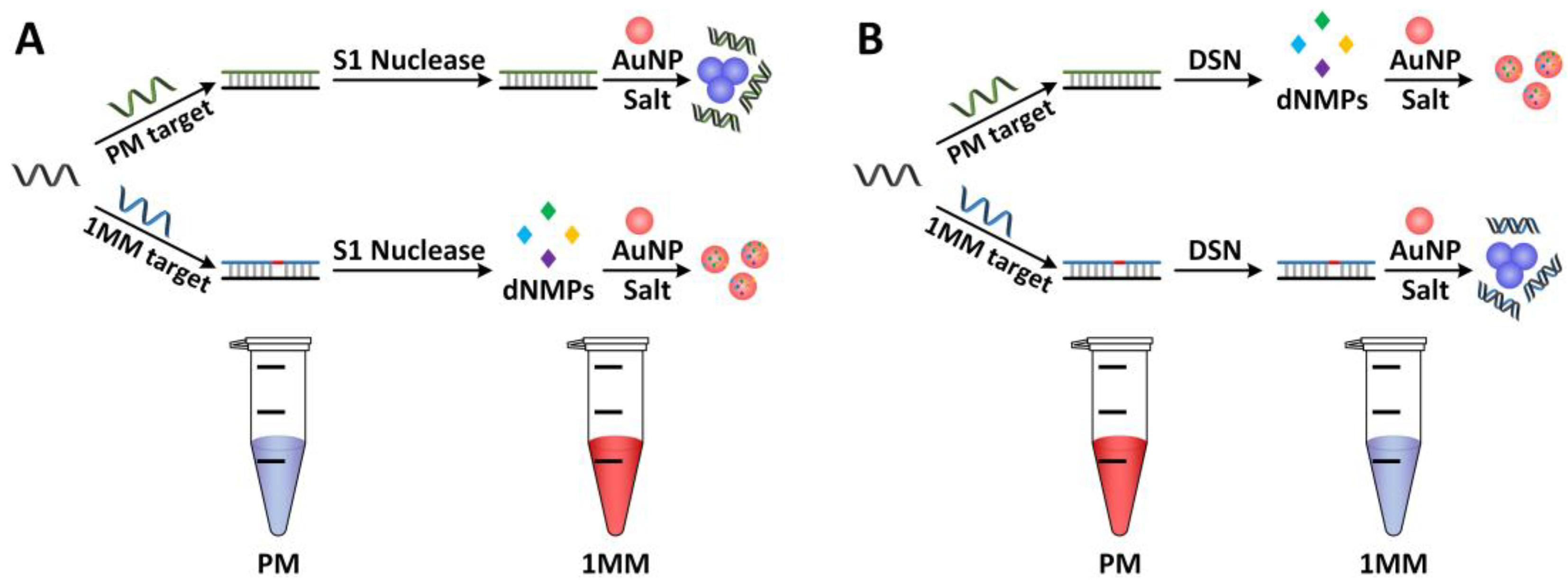
2.4. DSN-Based Additional Strategies for miRNA Detection
| Method | Target Analyte | Detection Limit | Real Sample | Reference |
|---|---|---|---|---|
| Fluorescence | miRNA-21 and miRNA-31 | 0.17 and 0.062 fM | CA549 and HeLa cells | [53] |
| Fluorescence | miRNA-21 | 258 pM | MCF-7 cells | [87] |
| Fluorescence | Let-7a | 0.26 pM | A549 cells | [88] |
| Fluorescence | miRNA-3188 | 25 fM | CNE-1 cells | [89] |
| Voltage-assisted liquid desorption electrospray ionization tandem mass spectrometry | miRNA-21 | 0.25 pM | Mouse peritoneal macrophage | [90] |
| SERS | miRNA-21 | 42 aM | A549, MCF-7, HeLa, and HepG cells | [91] |
| SERS | miRNA-21 | 0.1 fM | A549, 293T and HeLa cells | [92] |
| Electrochemiluminescent/electrochemical | miRNA-499 | 28.75 aM | Human serum | [93] |
| Electrochemical | miRNA-21 | 4.5 fM | HeLa, MCF-7 cells | [94] |
| Electrochemical | miRNA-21 | 0.28 fM | Human serum | [95] |
| Colorimetric | miRNA-21 | 29 fM | Human serum | [96] |
| Colorimetric | miRNA-122 | 7.7 fM | NA | [97] |
3. Diagnostic and Prognostic of SNP-Related Biomarkers
3.1. DSN Platforms for SNP-Related Cancer Biomarkers’ Detection
3.2. DSN Platforms for SNP-Related Neurodegeneration Biomarkers’ Detection
3.3. DSN Platforms for SNP-Related Virus Detection
4. Conclusions and Outlook
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Keats, B.J.B.; Sherman, S.L. Population Genetics. In Emery and Rimoin’s Principles and Practice of Medical Genetics, 6th ed.; Rimoin, D., Pyeritz, R., Korf, B., Eds.; Academic Press: London, UK, 2013; pp. 1–12. [Google Scholar] [CrossRef]
- Liu, X.; Han, Z.; Yang, C. Associations of microRNA single nucleotide polymorphisms and disease risk and pathophysiology. Clin. Genet. 2017, 92, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Gong, S.; Li, J.; Pan, W.; Li, N.; Tang, B. Duplex-specific nuclease-assisted CRISPR-Cas12a strategy for microRNA detection using a personal glucose meter. Anal. Chem. 2021, 93, 10719–10726. [Google Scholar] [CrossRef] [PubMed]
- Safdar, S.; Lammertyn, J.; Spasic, D. RNA-cleaving NAzymes: The next big thing in biosensing? Trends Biotechnol. 2020, 38, 1343–1359. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.X.; Lv, Z.; Yuan, Y.; Xu, Q. MiRNA polymorphisms and cancer prognosis: A systematic review and meta-analysis. Front. Oncol. 2018, 8, 596. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhou, Y.; Yin, H.; Ai, S. Recent advances on signal amplification strategies in photoelectrochemical sensing of microRNAs. Biosens. Bioelectron. 2020, 166, 112476. [Google Scholar] [CrossRef]
- Yang, C.T.; Pourhassan-Moghaddam, M.; Wu, L.; Bai, P.; Thierry, B. Ultrasensitive detection of cancer prognostic miRNA biomarkers based on surface plasmon enhanced light scattering. ACS Sens. 2017, 2, 635–640. [Google Scholar] [CrossRef]
- Qi, H.; Yue, S.; Bi, S.; Ding, C.; Song, W. Isothermal exponential amplification techniques: From basic principles to applications in electrochemical biosensors. Biosens. Bioelectron. 2018, 110, 207–217. [Google Scholar] [CrossRef]
- Basu, A. Loop-Seq: A High-Throughput Technique to Measure the Mesoscale Mechanical Properties of DNA. In Methods in Enzymology; Eichman, B.F., Ed.; Academic Press: Cambridge, MA, USA, 2021; Volume 661, pp. 305–326. [Google Scholar] [CrossRef]
- Cai, S.; Pataillot-Meakin, T.; Shibakawa, A.; Ren, R.; Bevan, C.L.; Ladame, S.; Ivanov, A.P.; Edel, J.B. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 2021, 12, 3515. [Google Scholar] [CrossRef]
- Luka, G.; Ahmadi, A.; Najjaran, H.; Alocilja, E.; Derosa, M.; Wolthers, K.; Malki, A.; Aziz, H.; Althani, A.; Hoorfar, M. Microfluidics integrated biosensors: A leading technology towards lab-on-a-chip and sensing applications. Sensors 2015, 15, 30011–30031. [Google Scholar] [CrossRef]
- Dave, V.P.; Ngo, T.A.; Pernestig, A.K.; Tilevik, D.; Kant, K.; Nguyen, T.; Wolff, A.; Bang, D.D. MicroRNA amplification and detection technologies: Opportunities and challenges for point of care diagnostics. Lab. Investig. 2019, 99, 452–469. [Google Scholar] [CrossRef]
- Haji Mohammadi, M.; Mulder, S.; Khashayar, P.; Kalbasi, A.; Azimzadeh, M.; Aref, A.R. Saliva lab-on-a-chip biosensors: Recent novel ideas and applications in disease detection. Microchem. J. 2021, 168, 106506. [Google Scholar] [CrossRef]
- Aziz, N.B.; Mahmudunnabi, R.G.; Umer, M.; Sharma, S.; Rashid, M.A.; Alhamhoom, Y.; Shim, Y.-B.; Salomon, C.; Shiddiky, M.J.A. MicroRNAs in ovarian cancer and recent advances in the development of microRNA-based biosensors. Analyst 2020, 145, 2038–2057. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; Zhang, K.; Li, J. Isothermal amplification for microRNA Detection: From the test tube to the cell. Acc. Chem. Res. 2017, 50, 1059–1068. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Liang, W.; Li, Y.; Sun, Y.; Xiang, Y.; Zhang, Y.; Dai, Z.; Duo, Y.; Wu, L.; Qi, K.; et al. Ultrasensitive detection of miRNA with an antimonene-based surface plasmon resonance sensor. Nat. Commun. 2019, 10, 28. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef]
- Kim, J.K.; Choi, K.J.; Lee, M.; Jo, M.H.; Kim, S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials 2012, 33, 207–217. [Google Scholar] [CrossRef]
- Ma, F.; Liu, W.J.; Zhang, Q.; Zhang, C.Y. Sensitive detection of microRNAs by duplex specific nuclease-assisted target recycling and pyrene excimer switching. Chem. Commun. 2017, 53, 10596–10599. [Google Scholar] [CrossRef]
- Yue, S.; Li, Y.; Qiao, Z.; Song, W.; Bi, S. Rolling circle replication for biosensing, bioimaging, and biomedicine. Trends Biotechnol. 2021, 39, 1160–1172. [Google Scholar] [CrossRef]
- Yin, B.C.; Liu, Y.Q.; Ye, B.C. One-Step, Multiplexed fluorescence detection of microRNAs based on duplex-specific nuclease signal amplification. J. Am. Chem. Soc. 2012, 134, 5064–5067. [Google Scholar] [CrossRef]
- Shi, H.; Yang, L.; Zhou, X.; Bai, J.; Gao, J.; Jia, H.; Li, Q. A gold nanoparticle-based colorimetric strategy coupled to duplex-specific nuclease signal amplification for the determination of microRNA. Microchim. Acta 2017, 184, 525–531. [Google Scholar] [CrossRef]
- Bhat, A.I.; Aman, R.; Mahfouz, M. Onsite detection of plant viruses using isothermal amplification assays. Plant Biotechnol. J. 2022, 11, 13871. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, G.; Zhong, Z.T.; Asif, M.; Aziz, A.; Song, L.; Zhang, S.; Liu, B.; Chen, W.; Zhao, Y. Di Extension of duplex specific nuclease sensing application with RNA aptamer. Talanta 2022, 242, 123314. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.; Zhang, H.; Yu, H.; Jiang, T.; Luo, Y. Duplex-specific nuclease-mediated bioanalysis. Trends Biotechnol. 2015, 33, 180–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Zheng, C.; Jiang, Y.Z.; Zheng, Z.; Lin, M.; Lin, Y.; Zhang, Z.L.; Wang, H.; Pang, D.W. One-step monitoring of multiple enterovirus 71 infection-related microRNAs using core-satellite structure of magnetic nanobeads and multicolor quantum dots. Anal. Chem. 2020, 92, 830–837. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Xiong, C.; Zheng, Y.; Liang, W.; Yuan, R.; Chai, Y. Ultrasensitive photoelectrochemical biosensor based on DNA tetrahedron as nanocarrier for efficient immobilization of CdTe QDs-methylene blue as signal probe with near-zero background noise. Anal. Chem. 2018, 90, 8211–8216. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.T.; Ashraf, G.; Chen, W.; Song, L.B.; Zhang, S.J.; Liu, B.; Zhao, Y.D. A new strategy based on duplex-specific nuclease and DNA aptamer with modified hairpin structure for various analytes detection. Microchem. J. 2022, 179, 107510. [Google Scholar] [CrossRef]
- Gao, Z.; Yuan, H.; Mao, Y.; Ding, L.; Effah, C.Y.; He, S.; He, L.; Liu, L.E.; Yu, S.; Wang, Y.; et al. In situ detection of plasma exosomal microRNA for lung cancer diagnosis using duplex-specific nuclease and MoS2 nanosheets. Analyst 2021, 146, 1924–1931. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.L.; Zhang, N.; Yang, H.; Xu, J.J.; Chen, H.Y. Electrochemiluminescence resonance energy transfer system for dual-wavelength ratiometric miRNA detection. Anal. Chem. 2018, 90, 13723–13728. [Google Scholar] [CrossRef]
- Kuang, Y.; Cao, J.; Xu, F.; Chen, Y. Duplex-specific nuclease-mediated amplification strategy for mass spectrometry quantification of miRNA-200c in breast cancer stem cells. Anal. Chem. 2019, 91, 8820–8826. [Google Scholar] [CrossRef]
- Ki, J.; Lee, H.Y.; Son, H.Y.; Huh, Y.M.; Haam, S. Sensitive plasmonic detection of miR-10b in biological samples using enzyme-assisted target recycling and developed LSPR probe. ACS Appl. Mater. Interfaces 2019, 11, 18923–18929. [Google Scholar] [CrossRef]
- Wu, Y.; Cui, S.; Li, Q.; Zhang, R.; Song, Z.; Gao, Y.; Chen, W.; Xing, D. Recent advances in duplex-specific nuclease-based signal amplification strategies for microRNA detection. Biosens. Bioelectron. 2020, 165, 112449. [Google Scholar] [CrossRef] [PubMed]
- Verduci, L.; Tarcitano, E.; Strano, S.; Yarden, Y.; Blandino, G. CircRNAs: Role in human diseases and potential use as biomarkers. Cell Death Dis. 2021, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Cheong, J.K.; Tang, Y.C.; Zhou, L.; Cheng, H.; Too, H.P. Advances in quantifying circulatory microRNA for early disease detection. Curr. Opin. Biotechnol. 2022, 74, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Poirier, E.Z.; Buck, M.D.; Chakravarty, P.; Carvalho, J.; Frederico, B.; Cardoso, A.; Healy, L.; Ulferts, R.; Beale, R.; Reis E Sousa, C. An isoform of dicer protects mammalian stem cells against multiple RNA viruses. Science 2021, 373, 231–236. [Google Scholar] [CrossRef]
- Xu, L.; Shoaie, N.; Jahanpeyma, F.; Zhao, J.; Azimzadeh, M.; Al-Jamal, K.T. Optical, electrochemical and electrical (nano)biosensors for detection of exosomes: A comprehensive overview. Biosens. Bioelectron. 2020, 161, 112222. [Google Scholar] [CrossRef]
- Ding, Y.; Howes, P.D.; DeMello, A.J. Recent advances in droplet microfluidics. Anal. Chem. 2020, 92, 132–149. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Yazdanparast, S.; Benvidi, A.; Azimzadeh, M.; Tezerjani, M.D.; Ghaani, M.R. Experimental and theoretical study for miR-155 detection through resveratrol interaction with nucleic acids using magnetic core-shell nanoparticles. Microchim. Acta 2020, 187, 479. [Google Scholar] [CrossRef]
- Shi, L.; Liu, W.; Li, B.; Yang, C.J.; Jin, Y. Multichannel paper chip-based gas pressure bioassay for simultaneous detection of multiple microRNAs. ACS Appl. Mater. Interfaces 2021, 13, 15008–15016. [Google Scholar] [CrossRef]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M.; et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.L.; Yang, H.; Zhao, W.; Xu, J.J.; Chen, H.Y. Nanopore-based electrochemiluminescence for detection of microRNAs via duplex-specific nuclease-assisted target recycling. ACS Appl. Mater. Interfaces 2017, 9, 33360–33367. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Rong, Y.; Yang, Z.; She, D.; Gong, M. Construction of dual-target recognition-based specific microRNA detection method for acute pancreatitis analysis. Appl. Biochem. Biotechnol. 2022, 194, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, Z.; Xie, J.; Zhao, Y.; Tian, G.; Jiang, H.; Tao, H.; Liu, J. Target invasion-triggered signal amplification based on duplex-specific nuclease for selective and sensitive detection of miRNAs. Anal. Chim. Acta 2022, 1189, 339182. [Google Scholar] [CrossRef] [PubMed]
- Yao, G.; Xiao, Z.; Yu, S.; Yao, K.; Liu, D.; Chen, K.; Wei, Z.; Li, Y.; Sun, F. Tetrahedral structure supported two stages DSN-assisted amplification strategy for sensitive detection of lung cancer related microRNA. Microchem. J. 2022, 174, 107035. [Google Scholar] [CrossRef]
- Djebbi, K.; Shi, B.; Weng, T.; Bahri, M.; Elaguech, M.A.; Liu, J.; Tlili, C.; Wang, D. Highly sensitive fluorescence assay for MiRNA detection: Investigation of the DNA spacer effect on the DSN enzyme activity toward magnetic-bead-tethered probes. ACS Omega 2022, 7, 2224–2233. [Google Scholar] [CrossRef]
- Xiao, M.; Chandrasekaran, A.R.; Ji, W.; Li, F.; Man, T.; Zhu, C.; Shen, X.; Pei, H.; Li, Q.; Li, L. Affinity-modulated molecular beacons on MoS2 nanosheets for microRNA detection. ACS Appl. Mater. Interfaces 2018, 10, 35794–35800. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, C.; Chen, H.; Li, X.; Li, Y.; Fan, X. A simple G-quadruplex molecular beacon-based biosensor for highly selective detection of microRNA. Biosens. Bioelectron. 2017, 87, 552–557. [Google Scholar] [CrossRef]
- Tian, T.; Xiao, H.; Zhang, Z.; Long, Y.; Peng, S.; Wang, S.; Zhou, X.; Liu, S.; Zhou, X. Sensitive and convenient detection of microRNAs based on cascade amplification by catalytic dnazymes. Chem. Eur. J. 2013, 19, 92–95. [Google Scholar] [CrossRef]
- Wu, Z.; Zhou, H.; He, J.; Li, M.; Ma, X.; Xue, J.; Li, X.; Fan, X. G-triplex based molecular beacon with duplex-specific nuclease amplification for the specific detection of microRNA. Analyst 2019, 144, 5201–5206. [Google Scholar] [CrossRef]
- Cai, X.; Zhang, H.; Yu, X.; Wang, W. A microfluidic paper-based laser-induced fluorescence sensor based on duplex-specific nuclease amplification for selective and sensitive detection of miRNAs in cancer cells. Talanta 2020, 216, 120996. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ji, B.; Cheng, B.; Chen, J.; Bai, B. Neuroprotection of microRNA in neurological disorders (Review). Biomed. Rep. 2014, 2, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, D.S.; Soreq, H. MicroRNA therapeutics in neurological disease. Curr. Pharm. Des. 2014, 20, 6022–6027. [Google Scholar] [CrossRef] [PubMed]
- Çakmak, H.A.; Demir, M. MicroRNA and cardiovascular diseases. Balk. Med. J. 2020, 37, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Liu, M.; Tang, B.; Zhang, C.Y. Sensitive quantification of microRNAs by isothermal helicase-dependent amplification. Anal. Chem. 2017, 89, 6182–6187. [Google Scholar] [CrossRef]
- Li, Z.; Li, Z.; Zhao, D.; Wen, F.; Jiang, J.; Xu, D. Smartphone-based visualized microarray detection for multiplexed harmful substances in milk. Biosens. Bioelectron. 2017, 87, 874–880. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, L.; Wang, H.; Ji, W.; Zhang, Z.; Zhang, Y.; Yang, Z.; Cao, Z.; Zhang, S.; Chang, J. Intelligent detection platform for simultaneous detection of multiple miRNAs based on smartphone. ACS Sens. 2019, 4, 1873–1880. [Google Scholar] [CrossRef]
- Chen, X.; Xu, K.; Li, J.; Yang, M.; Li, X.; Chen, Q.; Lu, C.; Yang, H. Switch-conversional ratiometric fluorescence biosensor for miRNA detection. Biosens. Bioelectron. 2020, 155, 112104. [Google Scholar] [CrossRef]
- Degliangeli, F.; Kshirsagar, P.; Brunetti, V.; Pompa, P.P.; Fiammengo, R. Absolute and direct microRNA quantification using DNA-gold nanoparticle probes. J. Am. Chem. Soc. 2014, 136, 2264–2267. [Google Scholar] [CrossRef]
- Xi, Q.; Zhou, D.M.; Kan, Y.Y.; Ge, J.; Wu, Z.K.; Yu, R.Q.; Jiang, J.H. Highly sensitive and selective strategy for microRNA detection based on WS2 nanosheet mediated fluorescence quenching and duplex-specific nuclease signal amplification. Anal. Chem. 2014, 86, 1361–1365. [Google Scholar] [CrossRef]
- Ma, Y.; Chen, J.; Chen, D.; Xu, Y.; Zhang, L.; Dai, Z.; Zou, X. Short-probe-based duplex-specific nuclease signal amplification strategy enables imaging of endogenous microRNAs in living cells with ultrahigh specificity. Talanta 2018, 186, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Colomina, A.; Fernández, V.; Chinnappa, K.; Borrell, V. miRNAs in early brain development and pediatric cancer: At the intersection between healthy and diseased embryonic development. BioEssays 2021, 43, 2100073. [Google Scholar] [CrossRef] [PubMed]
- Rajgor, D.; Hanley, J.G. The ins and outs of miRNA-mediated gene silencing during neuronal synaptic plasticity. Non Coding RNA 2016, 2, 1. [Google Scholar] [CrossRef]
- Reddy, P.H. Increased mitochondrial fission and neuronal dysfunction in Huntington’s disease: Implications for molecular inhibitors of excessive mitochondrial fission. Drug Discov. Today 2014, 19, 951–955. [Google Scholar] [CrossRef] [PubMed]
- Buonfiglioli, A.; Efe, I.E.; Guneykaya, D.; Ivanov, A.; Huang, Y.; Orlowski, E.; Krüger, C.; Deisz, R.A.; Markovic, D.; Flüh, C.; et al. Let-7 microRNAs regulate microglial function and suppress glioma growth through toll-like receptor 7. Cell Rep. 2019, 29, 3460–3471.e7. [Google Scholar] [CrossRef] [PubMed]
- Derkow, K.; Rössling, R.; Schipke, C.; Krüger, C.; Bauer, J.; Fähling, M.; Stroux, A.; Schott, E.; Ruprecht, K.; Peters, O.; et al. Distinct expression of the neurotoxic microRNA family let-7 in the cerebrospinal fluid of patients with alzheimer’s disease. PLoS ONE 2018, 13, e0200602. [Google Scholar] [CrossRef] [PubMed]
- Shanesazzade, Z.; Peymani, M.; Ghaedi, K.; Nasr Esfahani, M.H. miR-34a/BCL-2 signaling axis contributes to apoptosis in MPP+-induced SH-SY5Y cells. Mol. Genet. Genom. Med. 2018, 6, 975–981. [Google Scholar] [CrossRef]
- Juzenas, S.; Venkatesh, G.; Hübenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A.; et al. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Liu, H.; Fan, J.; Liu, W.; Tong, C.; Xie, Z.; Deng, R.; Long, X. A dual signal amplification method for miR-204 assay by combining chimeric molecular beacon with double-stranded nuclease. Anal. Methods 2018, 10, 5834–5841. [Google Scholar] [CrossRef]
- Zada, S.; Lu, H.; Dai, W.; Tang, S.; Khan, S.; Yang, F.; Qiao, Y.; Fu, P.; Dong, H.; Zhang, X. Multiple amplified microRNAs monitoring in living cells based on fluorescence quenching of Mo2B and hybridization chain reaction. Biosens. Bioelectron. 2022, 197, 113815. [Google Scholar] [CrossRef]
- Chen, X.; Rong, Y.; Wang, H.; Zong, H.; Li, W. A mismatch-suppressed, duplex-specific nuclease powered nanowalker for multiplexed sensing of microRNA. Anal. Chim. Acta 2021, 1182, 338937. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Qin, L.; Yang, D.; Chen, W.; Qian, Y.; Jin, J. Signal amplification method for miR-205 assay through combining graphene oxide with duplex-specific nuclease. RSC Adv. 2019, 9, 27341–27346. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Howes, P.D.; Kim, E.; Spicer, C.D.; Thomas, M.R.; Lin, Y.; Crowder, S.W.; Pence, I.J.; Stevens, M.M. Duplex-specific nuclease-amplified detection of microRNA using compact quantum dot-DNA conjugates. ACS Appl. Mater. Interfaces 2018, 10, 28290–28300. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, C.; Mou, C. Duplex-specific nuclease and Exo-III enzyme-assisted signal amplification cooperating DNA-templated silver nanoclusters for label-free and sensitive miRNA detection. J. Anal. Sci. Technol. 2022, 13, 26. [Google Scholar] [CrossRef]
- Li, X.; Guo, Z.; Luo, G.; Miao, P. Fluorescence DNA switch for highly sensitive detection of miRNA amplified by duplex-specific nuclease. Sensors 2022, 22, 3252. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Liang, L.; Zhao, P.; Lan, F.; Zhang, L.; Ge, S.; Yu, J. Double signal amplification based on palladium nanoclusters and nucleic acid cycles on a μPAD for dual-model detection of microRNAs. J. Mater. Chem. B 2018, 6, 5795–5801. [Google Scholar] [CrossRef]
- Huang, J.; Shangguan, J.; Guo, Q.; Ma, W.; Wang, H.; Jia, R.; Ye, Z.; He, X.; Wang, K. Colorimetric and fluorescent dual-mode detection of microRNA based on duplex-specific nuclease assisted gold nanoparticle amplification. Analyst 2019, 144, 4917–4924. [Google Scholar] [CrossRef]
- Na, H.K.; Shon, H.K.; Son, H.Y.; Jang, E.; Joh, S.; Huh, Y.M.; Castner, D.G.; Lee, T.G. Utilization of chromogenic enzyme substrates for signal amplification in multiplexed detection of biomolecules using surface mass spectrometry. Sens. Actuators B Chem. 2021, 332, 129452. [Google Scholar] [CrossRef]
- Ashraf, G.; Chen, W.; Asif, M.; Aziz, A.; Zhong, Z.T.; Iftikhar, T.; Zhao, Y.D. Topical advancements in electrochemical and optical signal amplification for biomolecules detection: A comparison. Mater. Today Chem. 2022, 26, 101119. [Google Scholar] [CrossRef]
- Li, X.; Zhao, J.; Xu, R.; Pan, L.; Liu, Y.M. Mass Spectrometric quantification of microRNAs in biological samples based on multistage signal amplification. Analyst 2020, 145, 1783–1788. [Google Scholar] [CrossRef]
- Ashraf, G.; Asif, M.; Aziz, A.; Iftikhar, T.; Liu, H. Rice-spikelet-like copper oxide decorated with platinum stranded in the CNT network for electrochemical in vitro detection of serotonin. ACS Appl. Mater. Interfaces 2021, 13, 6023–6033. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, S.; Wang, Y.; He, Y.; Chai, Y.; Yuan, R. Stimuli-responsive DNA microcapsules for SERS sensing of trace microRNA. ACS Appl. Mater. Interfaces 2018, 10, 12491–12496. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wang, C.; Lu, L.C.; Wang, C.; Sun, Z.; Xiao, R. Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer. Biosens. Bioelectron. 2019, 130, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Yao, C.; Sun, L.; Li, Z. Plasmon-enhanced biosensors for microRNA analysis and cancer diagnosis. Biosens. Bioelectron. 2022, 203, 114041. [Google Scholar] [CrossRef] [PubMed]
- He, J.L.; Mei, T.T.; Tang, L.; Liao, S.Q.; Cao, Z. DSN/TdT recycling digestion based cyclic amplification strategy for microRNA Assay. Talanta 2020, 219, 121173. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Chu, H.; Zhu, L.; Xu, W. Duplex-specific nuclease-resistant triple-helix DNA nanoswitch for single-base differentiation of miRNA in lung cancer cells. Anal. Bioanal. Chem. 2020, 412, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lou, Z. Nasopharyngeal carcinoma related miRNA detection through a DSN enzyme assisted tetrahedral probe for more accurate prognosis. RSC Adv. 2021, 11, 11398–11402. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Rout, P.; Xu, R.; Pan, L.; Tchounwou, P.B.; Ma, Y.; Liu, Y.M. Quantification of microRNAs by coupling cyclic enzymatic amplification with microfluidic voltage-assisted liquid desorption electrospray ionization mass spectrometry. Anal. Chem. 2018, 90, 13663–13669. [Google Scholar] [CrossRef]
- Yao, Y.; Zhang, H.; Tian, T.; Liu, Y.; Zhu, R.; Ji, J.; Liu, B. Iodide-modified Ag nanoparticles coupled with DSN-assisted cycling amplification for label-free and ultrasensitive SERS detection of microRNA-21. Talanta 2021, 235, 122728. [Google Scholar] [CrossRef]
- Li, M.; Li, J.; Zhang, X.; Yao, M.; Li, P.; Xu, W. Simultaneous detection of tumor-related mRNA and miRNA in cancer cells with magnetic SERS nanotags. Talanta 2021, 232, 122432. [Google Scholar] [CrossRef]
- Zhu, L.; Zhang, M.; Ye, J.; Yan, M.; Zhu, Q.; Huang, J.; Yang, X. Ratiometric electrochemiluminescent/electrochemical strategy for sensitive detection of microRNA based on duplex-specific nuclease and multilayer circuit of catalytic hairpin assembly. Anal. Chem. 2020, 92, 8614–8622. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Yang, Y.; Lin, Y.; Chen, Z.; Xing, C.; Lu, C.; Yang, H.; Zhang, S. An electrochemical sensor based on enzyme-free recycling amplification for sensitive and specific detection of miRNAs from cancer cells. Analyst 2020, 145, 3353–3358. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.; Zhang, C.; Wang, C.; Zhou, C.; Gong, N.; Wang, Q.; Zhu, Y. DNA self-assembled FeNxC nanocatalytic network for ultrasensitive electrochemical detection of microRNA. Anal. Chim. Acta 2022, 1223, 340218. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Na, H.-K.; Lee, S.; Kim, W.-K. Advanced graphene oxide-based paper sensor for colorimetric detection of miRNA. Microchim. Acta 2022, 189, 35. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Yang, Z.; Chen, Y.; Zhang, L. Visual Detection of heart failure associated miRNA with DSN enzyme-based recycling amplification strategy. RSC Adv. 2021, 11, 18068–18073. [Google Scholar] [CrossRef] [PubMed]
- Hacia, J.G.; Fan, J.-B.; Ryder, O.; Jin, L.; Edgemon, K.; Ghandour, G.; Mayer, R.A.; Sun, B.; Hsie, L.; Robbins, C.M.; et al. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 1999, 22, 164–167. [Google Scholar] [CrossRef]
- Phillips, C.; Fondevila, M.; García-Magariños, M.; Rodriguez, A.; Salas, A.; Carracedo, Á.; Lareu, M.V. Resolving relationship tests that show ambiguous STR results using autosomal SNPs as supplementary markers. Forensic Sci. Int. Genet. 2008, 2, 198–204. [Google Scholar] [CrossRef]
- Shagin, D.A.; Rebrikov, D.V.; Kozhemyako, V.B.; Altshuler, I.M.; Shcheglov, A.S.; Zhulidov, P.A.; Bogdanova, E.A.; Staroverov, D.B.; Rasskazov, V.A.; Lukyanov, S. A novel method for SNP detection using a new duplex-specific nuclease from Crab Hepatopancreas. Genome Res. 2002, 12, 1935–1942. [Google Scholar] [CrossRef]
- Brody, T. Biomarkers. In Clinical Trials, 2nd ed.; Academic Press: Cambridge, MA, USA, 2016; pp. 377–419. [Google Scholar] [CrossRef]
- Michailidou, K.; Hall, P.; Gonzalez-Neira, A.; Ghoussaini, M.; Dennis, J.; Milne, R.L.; Schmidt, M.K.; Chang-Claude, J.; Bojesen, S.E.; Bolla, M.K.; et al. Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat. Genet. 2013, 45, 353–361. [Google Scholar] [CrossRef]
- Altshuler, I.M.; Zhulidov, P.A.; Bogdanova, E.A.; Mudrik, N.N.; Shagin, D.A. Application of the duplex-specific nuclease preference method to the analysis of single nucleotide polymorphisms in human genes. Russ. J. Bioorg. Chem. 2005, 31, 567–575. [Google Scholar] [CrossRef]
- Camchong, J.; Dyckman, K.A.; Austin, B.P.; Clementz, B.A.; McDowell, J.E. Common neural circuitry supporting volitional saccades and its disruption in schizophrenia patients and relatives. Biol. Psychiatry 2008, 64, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Sanders, A.R.; Rusu, I.; Duan, J.; Vander Molen, J.E.; Hou, C.; Schwab, S.G.; Wildenauer, D.B.; Martinez, M.; Gejman, P.V. Haplotypic association spanning the 22q11.21 genes COMT and ARVCF with schizophrenia. Mol. Psychiatry 2005, 10, 353–365. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zou, Y.; Xiao, J.; Pan, C.; Jiang, S.; Zheng, Z.; Yan, Z.; Tang, K.; Tan, L.; Tang, M. COMT Val158Met polymorphism and Parkinson’s disease risk: A pooled analysis in different populations. Neurol. Res. 2019, 41, 319–325. [Google Scholar] [CrossRef]
- Shagina, I.A.; Bogdanova, E.A.; Altshuler, I.M.; Luk’yanov, S.A.; Shagin, D.A. The use of duplex-specific Crab nuclease for rapid analysis of single-nucleotide polymorphisms and the detection of DNA targets in complex PCR products. Russ. J. Bioorg. Chem. 2011, 37, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Goto, T.; Hirotsu, Y.; Moriyama, M.; Omata, M. Molecular mechanisms driving progression of liver cirrhosis towards hepatocellular carcinoma in chronic hepatitis B and C infections: A review. Int. J. Mol. Sci. 2019, 20, 1358. [Google Scholar] [CrossRef] [PubMed]
- Coppola, N.; Tonziello, G.; Colombatto, P.; Pisaturo, M.; Messina, V.; Moriconi, F.; Alessio, L.; Sagnelli, C.; Cavallone, D.; Brunetto, M.; et al. Lamivudine-resistant HBV strain rtM204V/I in acute hepatitis B. J. Infect. 2013, 67, 322–328. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.U.; Yoshimura, Y.; Mosharraf Hossain, M.; Tamiya, E.; Fujimoto, K. Construction of branched DNA for SNP determination on glass-chip using photochemical ligation. BioChip J. 2011, 5, 206–213. [Google Scholar] [CrossRef]
- Tonosaki, K.; Kudo, J.; Kitashiba, H.; Nishio, T. Allele-specific hybridization using streptavidin-coated magnetic beads for species identification, S genotyping, and SNP analysis in plants. Mol. Breed. 2013, 31, 419–428. [Google Scholar] [CrossRef]
- Liu, M.; Yuan, M.; Lou, X.; Mao, H.; Zheng, D.; Zou, R.; Zou, N.; Tang, X.; Zhao, J. Label-free optical detection of single-base mismatches by the combination of nuclease and gold nanoparticles. Biosens. Bioelectron. 2011, 26, 4294–4300. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ashraf, G.; Zhong, Z.-T.; Asif, M.; Aziz, A.; Iftikhar, T.; Chen, W.; Zhao, Y.-D. State-of-the-Art Fluorescent Probes: Duplex-Specific Nuclease-Based Strategies for Early Disease Diagnostics. Biosensors 2022, 12, 1172. https://doi.org/10.3390/bios12121172
Ashraf G, Zhong Z-T, Asif M, Aziz A, Iftikhar T, Chen W, Zhao Y-D. State-of-the-Art Fluorescent Probes: Duplex-Specific Nuclease-Based Strategies for Early Disease Diagnostics. Biosensors. 2022; 12(12):1172. https://doi.org/10.3390/bios12121172
Chicago/Turabian StyleAshraf, Ghazala, Zi-Tao Zhong, Muhammad Asif, Ayesha Aziz, Tayyaba Iftikhar, Wei Chen, and Yuan-Di Zhao. 2022. "State-of-the-Art Fluorescent Probes: Duplex-Specific Nuclease-Based Strategies for Early Disease Diagnostics" Biosensors 12, no. 12: 1172. https://doi.org/10.3390/bios12121172
APA StyleAshraf, G., Zhong, Z.-T., Asif, M., Aziz, A., Iftikhar, T., Chen, W., & Zhao, Y.-D. (2022). State-of-the-Art Fluorescent Probes: Duplex-Specific Nuclease-Based Strategies for Early Disease Diagnostics. Biosensors, 12(12), 1172. https://doi.org/10.3390/bios12121172






