Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses
2.2. Buffer Medium
2.3. SERS Substrates
2.4. Raman Spectroscopy Setup
3. Results
3.1. Spectra of A Pure Buffer Medium and Viruses in A Buffer Medium
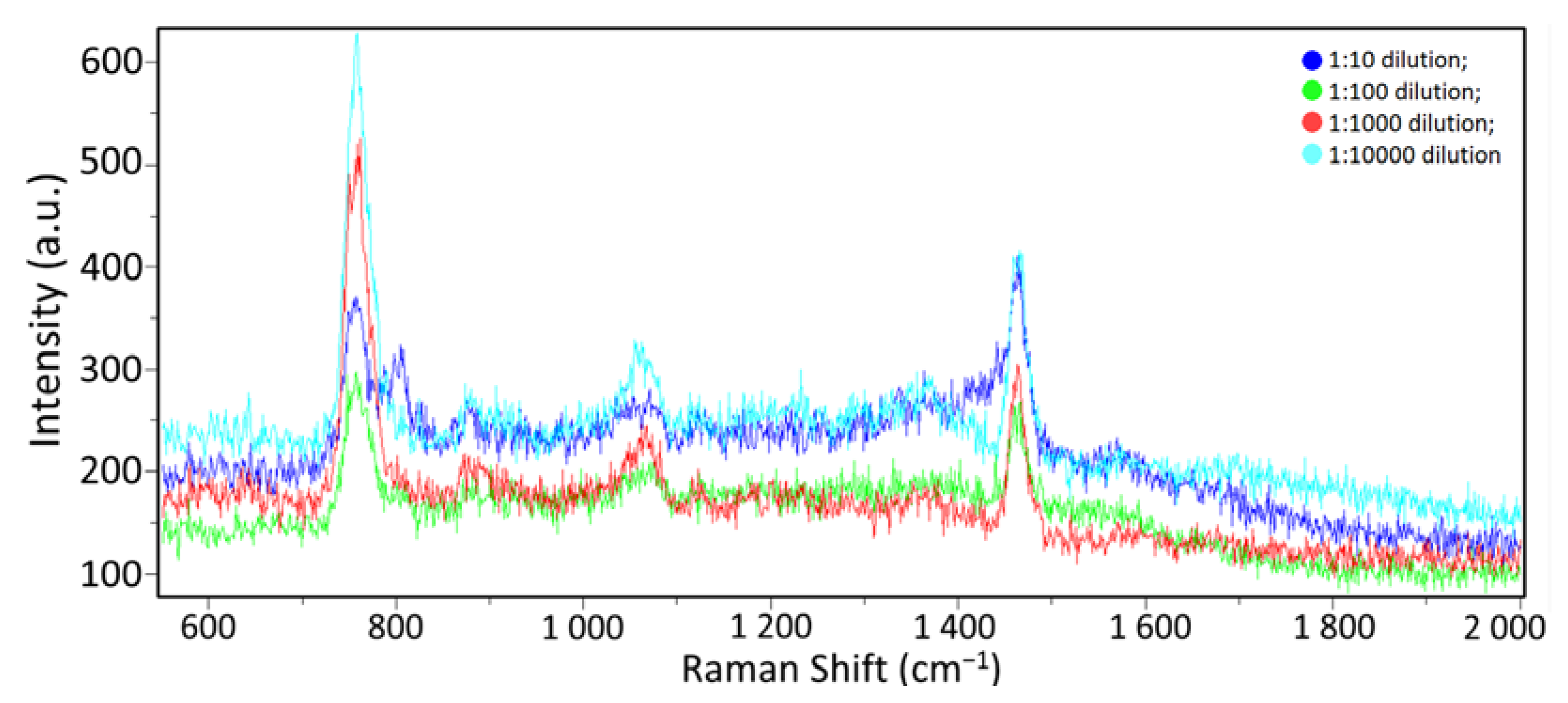
3.2. Spectra of Influenza A Virus in A Buffer Medium at Different Concentrations
3.3. Mathematical Processing of Spectra
3.3.1. Detection of Influenza A Virus in a Buffer Medium
3.3.2. Differentiation of Influenza A, Influenza B, and Pure Buffer Medium Spectra
3.3.3. Determination of the Minimum Allowable Concentration of Viral Particles for The Detection and Classification of the Influenza A Virus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sedlak, R.H.; Jerome, K.R. Viral diagnostics in the era of digital polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 2013, 75, 1–4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atmar, R.L. Viral Infections of Humans; Kaslow, R., Stanberry, L., Le Duc, J., Eds.; Springer: Boston, MA, USA, 2014; Immunological detection and characterization; pp. 47–62. ISBN 978-1-4899-7448-8. [Google Scholar]
- Yamada, A.; Imanishi, J.; Nakajima, E.; Nakajima, K.; Nakajima, S. Detection of influenza viruses in throat swab by using polymerase chain reaction. Microbiol. Immunol. 1991, 35, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Akylbayeva, K.K.; Shynybekova, G.O.; Tlenchiyeva, T.M.; Sultankulova, К.Т.; Sandybaev, N.T. Лабoратoрная диагнoстика гриппа типoв A и Б метoдoм ПЦР. Вестник КазНУ. Серия биoлoгическая 2017, 70, 100–108. [Google Scholar]
- Wikramaratna, P.S.; Paton, R.S.; Ghafari, M.; Lourenço, J. Estimating the false-negative test probability of SARS-CoV-2 by RT-PCR. Eurosurveillance 2020, 25, 2000568. [Google Scholar] [CrossRef] [PubMed]
- Boonham, N.; Kreuze, J.; Winter, S.; van der Vlugt, R.; Bergervoet, J.; Tomlinson, J.; Mumford, R. Methods in virus diagnostics: From ELISA to next generation sequencing. Virus Res. 2014, 186, 20–31. [Google Scholar] [CrossRef]
- Shanmukh, S.; Jones, L.; Driskell, J.; Zhao, Y.; Dluhy, R.; Tripp, R.A. Rapid and sensitive detection of respiratory virus molecular signatures using a silver nanorod array SERS substrate. Nano Lett. 2006, 6, 2630–2636. [Google Scholar] [CrossRef]
- Savinon-Flores, F.; Mendez, E.; Lopez-Castanos, M.; Carabarin-Lima, A.; Lopez-Castanos, K.A.; Gonzalez-Fuentes, M.A.; Mendez-Albores, A. A review on SERS-based detection of human virus infections: Influenza and coronavirus. Biosensors 2021, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Tabarov, A.T.; Vitkin, V.V.; Danilenko, D.M.; Andreeva, O.V.; Shemanaeva, A.A.; Popov, E.E.; Dobroslavin, A.A.; Kurikova, V.V.; Kuznetsova, O.B. Using the SERS method and machine learning technology to detect the influenza A virus. In Proceedings of the 2022 International Conference Laser Optics (ICLO), Saint Petersburg, Russia, 20–24 June 2022. [Google Scholar]
- Tabarov, A.T.; Vitkin, V.V.; Danilenko, D.M.; Andreeva, O.V.; Shemanaeva, A.A.; Popov, E.E.; Dobroslavin, A.A.; Kurikova, V.V.; Kuznetsova, O.B. Detection of Influenza A and B viruses by Raman spectroscopy. In Proceedings of the 2022 International Conference Laser Optics (ICLO), Saint Petersburg, Russia, 20–24 June 2022. [Google Scholar]
- Paria, D.; Kwok, K.S.; Raj, P.; Zheng, P.; Gracias, D.; Barman, I. Label-Free SARS-CoV-2 Detection on Flexible Substrates. medRxiv 2021. [Google Scholar] [CrossRef]
- Eom, G.; Hwang, A.; Lee, D.K.; Guk, K.; Moon, J.; Jeong, J.; Jung, J.; Kim, B.; Lim, E.K.; Kang, T. Superb specific, ultrasensitive, and rapid identification of the Oseltamivir-resistant H1N1 virus: Naked-eye and SERS dual-mode assay using functional gold nanoparticles. ACS Appl. Bio Mater. 2019, 2, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, C.; Wang, X.; Wang, K.; Zhu, Y.; Rong, Z.; Wang, W.; Xiao, R.; Wang, S. Magnetic SERS strip for sensitive and simultaneous detection of respiratory viruses. ACS Appl. Mater. Interfaces 2019, 11, 19495–19505. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Drelich, A.J.; Hopkins, C.M.; Mecozzi, S.; Li, L.; Kwon, G.; Hong, S. Gold nanoparticles in virus detection: Recent advances and potential considerations for SARS-CoV-2 testing development. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, 14, e1754. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Puebla, R.A.; Liz-Marzán, L.M. SERS-based diagnosis and biodetection. Small 2010, 6, 604–610. [Google Scholar] [CrossRef] [PubMed]
- Othman, N.H.; Lee, K.Y.; Radzol, A.R.M.; Mansor, W.; Ramlan, N.N.M. Linear discriminant analysis for detection of salivary NS1 from SERS spectra. In Proceedings of the TENCON 2017–2017 IEEE Region 10 Conference, Penang, Malaysia, 5–8 November 2017; pp. 2876–2879. [Google Scholar]
- Ralbovsky, N.M.; Lednev, I.K. Towards development of a novel universal medical diagnostic method: Raman spectroscopy and machine learning. Chem. Soc. Rev. 2020, 49, 7428–7453. [Google Scholar] [CrossRef] [PubMed]
- Sivashanmugan, K.; Liao, J.D.; You, J.W.; Wu, C.L. Focused-ion-beam-fabricated Au/Ag multilayered nanorod array as SERS-active substrate for virus strain detection. Sens. Actuators B Chem. 2013, 181, 361–367. [Google Scholar] [CrossRef]
- Chang, C.W.; Liao, J.D.; Shiau, A.L.; Yao, C.K. Non-labeled virus detection using inverted triangular Au nano-cavities arrayed as SERS-active substrate. Sens. Actuators B Chem. 2011, 156, 471–478. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Liao, J.D.; Yang, M.L.; Wu, C.L. Target-size embracing dimension for sensitive detection of viruses with various sizes and influenza virus strains. Biosens. Bioelectron. 2012, 35, 447–451. [Google Scholar] [CrossRef]
- Lin, Y.Y.; Liao, J.D.; Ju, Y.H.; Chang, C.W.; Shiau, A.L. Focused ion beam-fabricated Au micro/nanostructures used as a surface enhanced Raman scattering-active substrate for trace detection of molecules and influenza virus. Nanotechnology 2011, 22, 185308. [Google Scholar] [CrossRef]
- Lim, J.Y.; Nam, J.S.; Yang, S.E.; Shin, H.; Jang, Y.H.; Bae, G.U.; Kang, T.; Lim, K.I.; Choi, Y. Identification of newly emerging influenza viruses by surface-enhanced Raman spectroscopy. Anal. Chem. 2015, 87, 11652–11659. [Google Scholar] [CrossRef]
- Alexander, T.A. Development of methodology based on commercialized SERS-active substrates for rapid discrimination of Poxviridae virions. Anal. Chem. 2008, 80, 2817–2825. [Google Scholar] [CrossRef]
- Yeh, Y.T.; Gulino, K.; Zhang, Y.; Sabestien, A.; Chou, T.W.; Zhou, B.; Lin, Z.; Albert, I.; Lu, H.; Swaminathan, V.; et al. A rapid and label-free platform for virus capture and identification from clinical samples. Proc. Natl. Acad. Sci. USA 2020, 117, 895–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, J.Y.; Nam, J.S.; Shin, H.; Park, J.; Song, H.I.; Kang, M.; Lim, K.I.; Choi, Y. Identification of newly emerging influenza viruses by detecting the virally infected cells based on surface enhanced Raman spectroscopy and principal component analysis. Anal. Chem. 2019, 91, 5677–5684. [Google Scholar] [CrossRef]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. TrAC Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Huang, J.; Wen, J.; Zhou, M.; Ni, S.; Le, W.; Chen, G.; Wei, L.; Zeng, Y.; Qi, D.; Pan, M.; et al. On-site detection of SARS-CoV-2 antigen by deep learning-based surface-enhanced raman spectroscopy and its biochemical foundations. Anal. Chem. 2021, 93, 9174–9182. [Google Scholar] [CrossRef]
- LaValley, M.P. Logistic regression. Circulation 2008, 117, 2395–2399. [Google Scholar] [CrossRef] [Green Version]
- Abdi, H.; Williams, L.J. Principal component analysis. Wiley Interdiscip. Rev. Comput. Stat. 2010, 2, 433–459. [Google Scholar] [CrossRef]
- Zhang, Z.; Jiang, S.; Wang, X.; Dong, T.; Wang, Y.; Li, D.; Gao, X.; Qu, Z.; Li, Y. A novel enhanced substrate for label-free detection of SARS-CoV-2 based on surface-enhanced Raman scattering. Sens. Actuators B Chem. 2022, 359, 131568. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, D.; Wang, X.; Wang, Y.; Lin, J.; Jiang, S.; Wu, Z.; He, Y.; Gao, X.; Zhu, Z.; et al. Rapid detection of viruses: Based on silver nanoparticles modified with bromine ions and acetonitrile. Chem. Eng. J. 2022, 438, 135589. [Google Scholar] [CrossRef]
- Blei, D.M.; Ng, A.Y.; Jordan, M.I. Latent dirichlet allocation. J. Mach. Learn. Res. 2003, 3, 993–1022. [Google Scholar] [CrossRef]
- Durmanov, N.N.; Guliev, R.R.; Eremenko, A.V.; Boginskaya, I.A.; Ryzhikov, I.A.; Trifonova, E.A.; Putlyaev, E.V.; Mukhin, A.N.; Kalnov, S.L.; Balandina, M.V.; et al. Non-labeled selective virus detection with novel SERS-active porous silver nanofilms fabricated by Electron Beam Physical Vapor Deposition. Sens. Actuators B Chem. 2018, 257, 37–47. [Google Scholar] [CrossRef]
- Paria, D.; Kwok, K.S.; Raj, P.; Zheng, P.; Gracias, D.H.; Barman, I. Label-free spectroscopic SARS-CoV-2 detection on versatile nanoimprinted substrates. Nano Lett. 2022, 22, 3620–3627. [Google Scholar] [CrossRef]
- Song, C.; Driskell, J.D.; Tripp, R.A.; Cui, Y.; Zhao, Y. The use of a handheld Raman system for virus detection. In Proceedings of the Chemical, Biological, Radiological, Nuclear, and Explosives (CBRNE) Sensing XIII, Baltimore, MD, USA, 24–27 April 2012; Volume 8358, pp. 109–115. [Google Scholar]
- Modak, S.; Mehta, S.; Sehgal, D.; Valadi, J. Application of support vector machines in viral biology. In Global Virology III: Virology in the 21st Century; Shapshak, P., Balaji, S., Eds.; Springer: Cham, Switzerland, 2019; pp. 361–403. ISBN 978-3-030-29021-4. [Google Scholar]
- Yang, Y.; Xu, B.; Murray, J.; Haverstick, J.; Chen, X.; Tripp, R.A.; Zhao, Y. Rapid and quantitative detection of respiratory viruses using surface-enhanced Raman spectroscopy and machine learning. Biosens. Bioelectron. 2022, 217, 114721. [Google Scholar] [CrossRef]
- Johansson, B.E.; Brett, I.C. Recombinant influenza B virus HA and NA antigens administered in equivalent amounts are immunogenically equivalent and induce equivalent homotypic and broader heterovariant protection in mice than conventional and live influenza vaccines. Hum. Vaccines 2008, 4, 420–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- SERSitive. User Guide and Datasheed. Available online: https://sersitive.eu/wp-content/uploads/Manual_product-sheet.pdf (accessed on 19 October 2022).
- Waterborg, J.H.; Matthews, H.R. Basic Protein and Peptide Protocols; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 1994; The Lowry method for protein quantitation; Volume 32, pp. 1–4. ISBN 978-1-59259-519-8. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Griffel, L.M.; Delparte, D.; Edwards, J. Using Support Vector Machines classification to differentiate spectral signatures of potato plants infected with Potato Virus Y. Comput. Electron. Agric. 2018, 153, 318–324. [Google Scholar] [CrossRef]







| Iteration | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Test set size | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 5 | 5 | 5 |
| Accuracy | 100% | 94% | 94% | 100% | 94% | 94% | 94% | 88% | 100% | 94% |
| Iteration | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracy | 93% | 89% | 93% | 100% | 93% | 96% | 89% | 85% | 100% | 96% |
| SD | 4.8% | |||||||||
| Sample Dilution | Average Accuracy | SD of Accuracy | Average Spectrum RSD | Total Number of Samples | Training Set Size | Test Set Size |
|---|---|---|---|---|---|---|
| 1:10 | 100% | 0% | 24% | 20 | 15 | 5 |
| 1:100 | 100% | 0% | 42% | 20 | 15 | 5 |
| 1:1000 | 94.5% | 6.9% | 26% | 24 | 20 | 4 |
| 1:10,000 | 84% | 15.2% | 28% | 20 | 15 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabarov, A.; Vitkin, V.; Andreeva, O.; Shemanaeva, A.; Popov, E.; Dobroslavin, A.; Kurikova, V.; Kuznetsova, O.; Grigorenko, K.; Tzibizov, I.; et al. Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors 2022, 12, 1065. https://doi.org/10.3390/bios12121065
Tabarov A, Vitkin V, Andreeva O, Shemanaeva A, Popov E, Dobroslavin A, Kurikova V, Kuznetsova O, Grigorenko K, Tzibizov I, et al. Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors. 2022; 12(12):1065. https://doi.org/10.3390/bios12121065
Chicago/Turabian StyleTabarov, Artem, Vladimir Vitkin, Olga Andreeva, Arina Shemanaeva, Evgeniy Popov, Alexander Dobroslavin, Valeria Kurikova, Olga Kuznetsova, Konstantin Grigorenko, Ivan Tzibizov, and et al. 2022. "Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning" Biosensors 12, no. 12: 1065. https://doi.org/10.3390/bios12121065
APA StyleTabarov, A., Vitkin, V., Andreeva, O., Shemanaeva, A., Popov, E., Dobroslavin, A., Kurikova, V., Kuznetsova, O., Grigorenko, K., Tzibizov, I., Kovalev, A., Savchenko, V., Zheltuhina, A., Gorshkov, A., & Danilenko, D. (2022). Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors, 12(12), 1065. https://doi.org/10.3390/bios12121065





