Ultrasensitive Detection of C-Reactive Protein by a Novel Nanoplasmonic Immunoturbidimetry Assay
Abstract
1. Introduction
2. Experimental Methods
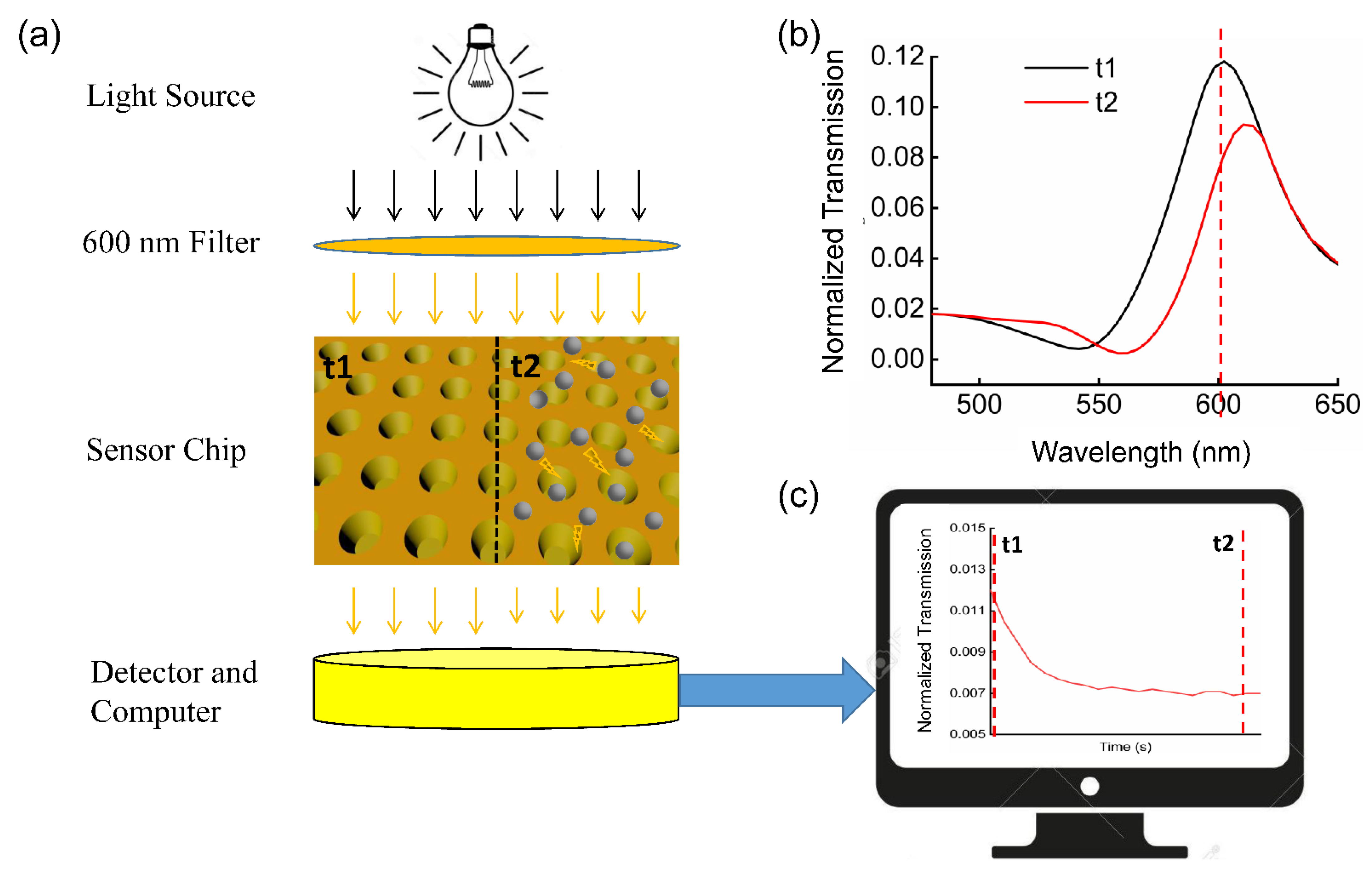
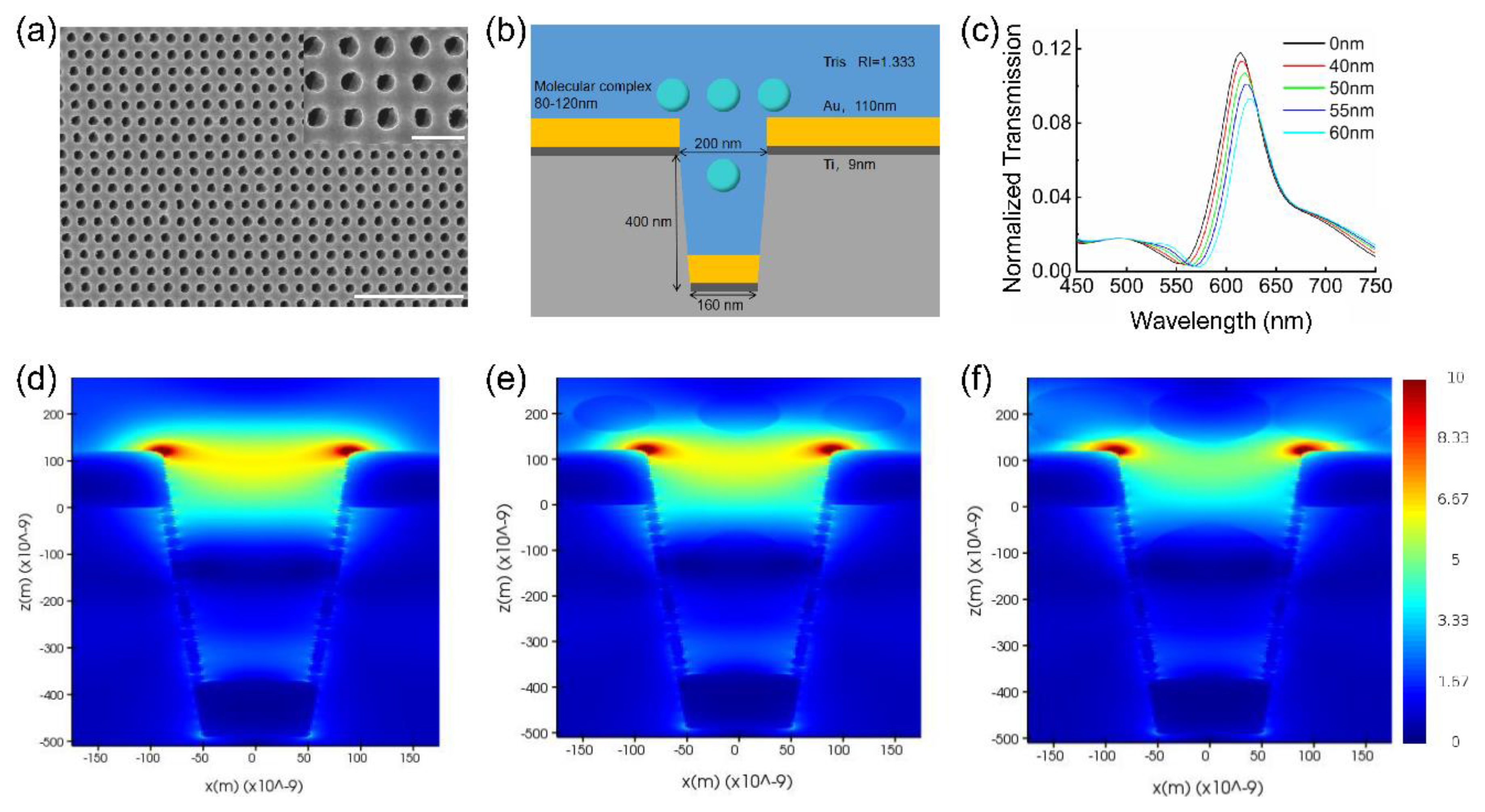
2.1. Immunoturbidimetry Sensor Fabrication
2.2. Detection of Calcium Carbonate (CaCO3) Trace Precipitation
2.3. Detection of Self-Aggregation of Human IAPP
2.4. NanoPITA of CRP
2.5. FDTD Simulations
2.6. Immune Molecular Complex Size Determination
3. Results and Discussions
3.1. Device Characterization
3.2. Detection of CaCO3 and Self-Aggregation of The IAPP
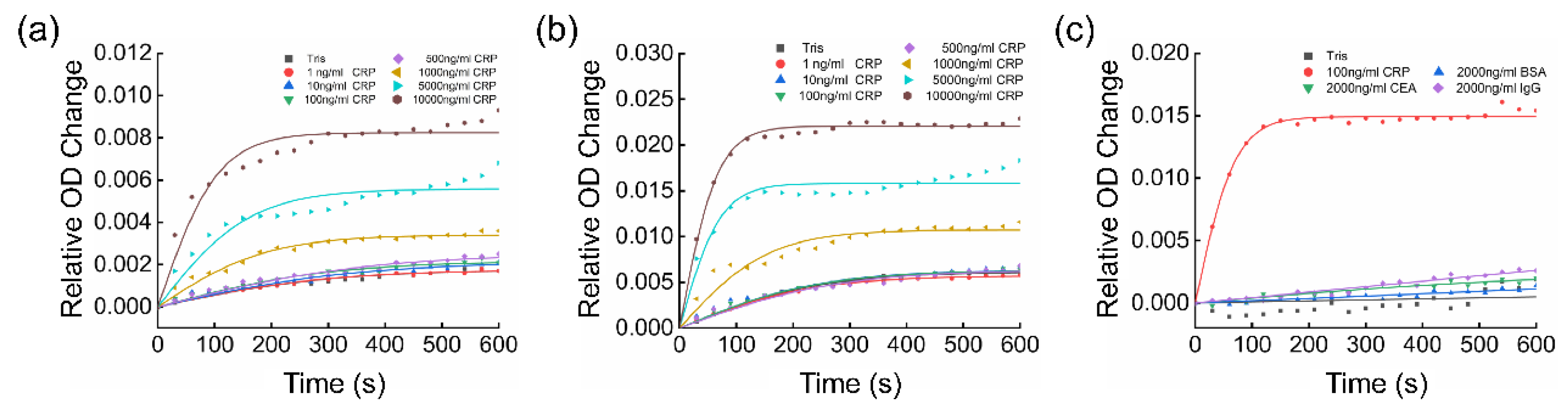
3.3. NanoPITA of CRP
3.4. Quantitative Analysis of the Kinetic Results
3.5. Evaluation of Enhancement and Cross-Reactivity
3.6. Evaluation of Blood Sample
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lim, C.T.; Han, J.; Guck, J.; Espinosa, H. Micro and nanotechnology for biological and biomedical applications. Med. Biol. Eng. Comput. 2010, 48, 941–943. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Durán, N.; Marcato, P.D. Nanobiotechnology perspectives. Role of nanotechnology in the food industry: A review. Int. J. Food Sci. Technol. 2013, 48, 1127–1134. [Google Scholar] [CrossRef]
- Qian, Y.; Qin, C.; Chen, M.; Lin, S. Nanotechnology in soil remediation−applications vs. implications. Ecotoxicol. Environ. Saf. 2020, 201, 110815. [Google Scholar] [CrossRef] [PubMed]
- Nagar, A.; Pradeep, T. Clean Water through Nanotechnology: Needs, Gaps, and Fulfillment. ACS Nano 2020, 14, 6420–6435. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, P.; Preece, J.A.; Mendes, P.M. Nanotechnology: The “Top-Down” and “Bottom-Up” Approaches. Supramol. Chem. Mol. Nanomater. 2012. [Google Scholar] [CrossRef]
- Talham, D.R. Conducting and Magnetic Langmuir? Blodgett Films. ChemInform 2005, 36. [Google Scholar] [CrossRef]
- Lee, K.-L.; Wu, T.-Y.; Hsu, H.-Y.; Yang, S.-Y.; Wei, P.-K. Low-Cost and Rapid Fabrication of Metallic Nanostructures for Sensitive Biosensors Using Hot-Embossing and Dielectric-Heating Nanoimprint Methods. Sensors 2017, 17, 1548. [Google Scholar] [CrossRef]
- Yeom, S.H.; Han, M.E.; Kang, B.H.; Kim, K.J.; Yuan, H.; Eum, N.S.; Kang, S.W. Enhancement of the sensitivity of LSPR-based CRP immunosensors by Au nanoparticle antibody conjugation. Sens. Actuators B Chem. 2013, 177, 376–383. [Google Scholar] [CrossRef]
- Sharma, P.; Semwal, V.; Gupta, B.D. A highly selective LSPR biosensor for the detection of taurine realized on optical fiber substrate and gold nanoparticles. Opt. Fiber Technol. 2019, 52, 101962. [Google Scholar] [CrossRef]
- El Barghouti, M.; Akjouj, A.; Mir, A. Design of silver nanoparticles with graphene coatings layers used for LSPR biosensor applications. Vacuum 2020, 180, 109497. [Google Scholar] [CrossRef]
- Baquedano, E.; González, M.U.; Paniagua-Domínguez, R.; Sánchez-Gil, J.A.; Postigo, P.A. Low-cost and large-size nanoplasmonic sensor based on Fano resonances with fast response and high sensitivity. Opt. Express 2017, 25, 15967–15976. [Google Scholar] [CrossRef] [PubMed]
- Ebbesen, T.W.; Lezec, H.J.; Ghaemi, H.F.; Thio, T.; Wolff, P.A. Extraordinary optical transmission through sub-wavelength hole arrays. Nature 1998, 391, 667–669. [Google Scholar] [CrossRef]
- Martín-Moreno, L.; Garcia-Vidal, F.; Lezec, H.J.; Pellerin, K.M.; Thio, T.; Pendry, J.; Ebbesen, T.W. Theory of Extraordinary Optical Transmission through Subwavelength Hole Arrays. Phys. Rev. Lett. 2001, 86, 1114–1117. [Google Scholar] [CrossRef] [PubMed]
- Pepys, M.B.; Hirschfield, G.M. C-reactive protein: A critical update. J. Clin. Investig. 2003, 111, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Gabay, C.; Kushner, I. Acute-Phase Proteins and Other Systemic Responses to Inflammation. N. Engl. J. Med. 1999, 340, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Ridker, P.M.; Hennekens, C.H.; Buring, J.E.; Rifai, N. C-Reactive Protein and Other Markers of Inflammation in the Prediction of Cardiovascular Disease in Women. N. Engl. J. Med. 2000, 342, 836–843. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sivakanesan, R. Does plasma fibrinogens and C-reactive protein predicts the incidence of myocardial infraction in patients with normal lipids profile? Pak. J. Med. Sci. 2008, 24, 336. [Google Scholar]
- di Napoli, M.; Papa, F.; Bocola, V. C-reactive protein in ischemic stroke: An independent prognostic factor. Stroke 2001, 32, 917–924. [Google Scholar] [CrossRef]
- Chalmers, J.D.; Singanayagam, A.; Hill, A.T. C-Reactive Protein Is an Independent Predictor of Severity in Community-acquired Pneumonia. Am. J. Med. 2008, 121, 219–225. [Google Scholar] [CrossRef]
- Ansar, W.; Ghosh, S. C-reactive protein and the biology of disease. Immunol. Res. 2013, 56, 131–142. [Google Scholar] [CrossRef]
- Guo, L.; Liu, S.; Zhang, S.; Chen, Q.; Zhang, M.; Quan, P.; Lu, J.; Sun, X. C-reactive protein and risk of breast cancer: A systematic review and meta-analysis. Sci. Rep. 2015, 5, 10508. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Zhang, D.; Jiang, F.; Yang, H.; Liu, S.; Li, P. Metal-enhanced chemiluminescence detection of C-reaction protein based on silver nanoparticle hybrid probes. Talanta 2019, 199, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Capua, L.; Sprunger, Y.; Elettro, H.; Grammoustianou, A.; Midahuen, R.; Ernst, T.; Barraud, S.; Gill, R.; Ionescu, A. Double-Gate Si Nanowire FET Sensor Arrays For Label-Free C-Reactive Protein detection enabled by antibodies fragments and pseu-do-super-Nernstian back-gate operation. In Proceedings of the 2021 IEEE International Electron Devices Meeting (IEDM), San Francisco, CA, USA, 11–15 December 2021; p. 16. [Google Scholar]
- McNaught, A.D.; Wilkinson, A. Compendium of Chemical Terminology; Blackwell Science: Oxford, UK, 1997. [Google Scholar]
- Nishimura, A.; Sawai, T. Determination of adiponectin in serum using a latex particle-enhanced turbidimetric immunoassay with an automated analyzer. Clin. Chim. Acta 2006, 371, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Shen, H.; Zhu, Y.; Xu, H.; Li, Z.; Si, J. A sensitive three monoclonal antibodies based automatic latex particle-enhanced turbidimetric immunoassay for Golgi protein 73 detection. Sci. Rep. 2017, 7, 40090. [Google Scholar] [CrossRef] [PubMed]
- Thanh, N.T.K.; Rosenzweig, Z. Development of an Aggregation-Based Immunoassay for Anti-Protein A Using Gold Nanoparticles. Anal. Chem. 2002, 74, 1624–1628. [Google Scholar] [CrossRef] [PubMed]
- Al-Turkmani, M.R.; Law, T.; Kellogg, M.D. Performance evaluation of a particle-enhanced turbidimetric cystatin C assay on the Hitachi 917 analyzer. Clin. Chim. Acta 2008, 398, 75–77. [Google Scholar] [CrossRef] [PubMed]
- Dang, T.; Hu, W.; Zhang, W.; Song, Z.; Wang, Y.; Chen, M.; Xu, H.; Liu, G.L. Protein binding kinetics quantification via coupled plasmonic-photonic resonance nanosensors in generic microplate reader. Biosens. Bioelectron. 2019, 142, 111494. [Google Scholar] [CrossRef]
- Frear, G.L.; Johnston, J. The solubility of calcium carbonate (calcite) in certain aqueous solutions AT 25°. J. Am. Chem. Soc. 1929, 51, 2082–2093. [Google Scholar] [CrossRef]
- Westermark, P.; Andersson, A.; Westermark, G.T. Islet amyloid polypeptide, islet amyloid, and diabetes mellitus. Physiol. Rev. 2011, 91, 795–826. [Google Scholar] [CrossRef]
- Buxbaum, J. Diseases of protein conformation: What do in vitro experiments tell us about in vivo diseases? Trends Biochem. Sci. 2003, 28, 585–592. [Google Scholar] [CrossRef]
- Dobson, C.M. Protein misfolding, evolution and disease. Trends Biochem. Sci. 1999, 24, 329–332. [Google Scholar] [CrossRef]
- Glenner, G.G. Amyloid deposits and amyloidosis. The beta-fibrilloses (first of two parts). N. Engl. J. Med. 1980, 302, 1283–1292. [Google Scholar] [CrossRef] [PubMed]
- Hand, D.B. The refractivity of protein solutions. J. Biol. Chem. 1935, 108, 703–707. [Google Scholar] [CrossRef]
- Shankaran, D.R.; Gobiand, K.V.; Miura, N. Recent advancements in surface plasmon resonance immunosensors for detection of small molecules of biomedical, food and environmental interest. Sens. Actuators B Chem. 2007, 121, 158–177. [Google Scholar] [CrossRef]
- Lim, C.Z.J.; Zhang, Y.; Chen, Y.; Zhao, H.; Stephenson, M.; Ho, N.R.Y.; Chen, Y.; Chung, J.; Reilhac, A.; Loh, T.P.; et al. Subtyping of circulating exosome-bound amyloid β reflects brain plaque deposition. Nat. Commun. 2019, 10, 1144. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Pérez-Luna, V.H. Dextran−Gold Nanoparticle Hybrid Material for Biomolecule Immobilization and Detection. Anal. Chem. 2005, 77, 7204–7211. [Google Scholar] [CrossRef]
- Nath, N.; Chilkoti, A. A Colorimetric Gold Nanoparticle Sensor To Interrogate Biomolecular Interactions in Real Time on a Surface. Anal. Chem. 2001, 74, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Piston, D.W. Choosing objective lenses: The importance of numerical aperture and magnification in digital optical microscopy. Biol. Bull. 1998, 195, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Holownia, P.; Perez-Amodio, S.; Price, C.P. Effect of Poly(ethylene glycol), Tetramethylammonium Hydroxide, and Other Surfactants on Enhancing Performance in a Latex Particle Immunoassay of C-Reactive Protein. Anal. Chem. 2001, 73, 3426–3431. [Google Scholar] [CrossRef]
- U.S. Department of Health and Human Services Food and Drug Administration. Guidance for Industry and FDA Staff: Review Criteria for Assessment of C-Reactive Protein (CRP), High Sensitivity C-Reactive Protein (hsCRP) and Cardiac C-Reactive Protein (cCRP) Assays; U.S. Department of Health and Human Services Food and Drug Administration: Silver Spring, MD, USA, 2005. [Google Scholar]
- Patel, N.; Belcher, J.; Thorpe, G.; Forsyth, N.R.; Spiteri, M.A. Measurement of C-reactive protein, procalcitonin and neutrophil elastase in saliva of COPD patients and healthy controls: Correlation to self-reported wellbeing parameters. Respir. Res. 2015, 16, 62. [Google Scholar] [CrossRef]
- Marmer, D.J.; Hurtubise, P.E. Nephelometric and turbidimetric immunoassay. Immunoassay 1996, 363–387. [Google Scholar] [CrossRef]
- Sridevi, S.; Vasu, K.; Asokan, S.; Sood, A. Sensitive detection of C-reactive protein using optical fiber Bragg gratings. Biosens. Bioelectron. 2015, 65, 251–256. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Prasad, B.B. Multiwalled carbon nanotubes embedded molecularly imprinted polymer-modified screen printed carbon electrode for the quantitative analysis of C-reactive protein. Sens. Actuators B Chem. 2012, 171–172, 1141–1150. [Google Scholar] [CrossRef]
- Zubiate, P.; Zamarreño, C.; Sánchez, P.; Matias, I.; Arregui, F. High sensitive and selective C-reactive protein detection by means of lossy mode resonance based optical fiber devices. Biosens. Bioelectron. 2017, 93, 176–181. [Google Scholar] [CrossRef]
- Fan, Y.-J.; Sheen, H.-J.; Liu, Y.-H.; Tsai, J.-F.; Wu, T.-H.; Wu, K.-C.; Lin, S. Detection of C-Reactive Protein in Evanescent Wave Field Using Microparticle-Tracking Velocimetry. Langmuir 2010, 26, 13751–13754. [Google Scholar] [CrossRef]
- Jeng, M.-J.; Sharma, M.; Li, Y.-C.; Lu, Y.-C.; Yu, C.-Y.; Tsai, C.-L.; Huang, S.-F.; Chang, L.-B.; Lai, C.-S. Surface Acoustic Wave Sensor for C-Reactive Protein Detection. Sensors 2020, 20, 6640. [Google Scholar] [CrossRef] [PubMed]
- Eckersall, P.D.; Conner, J.G.; Harvie, J. An immunoturbidimetric assay for canine C-reactive protein. Veter. Res. Commun. 1991, 15, 17–24. [Google Scholar] [CrossRef]
- Whicher, J.; Price, C.; Spencer, K.; Ward, A.M. Immunonephelometric and immunoturbidimetric assays for proteins. CRC Crit. Rev. Clin. Lab. Sci. 1982, 18, 213–260. [Google Scholar] [CrossRef] [PubMed]



| Experimental Method | Experimental Readout | LOD (ng/mL) | Linear Range (ng/mL) | Process Time | Biological Modification Time |
|---|---|---|---|---|---|
| Optical fiber Bragg gratings [45] | Bragg wavelength | 10 | 10–106 | 10 min | 11 h |
| Surface molecular imprinting [46] | Differential pulse voltammetry | 40 | 180–8510 | N/A | 9 h |
| Lossy mode resonances [47] | Wavelength shift | 62.5 | 62.5–1000 | 61 s | 26 min |
| Microparticle tracking velocimetry [48] | Brownian velocity | 100 | 100–104 | 10 min | 3 h |
| Surface acoustic wave [49] | Amplitude | 100 | 100–106 | 10 min | 48.5 h |
| NanoPITA (this study) | Optical density | 0.54 | 1–500 | 35 min | 10 min |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, T.; Li, Z.; Zhao, L.; Zhang, W.; Huang, L.; Meng, F.; Liu, G.L.; Hu, W. Ultrasensitive Detection of C-Reactive Protein by a Novel Nanoplasmonic Immunoturbidimetry Assay. Biosensors 2022, 12, 958. https://doi.org/10.3390/bios12110958
Dang T, Li Z, Zhao L, Zhang W, Huang L, Meng F, Liu GL, Hu W. Ultrasensitive Detection of C-Reactive Protein by a Novel Nanoplasmonic Immunoturbidimetry Assay. Biosensors. 2022; 12(11):958. https://doi.org/10.3390/bios12110958
Chicago/Turabian StyleDang, Tang, Zhenyu Li, Liyuan Zhao, Wei Zhang, Liping Huang, Fanling Meng, Gang Logan Liu, and Wenjun Hu. 2022. "Ultrasensitive Detection of C-Reactive Protein by a Novel Nanoplasmonic Immunoturbidimetry Assay" Biosensors 12, no. 11: 958. https://doi.org/10.3390/bios12110958
APA StyleDang, T., Li, Z., Zhao, L., Zhang, W., Huang, L., Meng, F., Liu, G. L., & Hu, W. (2022). Ultrasensitive Detection of C-Reactive Protein by a Novel Nanoplasmonic Immunoturbidimetry Assay. Biosensors, 12(11), 958. https://doi.org/10.3390/bios12110958







