Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion due to Lung Cancer
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
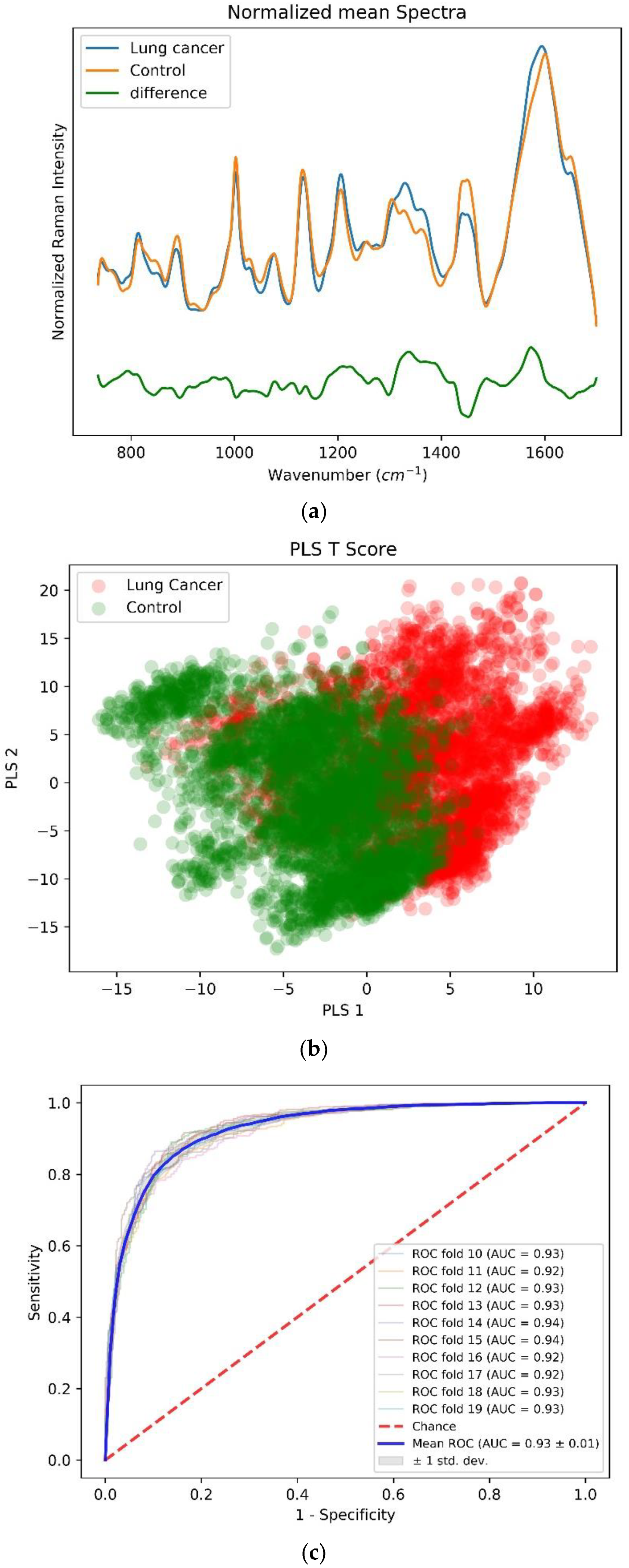
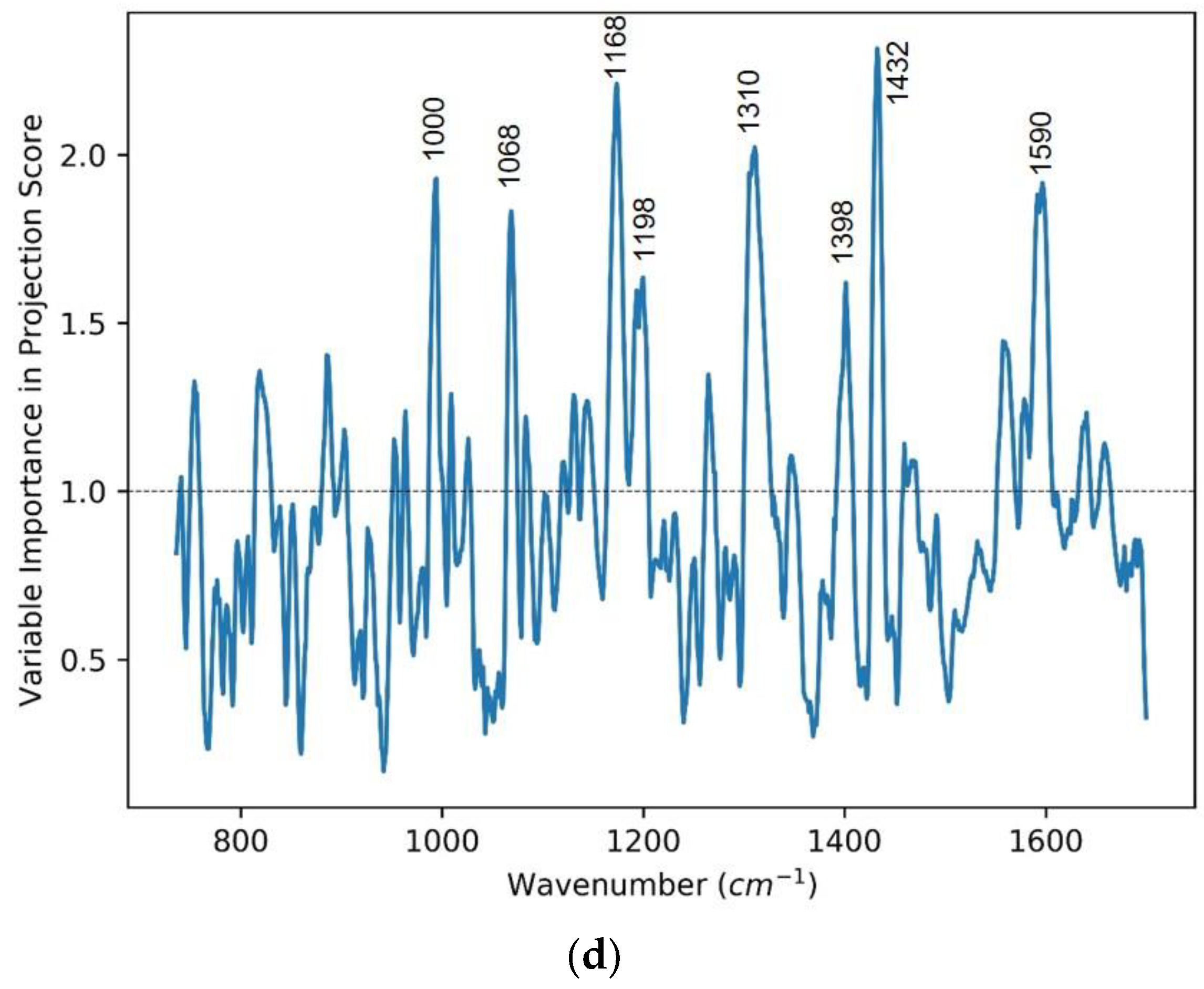
3.1. Case One: Lung Cancer vs. Control
3.2. Case 2: All Cancer vs. Control
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global Cancer Statistics, 2012. CA A Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef] [PubMed]
- McGuire, S. World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Adv. Nutr. 2016, 7, 418–419. [Google Scholar] [CrossRef] [PubMed]
- Goldstraw, P.; Crowley, J.; Chansky, K.; Giroux, D.J.; Groome, P.A.; Rami-Porta, R.; Postmus, P.E.; Rusch, V.; Sobin, L. The IASLC Lung Cancer Staging Project: Proposals for the Revision of the TNM Stage Groupings in the Forthcoming (Seventh) Edition of the TNM Classification of Malignant Tumours. J. Thorac. Oncol. 2007, 2, 706–714. [Google Scholar] [CrossRef]
- National Cancer Institute. Cancer Statistics Review 1975–2009 (Vintage 2009 Populations): Introduction; SEER Cancer Statistics; National Cancer Institute: Bethesda, MD, USA, 2010. [Google Scholar]
- Langwith, J. Diagnosis and Staging. In Lung Cancer; Chapter 6; Greenhaven Press: New York, NY, USA, 2007; pp. 71–77. [Google Scholar]
- Li, X.; Jin, H. Raman Spectroscopy of Serum for Cancer Detection. Proc. SPIE 2001, 3863, 77–78. [Google Scholar]
- Harris, A.T.; Lungari, A.; Needham, C.J.; Smith, S.L.; Lones, M.A.; Fisher, S.E.; Yang, X.B.; Cooper, N.; Kirkham, J.; Smith, D.A.; et al. Potential for Raman Spectroscopy to Provide Cancer Screening Using a Peripheral Blood Sample. Head Neck Oncol. 2009, 1, 34. [Google Scholar] [CrossRef]
- Kah, J.C.Y.; Kho, K.W.; Lee, C.G.L.; James, C.; Sheppard, R.; Shen, Z.X.; Soo, K.C.; Olivo, M.C. Early Diagnosis of Oral Cancer Based on the Surface Plasmon Resonance of Gold Nanoparticles. Int. J. Nanomed. 2007, 2, 785–798. [Google Scholar]
- Choi, S.; Park, H.-K.; Min, G.E.; Kim, Y.-H. Biochemical Investigations of Human Papillomavirus-Infected Cervical Fluids. Microsc. Res. Tech. 2014, 78, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Elumalai, B.; Prakasarao, A.; Ganesan, B.; Dornadula, K.; Ganesan, S. Raman Spectroscopic Characterization of Urine of Normal and Oral Cancer Subjects. J. Raman Spectrosc. 2014, 46, 84–93. [Google Scholar] [CrossRef]
- Nie, S. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 1997, 275, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Perumal, J.; Kong, K.V.; Dinish, U.S.; Bakker, R.M.; Olivo, M. Design and Fabrication of Random Silver Films as Substrate for SERS Based Nano-Stress Sensing of Proteins. RSC Adv. 2014, 4, 12995. [Google Scholar] [CrossRef]
- Lin, X.-M.; Cui, Y.; Xu, Y.-H.; Ren, B.; Tian, Z.-Q. Surface-Enhanced Raman Spectroscopy: Substrate-Related Issues. Anal. Bioanal. Chem. 2009, 394, 1729–1745. [Google Scholar] [CrossRef] [PubMed]
- Perumal, J.; Dinish, U.; Bendt, A.; Kazakeviciute, A.; Fu, C.Y.; Ong, I.L.H.; Olivo, M. Identification of Mycolic Acid Forms Using Surface-Enhanced Raman Scattering as a Fast Detection Method for Tuberculosis. Int. J. Nanomed. 2018, 13, 6029–6038. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-C.; Han, X.; Lee, Y.H.; Lee, H.K.; Phan-Quang, G.C.; Lay, C.L.; Sim, H.Y.F.; Phua, V.J.X.; Ng, L.S.; Ku, C.W.; et al. Multiplex Surface-Enhanced Raman Scattering Identification and Quantification of Urine Metabolites in Patient Samples within 30 Min. ACS Nano 2020, 14, 2542–2552. [Google Scholar] [CrossRef]
- Connolly, J.M.; Davies, K.; Kazakeviciute, A.; Wheatley, A.M.; Dockery, P.; Keogh, I.; Olivo, M. Non-Invasive and Label-Free Detection of Oral Squamous Cell Carcinoma Using Saliva Surface-Enhanced Raman Spectroscopy and Multivariate Analysis. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Huang, S.; Lin, D.; Chen, G.; Xu, Y.; Li, Y.; Huang, Z.; Pan, J.; Chen, R.; Zeng, H. Surface-Enhanced Raman Spectroscopy of Saliva Proteins for the Noninvasive Differentiation of Benign and Malignant Breast Tumors. Int. J. Nanomed. 2015, 10, 537. [Google Scholar] [CrossRef] [PubMed]
- Li, X. Spectral Analysis of Human Saliva for Detection of Lung Cancer Using Surface-Enhanced Raman Spectroscopy. J. Biomed. Opt. 2012, 17, 037003. [Google Scholar] [CrossRef] [PubMed]
- Rinnan, Å. Pre-Processing in Vibrational Spectroscopy—When, Why and How. Anal. Methods 2014, 6, 7124–7129. [Google Scholar] [CrossRef]
- Liland, K.H.; Kohler, A.; Afseth, N.K. Model-Based Pre-Processing in Raman Spectroscopy of Biological Samples. J. Raman Spectrosc. 2016, 47, 643–650. [Google Scholar] [CrossRef]
- Boelens, H.F.M.; Dijkstra, R.J.; Eilers, P.H.C.; Fitzpatrick, F.; Westerhuis, J.A. New Background Correction Method for Liquid Chromatography with Diode Array Detection, Infrared Spectroscopic Detection and Raman Spectroscopic Detection. J. Chromatogr. A 2004, 1057, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M.J.E. Smoothing and Differentiation of Data by Simplified Least Squares Procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification Tools in Chemistry. Part 1: Linear Models. PLS-DA. Anal. Methods 2013, 5, 3790. [Google Scholar] [CrossRef]
- de Almeida, M.R.; Correa, D.N.; Rocha, W.F.C.; Scafi, F.J.O.; Poppi, R.J. Discrimination between Authentic and Counterfeit Banknotes Using Raman Spectroscopy and PLS-DA with Uncertainty Estimation. Microchem. J. 2013, 109, 170–177. [Google Scholar] [CrossRef]
- Brereton, R.G.; Lloyd, G.R. Partial Least Squares Discriminant Analysis: Taking the Magic Away. J. Chemom. 2014, 28, 213–225. [Google Scholar] [CrossRef]
- Surmacki, J.M.; Woodhams, B.J.; Haslehurst, A.; Ponder, B.A.J.; Bohndiek, S.E. Raman Micro-Spectroscopy for Accurate Identification of Primary Human Bronchial Epithelial Cells. Sci. Rep. 2018, 8, 12604. [Google Scholar] [CrossRef]
- Huang, Z.; McWilliams, A.; Lui, H.; McLean, D.I.; Lam, S.; Zeng, H. Near-Infrared Raman Spectroscopy for Optical Diagnosis of Lung Cancer. Int. J. Cancer 2003, 107, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Moisoiu, V.; Stefancu, A.; Gulei, D.; Boitor, R.; Magdo, L.; Raduly, L.; Pasca, S.; Kubelac, P.; Mehterov, N.; Chis, V.; et al. SERS-Based Differential Diagnosis between Multiple Solid Malignancies: Breast, Colorectal, Lung, Ovarian and Oral Cancer. Int. J. Nanomed. 2019, 14, 6165–6178. [Google Scholar] [CrossRef]
- Liu, K.; Jin, S.; Song, Z.; Jiang, L. High Accuracy Detection of Malignant Pleural Effusion Based on Label-Free Surface-Enhanced Raman Spectroscopy and Multivariate Statistical Analysis. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 226, 117632. [Google Scholar] [CrossRef]

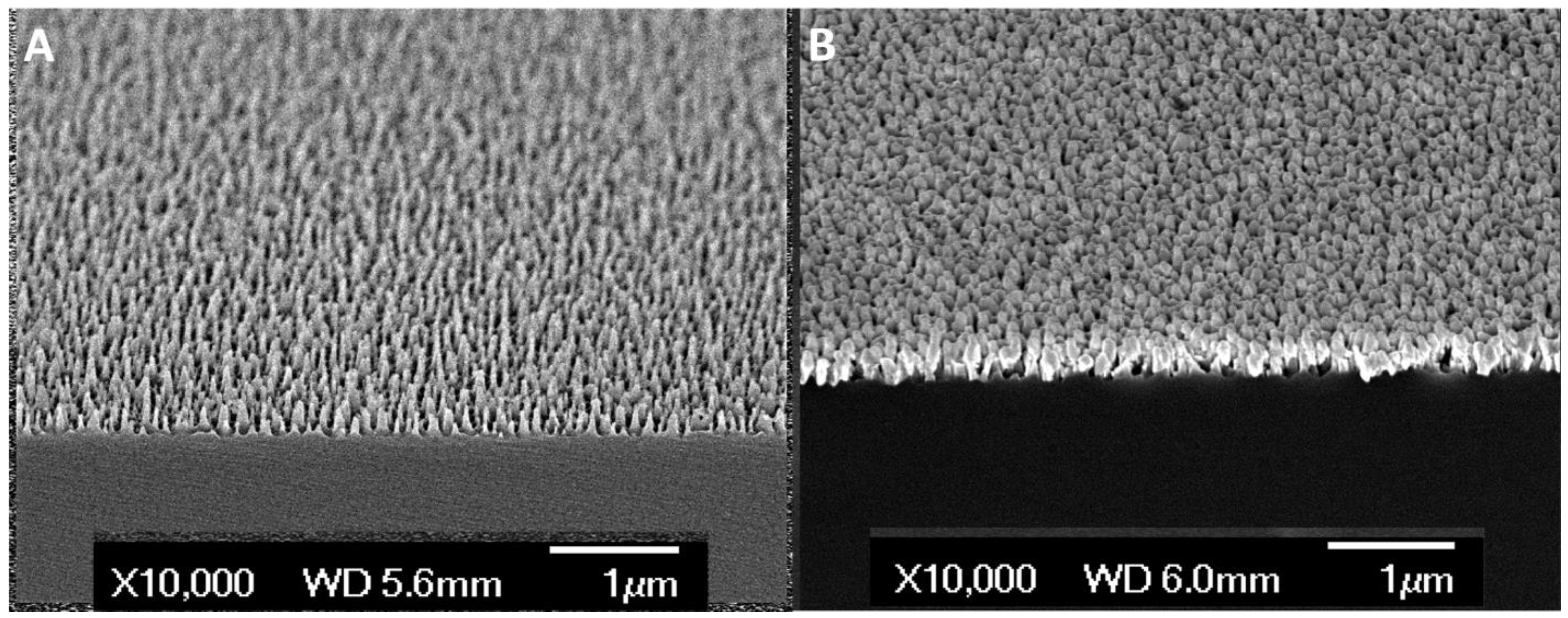


| Cancer Types | No. of Subjects |
|---|---|
| LUNG CANCER | 15 |
| OTHER CANCERS | |
| 1 |
| 1 |
| 1 |
| 2 |
| 1 |
| 1 |
| NON-CANCER (CONTROL) | |
| 8 |
| 4 |
| TOTAL | 34 |
| Peak Position (cm−1) | Vibrational Mode Assignment [26] |
|---|---|
| 1002 | Phenylalanine, C-C aromatic ring stretching |
| 1068 | C-C skeletal stretching (lipids) |
| 1168 | ν (C═C) δ(COH) (lipid) |
| 1198 | Nucleic acids and phosphates |
| 1310 | CH3/CH2 twisting or bending mode of lipid/protein assignment |
| 1398 | C═O symmetric stretch, CH2 deformation |
| 1432 | CH2 scissoring vibration (lipid) |
| 1590 | C═C olefinic stretch (protein) |
| Peak Position (cm−1) | Vibrational Mode Assignment [26] |
|---|---|
| 887 | Protein bands, Structural protein modes of tumors |
| 959 | Single bond stretching vibrations for the amino acids proline, valine and polysaccharides |
| 1008 | Phenylalanine ν(CO), ν(CC), δ(OCH) |
| 1078 | ν(C-C) or ν(C-O), phospholipids and nucleic acid |
| 1120–1170 | ν(C-N), C-C skeletal trans conformation, phospholipidsνC-C of proteins (also carotenoids) |
| 1222 | Amide III (β-sheet) |
| 1325 | CH3CH2 wagging mode in purine bases of nucleic acids |
| 1655 | Amide I (protein) and C═C stretch (lipid) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perumal, J.; Lee, P.; Dev, K.; Lim, H.Q.; Dinish, U.S.; Olivo, M. Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion due to Lung Cancer. Biosensors 2022, 12, 940. https://doi.org/10.3390/bios12110940
Perumal J, Lee P, Dev K, Lim HQ, Dinish US, Olivo M. Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion due to Lung Cancer. Biosensors. 2022; 12(11):940. https://doi.org/10.3390/bios12110940
Chicago/Turabian StylePerumal, Jayakumar, Pyng Lee, Kapil Dev, Hann Qian Lim, U. S. Dinish, and Malini Olivo. 2022. "Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion due to Lung Cancer" Biosensors 12, no. 11: 940. https://doi.org/10.3390/bios12110940
APA StylePerumal, J., Lee, P., Dev, K., Lim, H. Q., Dinish, U. S., & Olivo, M. (2022). Machine Learning Assisted Real-Time Label-Free SERS Diagnoses of Malignant Pleural Effusion due to Lung Cancer. Biosensors, 12(11), 940. https://doi.org/10.3390/bios12110940





