Optical Methods for Label-Free Detection of Bacteria
Abstract
:1. Introduction
2. Surface Plasmon Resonance for Bacteria Detection
2.1. Principle of Surface Plasmon Resonance
2.2. Method and Application of SPR Technology for Label-Free Detection of Bacteria
3. Raman Spectroscopy for Pathogen Bacteria Detection
3.1. The Principle of Raman Spectroscopy
3.2. Label-Free Detection of Bacteria by Raman Spectroscopy
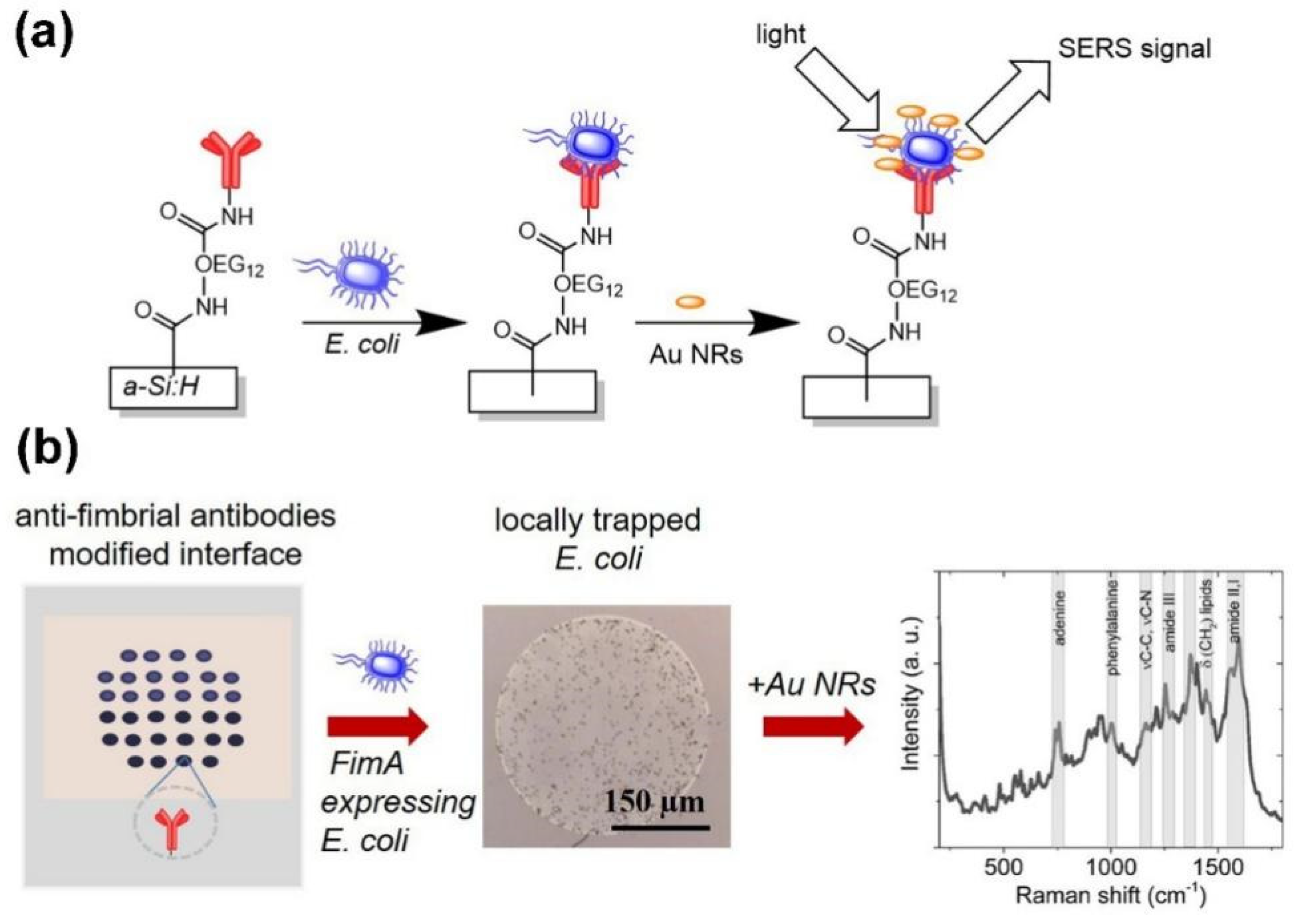
3.3. Label-Free Detection of Bacteria by SERS
3.3.1. In Situ Formation of Colloidal Silver/Gold on the Surface of Bacteria
3.3.2. Direct Bacteria Detection on a Planar SERS Surface
3.3.3. Direct Bacteria Detection in SERS Suspension
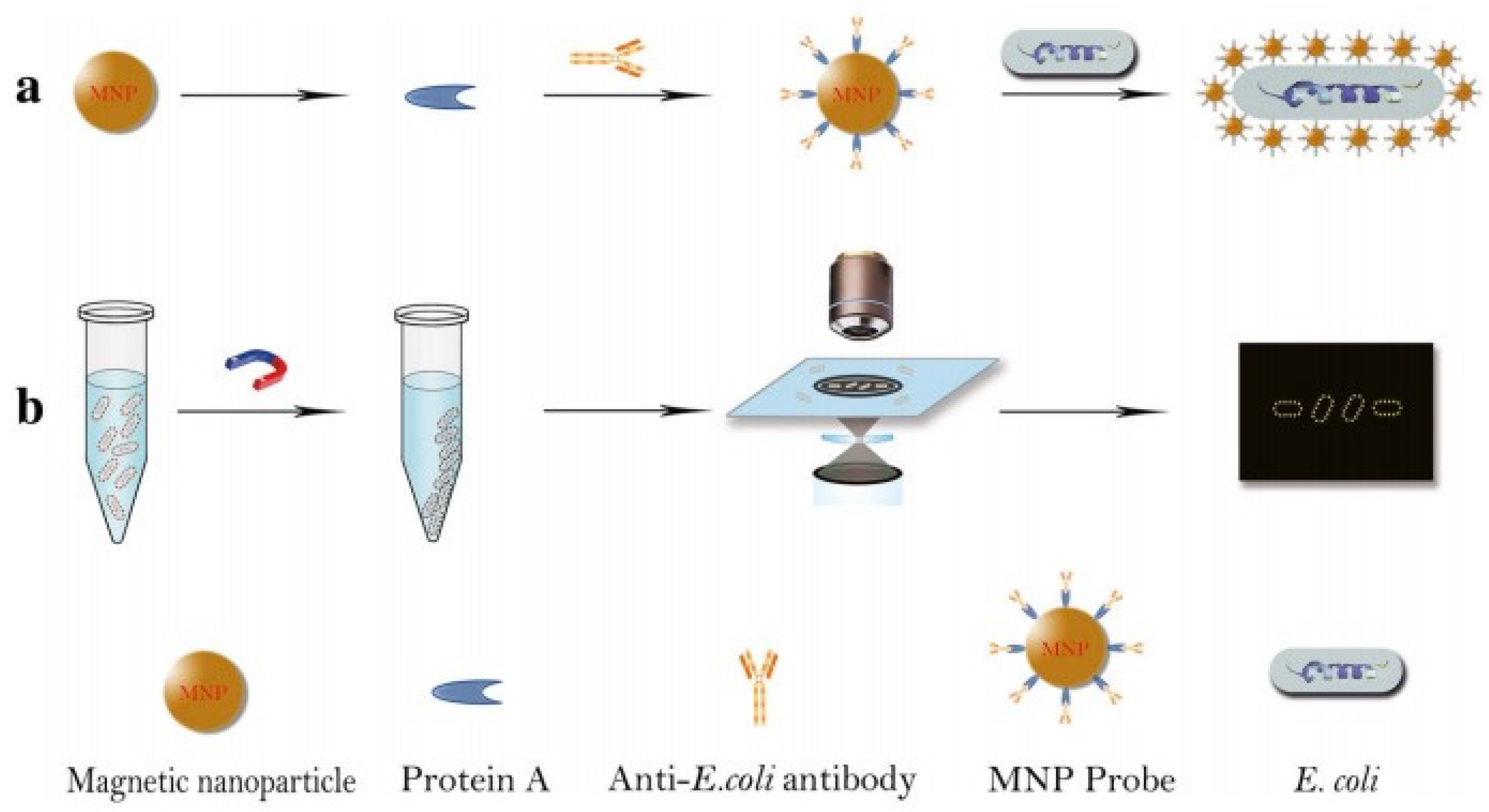
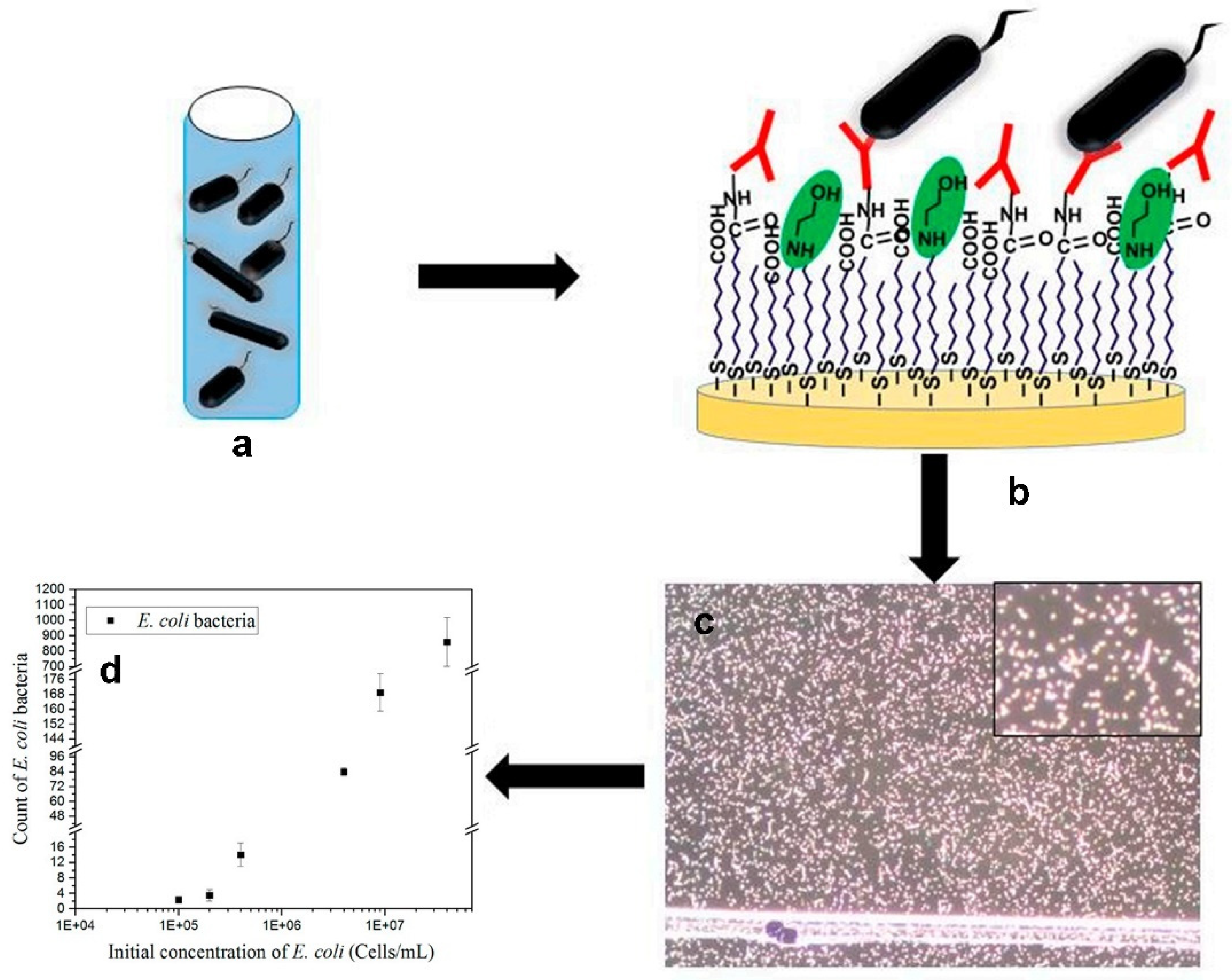
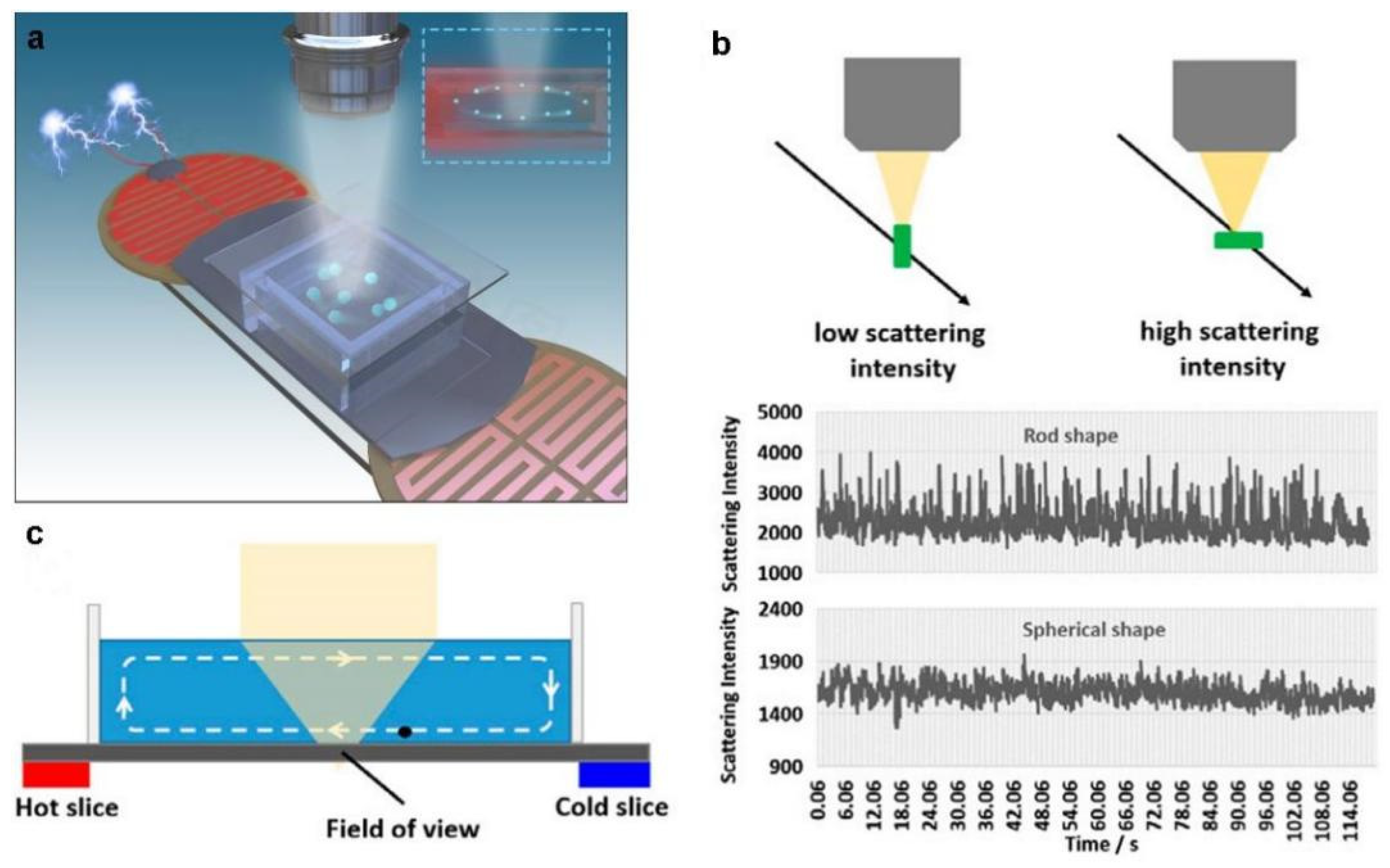
4. Label-Free Detection of Bacteria by Dark-Field Microscopy
4.1. Dark-Field Microscopy Imaging Principle
4.2. Label and Label-Free Detection of Bacteria by Dark-Field Microscopy
5. Other Methods for Label-Free Detection of Bacteria
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drlica, K.; Zhao, X. Bacterial death from treatment with fluoroquinolones and other lethal stressors. Expert Rev. Anti-Infect. Ther. 2021, 19, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, S.H.E.; Dorhoi, A.; Hotchkiss, R.S.; Bartenschlager, R. Host-directed therapies for bacterial and viral infections. Nat. Rev. Drug Discov. 2018, 17, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; de Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, e1001923. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Han, C.; Chen, J.; Huang, Y.-W.; Zhao, Y. Rapid Detection of Pathogenic Bacteria from Fresh Produce by Filtration and Surface-Enhanced Raman Spectroscopy. J. Miner. 2016, 68, 1156–1162. [Google Scholar] [CrossRef]
- Tsalik, E.L.; Bonomo, R.A.; Fowler, V.G., Jr. New Molecular Diagnostic Approaches to Bacterial Infections and Antibacterial Resistance. Annu. Rev. Med. 2018, 69, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Piano, S.; Bartoletti, M.; Tonon, M.; Baldassarre, M.; Chies, G.; Romano, A.; Viale, P.; Vettore, E.; Domenicali, M.; Stanco, M.; et al. Assessment of Sepsis-3 criteria and quick SOFA in patients with cirrhosis and bacterial infections. Gut 2018, 67, 1892–1899. [Google Scholar] [CrossRef]
- Seale, A.C.; Gordon, N.C.; Islam, J.; Peacock, S.J.; Scott, J.A.G. AMR Surveillance in low and middle-income settings—A roadmap for participation in the Global Antimicrobial Surveillance System (GLASS). Wellcome Open Res. 2017, 2, 92. [Google Scholar] [CrossRef]
- Pang, X.; Xiao, Q.; Cheng, Y.; Ren, E.; Lian, L.; Zhang, Y.; Gao, H.; Wang, X.; Leung, W.; Chen, X.; et al. Bacteria-Responsive Nanoliposomes as Smart Sonotheranostics for Multidrug Resistant Bacterial Infections. ACS Nano 2019, 13, 2427–2438. [Google Scholar] [CrossRef]
- Maragakis, L.L.; Perencevich, E.N.; Cosgrove, S.E. Clinical and economic burden of antimicrobial resistance. Expert Rev. Anti-Infect. Ther. 2008, 6, 751–763. [Google Scholar] [CrossRef]
- Grant, S.S.; Hung, D.T. Persistent bacterial infections, antibiotic tolerance, and the oxidative stress response. Virulence 2013, 4, 273–283. [Google Scholar] [CrossRef]
- Sheikhzadeh, E.; Chamsaz, M.; Turner, A.P.F.; Jager, E.W.H.; Beni, V. Label-free impedimetric biosensor for Salmonella Typhimurium detection based on poly [pyrrole-co-3-carboxyl-pyrrole] copolymer supported aptamer. Biosens. Bioelectron. 2016, 80, 194–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakvetadze, C.; Purcarea, A.; Pitsch, A.; Chelly, J.; Diamantis, S. Detection of Fusobacterium nucleatum in culture-negative brain abscess by broad-spectrum bacterial 16S rRNA Gene PCR. IDCases 2017, 8, 94–95. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Yuan, Y.H.; Niu, C.; Qiu, Y.; Wei, J.P.; Yue, T.L. Development of an indirect enzyme-linked immunosorbent assay for the detection of osmotolerant yeast Zygosaccharomyces rouxii in different food. Food Agric. Immunol. 2018, 29, 976–988. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.; Wu, T.Z.; Adams-Sapper, S.; Satoorian, T.; Geisberg, M.; Murthy, N.; Lee, L.; Riley, L.W. A multiplexed, indirect enzyme-linked immunoassay for the detection and differentiation of E. coli from other Enterobacteriaceae and P. aeruginosa from other glucose non-fermenters. J. Microbiol. Methods 2019, 158, 52–58. [Google Scholar] [CrossRef]
- Szunerits, S. Editorial Overview: Sensors and Biosensors (2021) Opening up the field to medical biosensors. Curr. Opin. Electrochem. 2021, 30, 100864. [Google Scholar] [CrossRef]
- Das, R.; Chaterjee, B.; Kapil, A.; Sharma, T.K. Aptamer-NanoZyme mediated sensing platform for the rapid detection of Escherichia coli in fruit juice. Sens. Bio-Sens. Res. 2020, 27, 100313. [Google Scholar] [CrossRef]
- Dong, X.; Shi, Z.; Xu, C.; Yang, C.; Chen, F.; Lei, M.; Wang, J.; Cui, Q. CdS quantum dots/Au nanoparticles/ZnO nanowire array for self-powered photoelectrochemical detection of Escherichia coli O157:H7. Biosens. Bioelectron. 2020, 149, 111843. [Google Scholar] [CrossRef]
- Bai, H.; Bu, S.; Liu, W.; Wang, C.; Li, Z.; Hao, Z.; Wan, J.; Han, Y. An electrochemical aptasensor based on cocoon-like DNA nanostructure signal amplification for the detection of Escherichia coli O157:H7. Analyst 2020, 145, 7340–7348. [Google Scholar] [CrossRef]
- Zhang, J.; Oueslati, R.; Cheng, C.; Zhao, L.; Chen, J.; Almeida, R.; Wu, J. Rapid, highly sensitive detection of Gram-negative bacteria with lipopolysaccharide based disposable aptasensor. Biosens. Bioelectron. 2018, 112, 48–53. [Google Scholar] [CrossRef] [Green Version]
- Kaur, H.; Shorie, M.; Sharma, M.; Ganguli, A.K.; Sabherwal, P. Bridged Rebar Graphene functionalized aptasensor for pathogenic E. coil O78:K80:H11 detection. Biosens. Bioelectron. 2017, 98, 486–493. [Google Scholar] [CrossRef]
- Cai, R.; Zhang, Z.; Chen, H.; Tian, Y.; Zhou, N. A versatile signal-on electrochemical biosensor for Staphylococcus aureus based on triple-helix molecular switch. Sens. Actuators B-Chem. 2021, 326, 128842. [Google Scholar] [CrossRef]
- Ranjbar, S.; Shahrokhian, S. Design and fabrication of an electrochemical aptasensor using Au nanoparticles/carbon nanoparticles/cellulose nanofibers nanocomposite for rapid and sensitive detection of Staphylococcus aureus. Bioelectrochemistry 2018, 123, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Sun, Z.; Guo, Q.; Weng, X. Microfluidic thread-based electrochemical aptasensor for rapid detection of Vibrio parahaemolyticus. Biosens. Bioelectron. 2021, 182, 113191. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yao, C.; Yang, C.; Liu, Z.; Wan, S. Development of an in-situ signal amplified electrochemical assay for detection of Listeria monocytogenes with label-free strategy. Food Chem. 2021, 358, 129894. [Google Scholar] [CrossRef]
- Pham, T.-T.D.; Phan, L.M.T.; Park, J.; Cho, S. Review-Electrochemical Aptasensor for Pathogenic Bacteria Detection. J. Electrochem. Soc. 2022, 169, 087501. [Google Scholar] [CrossRef]
- Jones, L.M.; Dunham, D.; Rennie, M.Y.; Kirman, J.; Lopez, A.J.; Keim, K.C.; Little, W.; Gomez, A.; Bourke, J.; Ng, H.; et al. In vitro detection of porphyrin-producing wound bacteria with real-time fluorescence imaging. Future Microbiol. 2020, 15, 319–332. [Google Scholar] [CrossRef] [Green Version]
- Sparks, W.B.; Hough, J.; Germer, T.A.; Chen, F.; DasSarma, S.; DasSarma, P.; Robb, F.T.; Manset, N.; Kolokolova, L.; Reid, N.; et al. Detection of circular polarization in light scattered from photosynthetic microbes. Proc. Natl. Acad. Sci. USA 2009, 106, 7816–7821. [Google Scholar] [CrossRef] [Green Version]
- Martak, D.; Valot, B.; Sauget, M.; Cholley, P.; Thouverez, M.; Bertrand, X.; Hocquet, D. Fourier-Transform InfraRed Spectroscopy Can Quickly Type Gram-Negative Bacilli Responsible for Hospital Outbreaks. Front. Microbiol. 2019, 10, 1440. [Google Scholar] [CrossRef] [Green Version]
- Bagcioglu, M.; Fricker, M.; Johler, S.; Ehling-Schulz, M. Detection and Identification of Bacillus cereus, Bacillus cytotoxicus, Bacillus thuringiensis, Bacillus mycoides and Bacillus weihenstephanensis via Machine Learning Based FTIR Spectroscopy. Front. Microbiol. 2019, 10, 902. [Google Scholar] [CrossRef]
- Vogt, S.; Loeffler, K.; Dinkelacker, A.G.; Bader, B.; Autenrieth, I.B.; Peter, S.; Liese, J. Fourier-Transform Infrared (FTIR) Spectroscopy for Typing of Clinical Enterobacter cloacae Complex Isolates. Front. Microbiol. 2019, 10, 2582. [Google Scholar] [CrossRef]
- Lechowicz, L.; Urbaniak, M.; Adamus-Bialek, W.; Kaca, W. The use of infrared spectroscopy and artificial neural networks for detection of uropathogenic Escherichia coli strains’ susceptibility to cephalothin. Acta Biochim. Pol. 2013, 60, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Ayala, O.D.; Doster, R.S.; Manning, S.D.; O’Brien, C.M.; Aronoff, D.M.; Gaddy, J.A.; Mahadevan-Jansen, A. Raman microspectroscopy differentiates perinatal pathogens on ex vivo infected human fetal membrane tissues. J. Biophotonics 2019, 12, e201800449. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, U.-C.; Bokeloh, F.; O’Sullivan, M.; Glaser, U.; Wolf, K.; Pfister, W.; Popp, J.; Ducree, J.; Neugebauer, U. Rapid, culture-independent, optical diagnostics of centrifugally captured bacteria from urine samples. Biomicrofluidics 2015, 9, 044118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, O.D.; Wakeman, C.A.; Pence, I.J.; Gaddy, J.A.; Slaughter, J.C.; Skaar, E.P.; Mahadevan-Jansen, A. Drug-Resistant Staphylococcus aureus Strains Reveal Distinct Biochemical Features with Raman Microspectroscopy. ACS Infect. Dis. 2018, 4, 1197–1210. [Google Scholar] [CrossRef] [PubMed]
- Neugebauer, U.; Schmid, U.; Baumann, K.; Holzgrabe, U.; Ziebuhr, W.; Kozitskaya, S.; Kiefer, W.; Schmitt, M.; Popp, J. Characterization of bacterial growth and the influence of antibiotics by means of UV resonance Raman spectroscopy. Biopolymers 2006, 82, 306–311. [Google Scholar] [CrossRef]
- Wang, Y.; Lee, K.; Irudayaraj, J. Silver Nanosphere SERS Probes for Sensitive Identification of Pathogens. J. Phys. Chem. C 2010, 114, 16122–16128. [Google Scholar] [CrossRef]
- Premasiri, W.R.; Chen, Y.; Williamson, P.M.; Bandarage, D.C.; Pyles, C.; Ziegler, L.D. Rapid urinary tract infection diagnostics by surface-enhanced Raman spectroscopy (SERS): Identification and antibiotic susceptibilities. Anal. Bioanal. Chem. 2017, 409, 3043–3054. [Google Scholar] [CrossRef]
- Boardman, A.K.; Wong, W.S.; Premasiri, W.R.; Ziegler, L.D.; Lee, J.C.; Miljkovic, M.; Klapperich, C.M.; Sharon, A.; Sauer-Budge, A.F. Rapid Detection of Bacteria from Blood with Surface-Enhanced Raman Spectroscopy. Anal. Chem. 2016, 88, 8026–8035. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, X.; Wang, C.; Rong, Z.; Ding, H.; Li, H.; Li, S.; Shao, N.; Dong, P.; Xiao, R.; et al. Facile Synthesis of Au-Coated Magnetic Nanoparticles and Their Application in Bacteria Detection via a SERS Method. ACS Appl. Mater. Interfaces 2016, 8, 19958–19967. [Google Scholar] [CrossRef]
- La Spina, R.; António, D.C.; Desmet, C.; Valsesia, A.; Bombera, R.; Norlén, H.; Lettieri, T.; Colpo, P. Dark Field Microscopy-Based Biosensors for the Detection of E. coli in Environmental Water Samples. Sensors 2019, 19, 4652. [Google Scholar] [CrossRef]
- Wang, B.; Park, B. Immunoassay Biosensing of Foodborne Pathogens with Surface Plasmon Resonance Imaging: A Review. J. Agric. Food Chem. 2020, 68, 12927–12939. [Google Scholar] [CrossRef] [PubMed]
- Bodelón, G.; Montes-García, V.; Pérez-Juste, J.; Pastoriza-Santos, I. Surface-Enhanced Raman Scattering Spectroscopy for Label-Free Analysis of P. aeruginosa Quorum Sensing. Front. Cell. Infect. Microbiol. 2018, 8, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, Y.P.; Wang, W.; Shen, Y.; Guo, J.S. Surface plasmon resonance for water pollutant detection and water process analysis. Trac-Trends Anal. Chem. 2016, 85, 153–165. [Google Scholar] [CrossRef]
- Taghipour, P.; Zakariazadeh, M.; Sharifi, M.; Ezzati Nazhad Dolatabadi, J.; Barzegar, A. Bovine serum albumin binding study to erlotinib using surface plasmon resonance and molecular docking methods. J. Photochem. Photobiol. B Biol. 2018, 183, 11–15. [Google Scholar] [CrossRef]
- Lewis, T.; Giroux, E.; Jovic, M.; Martic-Milne, S. Localized surface plasmon resonance aptasensor for selective detection of SARS-CoV-2 S1 protein. Analyst 2021, 146, 7207–7217. [Google Scholar] [CrossRef]
- Xue, J.; Bai, Y.; Liu, H. Hybrid methods of surface plasmon resonance coupled to mass spectrometry for biomolecular interaction analysis. Anal. Bioanal. Chem. 2019, 411, 3721–3729. [Google Scholar] [CrossRef]
- Stojanović, I.; Ruivo, C.F.; van der Velden, T.J.G.; Schasfoort, R.B.M.; Terstappen, L. Multiplex Label Free Characterization of Cancer Cell Lines Using Surface Plasmon Resonance Imaging. Biosensors 2019, 9, 70. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Qi, Q.; Wang, C.; Qian, Y.; Liu, G.; Wang, Y.; Fu, L. Surface plasmon resonance (SPR) biosensors for food allergen detection in food matrices. Biosens. Bioelectron. 2019, 142, 111449. [Google Scholar] [CrossRef]
- Choi, H.J.; Chung, B.H.; Kim, Y. Analysis of Protein-Protein Interactions by Surface Plasmon Resonance Imaging-based Microwell and Microfluidic Chip. Bull. Korean Chem. Soc. 2016, 37, 752–755. [Google Scholar] [CrossRef]
- Day, C.J.; Poole, J.; Pluschke, G.; Jennings, M.P. Investigation of Mycobacterium ulcerans Glycan Interactions Using Glycan Array and Surface Plasmon Resonance. Methods Mol. Biol. 2022, 2387, 29–40. [Google Scholar]
- Syal, K.; Iriya, R.; Yang, Y.; Yu, H.; Wang, S.; Haydel, S.E.; Chen, H.Y.; Tao, N. Antimicrobial Susceptibility Test with Plasmonic Imaging and Tracking of Single Bacterial Motions on Nanometer Scale. ACS Nano 2016, 10, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, H.; Sakai, Y.; Mir, T.A. Real-time monitoring of intracellular signal transduction in PC12 cells by two-dimensional surface plasmon resonance imager. Anal. Biochem. 2013, 441, 185–189. [Google Scholar] [CrossRef] [PubMed]
- Yanase, Y.; Hiragun, T.; Yanase, T.; Kawaguchi, T.; Ishii, K.; Hide, M. Evaluation of peripheral blood basophil activation by means of surface plasmon resonance imaging. Biosens. Bioelectron. 2012, 32, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Prakash, A.; Tripathi, R. Performance analysis of graphene based surface plasmon resonance biosensors for detection of pseudomonas-like bacteria. Opt. Quantum Electron. 2015, 47, 1197–1205. [Google Scholar] [CrossRef]
- Chen, J.; Park, B. Label-free screening of foodborne Salmonella using surface plasmon resonance imaging. Anal. Bioanal. Chem. 2018, 410, 5455–5464. [Google Scholar] [CrossRef]
- Park, B.; Wang, B.; Chen, J. Label-Free Immunoassay for Multiplex Detections of Foodborne Bacteria in Chicken Carcass Rinse with Surface Plasmon Resonance Imaging. Foodborne Pathog. Dis. 2021, 18, 202–209. [Google Scholar] [CrossRef]
- Skottrup, P.D.; Nicolaisen, M.; Justesen, A.F. Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron. 2008, 24, 339–348. [Google Scholar] [CrossRef]
- Jenkins, A.T.A.; Ffrench-Constant, R.; Buckling, A.; Clarke, D.J.; Jarvis, K. Study of the attachment of Pseudomonas aeruginosa on gold and modified gold surfaces using surface plasmon resonance. Biotechnol. Prog. 2004, 20, 1233–1236. [Google Scholar] [CrossRef]
- Bulard, E.; Bouchet-Spinelli, A.; Chaud, P.; Roget, A.; Calemczuk, R.; Fort, S.; Livache, T. Carbohydrates as New Probes for the Identification of Closely Related Escherichia coli Strains Using Surface Plasmon Resonance Imaging. Anal. Chem. 2015, 87, 1804–1811. [Google Scholar] [CrossRef]
- Shankaran, D.R.; Gobi, K.V.A.; Miura, N. Recent advancements in surface plasmon resonance immunosensors for detection of small molecules of biomedical, food and environmental interest. Sens. Actuators B-Chem. 2007, 121, 158–177. [Google Scholar] [CrossRef]
- Puttharugsa, C.; Wangkam, T.; Huangkamhang, N.; Gajanandana, O.; Himananto, O.; Sutapun, B.; Amarit, R.; Somboonkaew, A.; Srikhirin, T. Development of surface plasmon resonance imaging for detection of Acidovorax avenae subsp. citrulli (Aac) using specific monoclonal antibody. Biosens. Bioelectron. 2011, 26, 2341–2346. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Arya, S.K.; Glass, N.; Hanifi-Moghaddam, P.; Naidoo, R.; Szymanski, C.M.; Tanha, J.; Evoy, S. Bacteriophage tailspike proteins as molecular probes for sensitive and selective bacterial detection. Biosens. Bioelectron. 2010, 26, 131–138. [Google Scholar] [CrossRef] [PubMed]
- Syal, K.; Wang, W.; Shan, X.; Wang, S.; Chen, H.Y.; Tao, N. Plasmonic imaging of protein interactions with single bacterial cells. Biosens. Bioelectron. 2015, 63, 131–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morlay, A.; Piat, F.; Mercey, T.; Roupioz, Y. Immunological detection of Cronobacter and Salmonella in powdered infant formula by plasmonic label-free assay. Lett. Appl. Microbiol. 2016, 62, 459–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouguelia, S.; Roupioz, Y.; Slimani, S.; Mondani, L.; Casabona, M.G.; Durmort, C.; Vernet, T.; Calemczuk, R.; Livache, T. On-chip microbial culture for the specific detection of very low levels of bacteria. Lab. Chip 2013, 13, 4024–4032. [Google Scholar] [CrossRef] [Green Version]
- Mondani, L.; Roupioz, Y.; Delannoy, S.; Fach, P.; Livache, T. Simultaneous enrichment and optical detection of low levels of stressed Escherichia coli O157:H7 in food matrices. J. Appl. Microbiol. 2014, 117, 537–546. [Google Scholar] [CrossRef]
- Raikwar, S.; Prajapati, Y.K.; Srivastava, D.K.; Maurya, J.B.; Saini, J.P. Detection of Leptospirosis Bacteria in Rodent Urine by Surface Plasmon Resonance Sensor Using Graphene. Photonic Sens. 2021, 11, 305–313. [Google Scholar] [CrossRef]
- Maurya, J.B.; Prajapati, Y.K.; Tripathi, R. Effect of Molybdenum Disulfide Layer on Surface Plasmon Resonance Biosensor for the Detection of Bacteria. Silicon 2018, 10, 245–256. [Google Scholar] [CrossRef]
- Arya, S.K.; Singh, A.; Naidoo, R.; Wu, P.; McDermott, M.T.; Evoy, S. Chemically immobilized T4-bacteriophage for specific Escherichia coli detection using surface plasmon resonance. Analyst 2011, 136, 486–492. [Google Scholar] [CrossRef]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Surface plasmon resonance detection of E. coli and methicillin-resistant S. aureus using bacteriophages. Biosens. Bioelectron. 2012, 37, 24–29. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Zhang, H.; Liu, W.; Ge, J.; Wu, J.; Wang, P. High sensitivity gram-negative bacteria biosensor based on a small-molecule modified surface plasmon resonance chip studied using a laser scanning confocal imaging-surface plasmon resonance system. Sens. Actuators B-Chem. 2018, 259, 492–497. [Google Scholar] [CrossRef]
- Dursun, A.D.; Borsa, B.A.; Bayramoglu, G.; Arica, M.Y.; Ozalp, V.C. Surface plasmon resonance aptasensor for Brucella detection in milk. Talanta 2022, 239, 123074. [Google Scholar] [CrossRef] [PubMed]
- Rebrosova, K.; Siler, M.; Samek, O.; Ruzicka, F.; Bernatova, S.; Hola, V.; Jezek, J.; Zemanek, P.; Sokolova, J.; Petras, P. Rapid identification of staphylococci by Raman spectroscopy. Sci. Rep. 2017, 7, 14846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willemse-Erix, D.F.M.; Scholtes-Timmerman, M.J.; Jachtenberg, J.-W.; van Leeuwen, W.B.; Horst-Kreft, D.; Schut, T.C.B.; Deurenberg, R.H.; Puppels, G.J.; van Belkum, A.; Vos, M.C.; et al. Optical Fingerprinting in Bacterial Epidemiology: Raman Spectroscopy as a Real-Time Typing Method. J. Clin. Microbiol. 2009, 47, 652–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayala, O.D.; Wakeman, C.A.; Pence, I.J.; O’Brien, C.M.; Werkhaven, J.A.; Skaar, E.P.; Mahadevan-Jansen, A. Characterization of bacteria causing acute otitis media using Raman microspectroscopy. Anal. Methods 2017, 9, 1864–1871. [Google Scholar] [CrossRef] [PubMed]
- Kloss, S.; Lorenz, B.; Dees, S.; Labugger, I.; Roesch, P.; Popp, J. Destruction-free procedure for the isolation of bacteria from sputum samples for Raman spectroscopic analysis. Anal. Bioanal. Chem. 2015, 407, 8333–8341. [Google Scholar] [CrossRef]
- Kloss, S.; Kampe, B.; Sachse, S.; Roesch, P.; Straube, E.; Pfister, W.; Kiehntopf, M.; Popp, J. Culture Independent Raman Spectroscopic Identification of Urinary Tract Infection Pathogens: A Proof of Principle Study. Anal. Chem. 2013, 85, 9610–9616. [Google Scholar] [CrossRef]
- Harz, M.; Kiehntopf, M.; Stoeckel, S.; Roesch, P.; Straube, E.; Deufel, T.; Popp, J. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophotonics 2009, 2, 70–80. [Google Scholar] [CrossRef]
- Hakonen, A.; Andersson, P.O.; Schmidt, M.S.; Rindzevicius, T.; Kall, M. Explosive and chemical threat detection by surface-enhanced Raman scattering: A review. Anal. Chim. Acta 2015, 893, 1–13. [Google Scholar] [CrossRef]
- Popp, J.; Mayerhöfer, T. Surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2009, 394, 1717–1718. [Google Scholar] [CrossRef] [Green Version]
- Nikoobakht, B.; Wang, J.P.; El-Sayed, M.A. Surface-enhanced Raman scattering of molecules adsorbed on gold nanorods: Off-surface plasmon resonance condition. Chem. Phys. Lett. 2002, 366, 17–23. [Google Scholar] [CrossRef]
- Fromm, D.P.; Sundaramurthy, A.; Kinkhabwala, A.; Schuck, P.J.; Kino, G.S.; Moerner, W.E. Exploring the chemical enhancement for surface-enhanced Raman scattering with Au bowtie nanoantennas. J. Chem. Phys. 2006, 124, 61101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.L.; Jensen, L.; Schatz, G.C. Surface-enhanced Raman scattering of pyrazine at the junction between two Ag-20 nanoclusters. Nano Lett. 2006, 6, 1229–1234. [Google Scholar] [CrossRef] [PubMed]
- Ko, H.; Singamaneni, S.; Tsukruk, V.V. Nanostructured Surfaces and Assemblies as SERS Media. Small 2008, 4, 1576–1599. [Google Scholar] [CrossRef]
- Halas, N.J.; Lal, S.; Chang, W.-S.; Link, S.; Nordlander, P. Plasmons in Strongly Coupled Metallic Nanostructures. Chem. Rev. 2011, 111, 3913–3961. [Google Scholar] [CrossRef]
- Schlücker, S. Surface-Enhanced Raman spectroscopy: Concepts and chemical applications. Angew. Chem. Int. Ed. 2014, 53, 4756–4795. [Google Scholar] [CrossRef]
- Prucek, R.; Ranc, V.; Kvítek, L.; Panáček, A.; Zbořil, R.; Kolář, M. Reproducible discrimination between gram-positive and gram-negative bacteria using surface enhanced Raman spectroscopy with infrared excitation. Analyst 2012, 137, 2866–2870. [Google Scholar] [CrossRef]
- Yu, R.; Liz-Marzán, L.M.; García de Abajo, F.J. Universal analytical modeling of plasmonic nanoparticles. Chem. Soc. Rev. 2017, 46, 6710–6724. [Google Scholar] [CrossRef] [Green Version]
- Gwo, S.; Chen, H.Y.; Lin, M.H.; Sun, L.; Li, X. Nanomanipulation and controlled self-assembly of metal nanoparticles and nanocrystals for plasmonics. Chem. Soc. Rev. 2016, 45, 5672–5716. [Google Scholar] [CrossRef]
- Hamon, C.; Liz-Marzán, L.M. Colloidal design of plasmonic sensors based on surface enhanced Raman scattering. J. Colloid Interface Sci. 2018, 512, 834–843. [Google Scholar] [CrossRef] [Green Version]
- Mosier-Boss, P.A. Review of SERS Substrates for Chemical Sensing. Nanomaterials 2017, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- Kitahama, Y.; Itoh, T.; Ishido, T.; Hirano, K.; Ishikawa, M. Surface-Enhanced Raman Scattering from Photoreduced Ag Nanoaggregates on an Optically Trapped Single Bacterium. Bull. Chem. Soc. Jpn. 2011, 84, 976–978. [Google Scholar] [CrossRef]
- Kogler, M.; Ryabchikov, Y.V.; Uusitalo, S.; Popov, A.; Popov, A.; Tselikov, G.; Valimaa, A.-L.; Al-Kattan, A.; Hiltunen, J.; Laitinen, R.; et al. Bare laser-synthesized Au-based nanoparticles as nondisturbing surface-enhanced Raman scattering probes for bacteria identification. J. Biophotonics 2018, 11, e201700225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahraman, M.; Yazici, M.M.; Sahin, F.; Bayrak, O.F.; Culha, M. Reproducible surface-enhanced Raman scattering spectra of bacteria on aggregated silver nanoparticles. Appl. Spectrosc. 2007, 61, 479–485. [Google Scholar] [CrossRef]
- Zhu, A.; Ali, S.; Xu, Y.; Qin, O.; Wang, Z.; Chen, Q. SERS-based Au@Ag NPs Solid-phase substrate combined with chemometrics for rapid discrimination of multiple foodborne pathogens. Spectrochim. Acta Part A-Mol. Biomol. Spectrosc. 2022, 270, 120814. [Google Scholar] [CrossRef]
- Liu, T.Y.; Chen, Y.; Wang, H.H.; Huang, Y.L.; Chao, Y.C.; Tsai, K.T.; Cheng, W.C.; Chuang, C.Y.; Tsai, Y.H.; Huang, C.Y.; et al. Differentiation of bacteria cell wall using Raman scattering enhanced by nanoparticle array. J. Nanosci. Nanotechnol. 2012, 12, 5004–5008. [Google Scholar] [CrossRef]
- Andrei, C.-C.; Moraillon, A.; Lau, S.; Felidj, N.; Yamakawa, N.; Bouckaert, J.; Larquet, E.; Boukherroub, R.; Ozanam, F.; Szunerits, S.; et al. Rapid and sensitive identification of uropathogenic Escherichia coli using a surface-enhanced-Raman-scattering-based biochip. Talanta 2020, 219, 121174. [Google Scholar] [CrossRef]
- Cao, Y.; Lv, M.; Xu, H.; Svec, F.; Tan, T.; Lv, Y. Planar monolithic porous polymer layers functionalized with gold nanoparticles as large-area substrates for sensitive surface-enhanced Raman scattering sensing of bacteria. Anal. Chim. Acta 2015, 896, 111–119. [Google Scholar] [CrossRef]
- Sengupta, A.; Mujacic, M.; Davis, E.J. Detection of bacteria by surface-enhanced Raman spectroscopy. Anal. Bioanal. Chem. 2006, 386, 1379–1386. [Google Scholar] [CrossRef]
- Tadesse, L.F.; Ho, C.S.; Chen, D.H.; Arami, H.; Banaei, N.; Gambhir, S.S.; Jeffrey, S.S.; Saleh, A.A.E.; Dionne, J. Plasmonic and Electrostatic Interactions Enable Uniformly Enhanced Liquid Bacterial Surface-Enhanced Raman Scattering (SERS). Nano Lett. 2020, 20, 7655–7661. [Google Scholar] [CrossRef]
- Fakhrullin, R.; Nigamatzyanova, L.; Fakhrullina, G. Dark-field/hyperspectral microscopy for detecting nanoscale particles in environmental nanotoxicology research. Sci. Total Environ. 2021, 772, 145478. [Google Scholar] [CrossRef] [PubMed]
- Belini, V.L.; Souza Freitas, B.L.; Sabogal-Paz, L.P.; Branco, N.; Bueno Franco, R.M. Label-Free Darkfield-Based Technique to Assist in the Detection of Giardia Cysts. Water Air Soil Pollut. 2018, 229, 195. [Google Scholar] [CrossRef]
- Mock, J.J.; Barbic, M.; Smith, D.R.; Schultz, D.A.; Schultz, S. Shape effects in plasmon resonance of individual colloidal silver nanoparticles. J. Chem. Phys. 2002, 116, 6755–6759. [Google Scholar] [CrossRef]
- Xu, X.; Chen, Y.; Wei, H.; Xia, B.; Liu, F.; Li, N. Counting Bacteria Using Functionalized Gold Nanoparticles as the Light-Scattering Reporter. Anal. Chem. 2012, 84, 9721–9728. [Google Scholar] [CrossRef]
- Liao, X.-W.; Xu, Q.-Y.; Tan, Z.; Liu, Y.; Wang, C. Recent Advances in Plasmonic Nanostructures Applied for Label-free Single-cell Analysis. Electroanalysis 2022, 34, 923–936. [Google Scholar] [CrossRef]
- Hernandez-Rodriguez, P.; Diaz, C.A.; Dalmau, E.A.; Quintero, G.M. A comparison between polymerase chain reaction (PCR) and traditional techniques for the diagnosis of leptospirosis in bovines. J. Microbiol. Methods 2011, 84, 1–7. [Google Scholar] [CrossRef]
- Cameron, C.E. Leptospiral Structure, Physiology, and Metabolism. In Leptospira and Leptospirosis; Springer: Berlin/Heidelberg, Germany, 2015; Volume 387, pp. 21–41. [Google Scholar]
- Gupta, S.; Jain, P.K.; Kumra, M.; Rehani, S.; Mathias, Y.; Gupta, R.; Mehendiratta, M.; Chander, A. Bacterial Viability within Dental Calculus: An Untrodden, Inquisitive Clinico-Patho- Microbiological Research. J. Clin. Diagn. Res. JCDR 2016, 10, ZC71-5. [Google Scholar] [CrossRef]
- Rodriguez-Fajardo, V.; Sanz, V.; de Miguel, I.; Berthelot, J.; Acimovic, S.S.; Porcar-Guezenec, R.; Quidant, R. Two-color dark-field (TCDF) microscopy for metal nanoparticle imaging inside cells. Nanoscale 2018, 10, 4019–4027. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Wen, Y.; Ren, W.; Duan, H.; Lin, J.; Irudayaraj, J. Hyperspectral dark-field microscopy for pathogen detection based on spectral angle mapping. Sens. Actuators B-Chem. 2022, 367, 132042. [Google Scholar] [CrossRef]
- Benacer, D.; Woh, P.Y.; Zain, S.N.M.; Amran, F.; Thong, K.L. Pathogenic and Saprophytic Leptospira Species in Water and Soils from Selected Urban Sites in Peninsular Malaysia. Microbes Environ. 2013, 28, 135–140. [Google Scholar] [CrossRef] [Green Version]
- Imai, M.; Mine, K.; Tomonari, H.; Uchiyama, J.; Matuzaki, S.; Niko, Y.; Hadano, S.; Watanabe, S. Dark-Field Microscopic Detection of Bacteria using Bacteriophage-Immobilized SiO2@AuNP Core-Shell Nanoparticles. Anal. Chem. 2019, 91, 12352–12357. [Google Scholar] [CrossRef] [PubMed]
- Nakao, H.; Saito, K.; Tomita, S.; Magariyama, Y.; Kaizuka, Y.; Takeda, Y. Direct Imaging of Carboxymethyl Cellulose-mediated Aggregation of Lactic Acid Bacteria Using Dark-field Microscopy. Anal. Sci. 2016, 32, 1047–1051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaki, A.M.; Hod, R.; Shamsusah, N.A.; Isa, Z.M.; Bejo, S.K.; Agustar, H.K. Detection ofLeptospira kmetyiat recreational areas in Peninsular Malaysia. Environ. Monit. Assess. 2020, 192, 703. [Google Scholar] [CrossRef] [PubMed]
- Fotso Fotso, A.; Drancourt, M. Laboratory Diagnosis of Tick-Borne African Relapsing Fevers: Latest Developments. Front. Public Health 2015, 3, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Cui, Y.; Irudayaraj, J. Single-Cell Quantification of Cytosine Modifications by Hyperspectral Dark-Field Imaging. ACS Nano 2015, 9, 11924–11932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanabe, S.; Itagaki, S.; Sun, S.; Matsui, K.; Kinoshita, T.; Nishii, S.; Yamamoto, Y.; Sadanaga, Y.; Shiigi, H. Quantification of Enterohemorrhagic Escherichia coli via Optical Nanoantenna and Temperature-responsive Artificial Antibodies. Anal. Sci. 2021, 37, 1597–1601. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Simon, T.; Tatar, A.-S.; Craciun, A.-M.; Vulpoi, A.; Jurj, M.-A.; Florea, A.; Tomuleasa, C.; Berindan-Neagoe, I.; Astilean, S.; Boca, S. Antibody Conjugated, Raman Tagged Hollow Gold-Silver Nanospheres for Specific Targeting and Multimodal Dark-Field/SERS/Two Photon-FLIM Imaging of CD19(+) B Lymphoblasts. ACS Appl. Mater. Interfaces 2017, 9, 21155–21168. [Google Scholar] [CrossRef]
- Xu, H.; Tang, F.; Dai, J.; Wang, C.; Zhou, X. Ultrasensitive and rapid count of Escherichia coli using magnetic nanoparticle probe under dark-field microscope. BMC Microbiol. 2018, 18, 100. [Google Scholar] [CrossRef] [Green Version]
- Theel, E.S.; Katz, S.S.; Pillay, A. Molecular and Direct Detection Tests for Treponema pallidum Subspecies pallidum: A Review of the Literature, 1964–2017. Clin. Infect. Dis. 2020, 71, S4–S12. [Google Scholar] [CrossRef]
- Chen, S.; Su, Y.-W.; Sun, J.; Chen, T.; Zheng, Y.; Sui, L.-J.; Yang, S.; Liu, C.; Wang, P.; Li, T.; et al. Label-free single-particle imaging approach for ultra-rapid detection of pathogenic bacteria in clinical samples. Proc. Natl. Acad. Sci. USA 2022, 119, e2206990119. [Google Scholar] [CrossRef]
- Zhang, F.; Mo, M.; Jiang, J.; Zhou, X.; McBride, M.; Yang, Y.; Reilly, K.S.; Grys, T.E.; Haydel, S.E.; Tao, N.; et al. Rapid Detection of Urinary Tract Infection in 10 min by Tracking Multiple Phenotypic Features in a 30 s Large-Volume Scattering Video of Urine Microscopy. ACS Sens. 2022, 7, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Smietana, M.; Bock, W.J.; Mikulic, P.; Ng, A.; Chinnappan, R.; Zourob, M. Detection of bacteria using bacteriophages as recognition elements immobilized on long-period fiber gratings. Opt. Express 2011, 19, 7971–7978. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.M.; Bock, W.J.; Mikulic, P.; Chinnappan, R.; Ng, A.; Tolba, M.; Zourob, M. Long period grating based biosensor for the detection of Escherichia coli bacteria. Biosens. Bioelectron. 2012, 35, 308–312. [Google Scholar] [CrossRef] [PubMed]
- Zuppolini, S.; Quero, G.; Consales, M.; Diodato, L.; Vaiano, P.; Venturelli, A.; Santucci, M.; Spyrakis, F.; Costi, M.P.; Giordano, M.; et al. Label-free fiber optic optrode for the detection of class C β-lactamases expressed by drug resistant bacteria. Biomed. Opt. Express 2017, 8, 5191–5205. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, D.; Massaroni, C.; Jorge Leitao, C.S.; De Fatima Domingues, M.; Sypabekova, M.; Barrera, D.; Floris, I.; Massari, L.; Oddo, C.M.; Sales, S.; et al. Fiber Bragg Gratings for Medical Applications and Future Challenges: A Review. IEEE Access 2020, 8, 156863–156888. [Google Scholar] [CrossRef]
- Srinivasan, R.; Umesh, S.; Murali, S.; Asokan, S.; Gorthi, S.S. Bare fiber Bragg grating immunosensor for real-time detection of Escherichia coli bacteria. J. Biophotonics 2017, 10, 224–230. [Google Scholar] [CrossRef]
- Manago, S.; Quero, G.; Zito, G.; Tullii, G.; Galeotti, F.; Pisco, M.; De Luca, A.C.; Cusano, A. Tailoring lab-on-fiber SERS optrodes towards biological targets of different sizes. Sens. Actuators B-Chem. 2021, 339, 129321. [Google Scholar] [CrossRef]
- Kim, J.A.; Wales, D.J.; Thompson, A.J.; Yang, G.-Z. Fiber-Optic SERS Probes Fabricated Using Two-Photon Polymerization For Rapid Detection of Bacteria. Adv. Opt. Mater. 2020, 8, 1901934. [Google Scholar] [CrossRef] [Green Version]
- Hunter, R.; Sohi, A.N.; Khatoon, Z.; Berthiaume, V.R.; Alarcon, E.I.; Godin, M.; Anis, H. Optofluidic label-free SERS platform for rapid bacteria detection in serum. Sens. Actuators B-Chem. 2019, 300, 126907. [Google Scholar] [CrossRef]
- Zang, C.-L.; Zhang, M.-D.; Zhang, Y.; Li, Y.-S.; Liu, K.; Xie, N.-N.; Sun, C.-Y.; Zhang, X.-G. Rapid label-free detection of Salmonella enterica with biolayer interferometry. J. Food Saf. 2021, 41, e12896. [Google Scholar] [CrossRef]
- Kim, D.M.; Yoo, S.M. Colorimetric Systems for the Detection of Bacterial Contamination: Strategy and Applications. Biosensors 2022, 12, 532. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, P.; Sun, H.; Yang, W.; Fang, Y. Optical Methods for Label-Free Detection of Bacteria. Biosensors 2022, 12, 1171. https://doi.org/10.3390/bios12121171
Wang P, Sun H, Yang W, Fang Y. Optical Methods for Label-Free Detection of Bacteria. Biosensors. 2022; 12(12):1171. https://doi.org/10.3390/bios12121171
Chicago/Turabian StyleWang, Pengcheng, Hao Sun, Wei Yang, and Yimin Fang. 2022. "Optical Methods for Label-Free Detection of Bacteria" Biosensors 12, no. 12: 1171. https://doi.org/10.3390/bios12121171
APA StyleWang, P., Sun, H., Yang, W., & Fang, Y. (2022). Optical Methods for Label-Free Detection of Bacteria. Biosensors, 12(12), 1171. https://doi.org/10.3390/bios12121171




